Sensitivity Analysis of Mathematical Model to Study the Effect of T Cells Infusion in Treatment of CLL
Abstract
1. Introduction
2. Materials and Methods
2.1. Nomenclature
- Employment rate of naive blood cells inflowing into circulatory blood from different sections as well as from blood transfusion;
- Natural death rate of susceptible blood cells;
- Decay rate of naive cells killed upon contact with tumor cells and becoming dysfunctional;
- Natural death rate of infected cells;
- Recruitment rate of cancer cells into blood system;
- Normal death rate of malignant cells;
- Loss of cancer cells due to encounter with immune cells;
- Rate of external intravenous re-infusion of T cells;
- Propagation rate of resistant cells in case of cancer setback;
- Natural death rate of immune cells;
- Loss rate of immune cells due to encounter with cancer cells.
2.2. Sensitivity Analysis
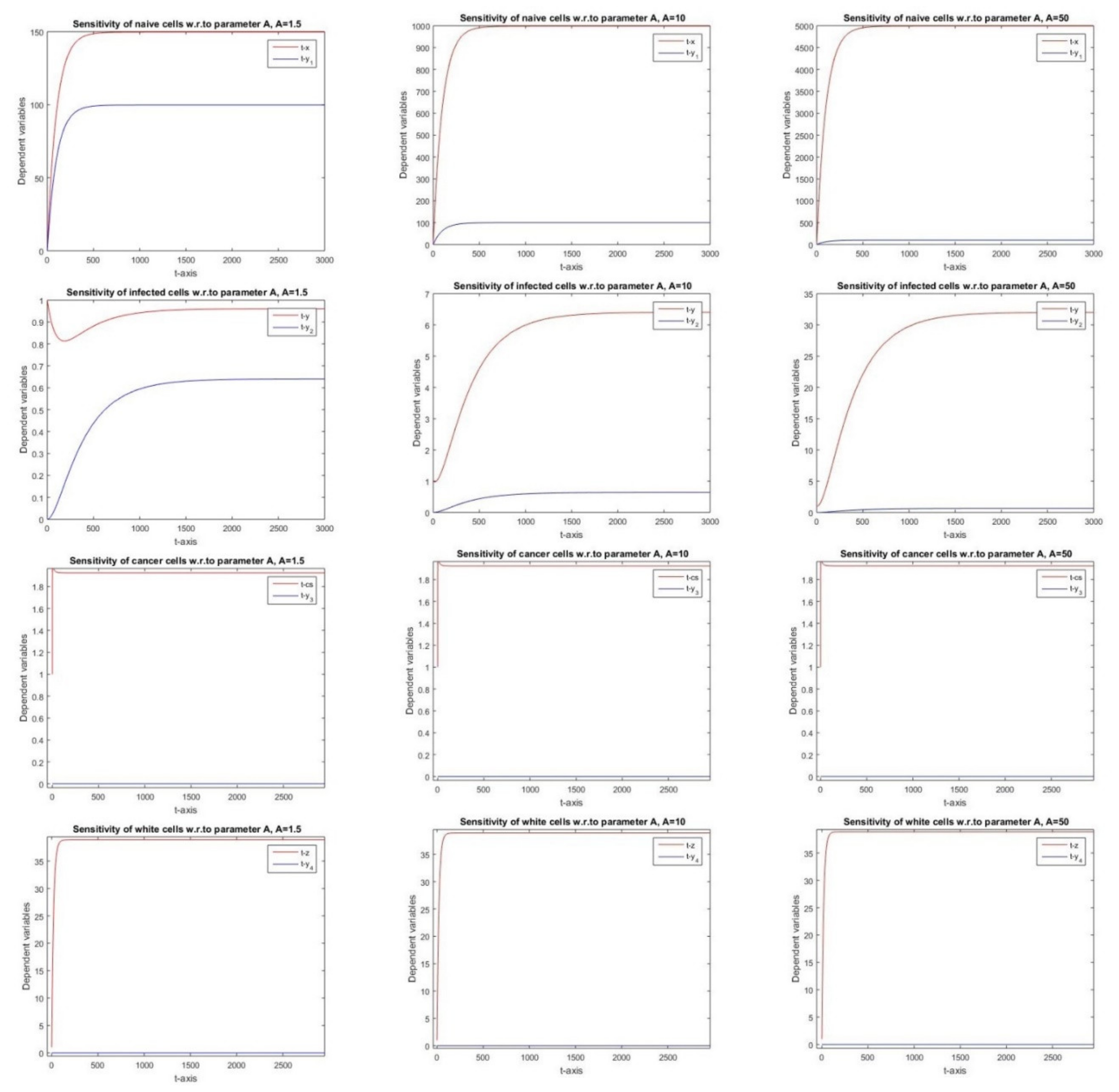
2.2.1. Sensitivity Analysis with Respect to Parameter A
2.2.2. Sensitivity with Respect to Parameter a0
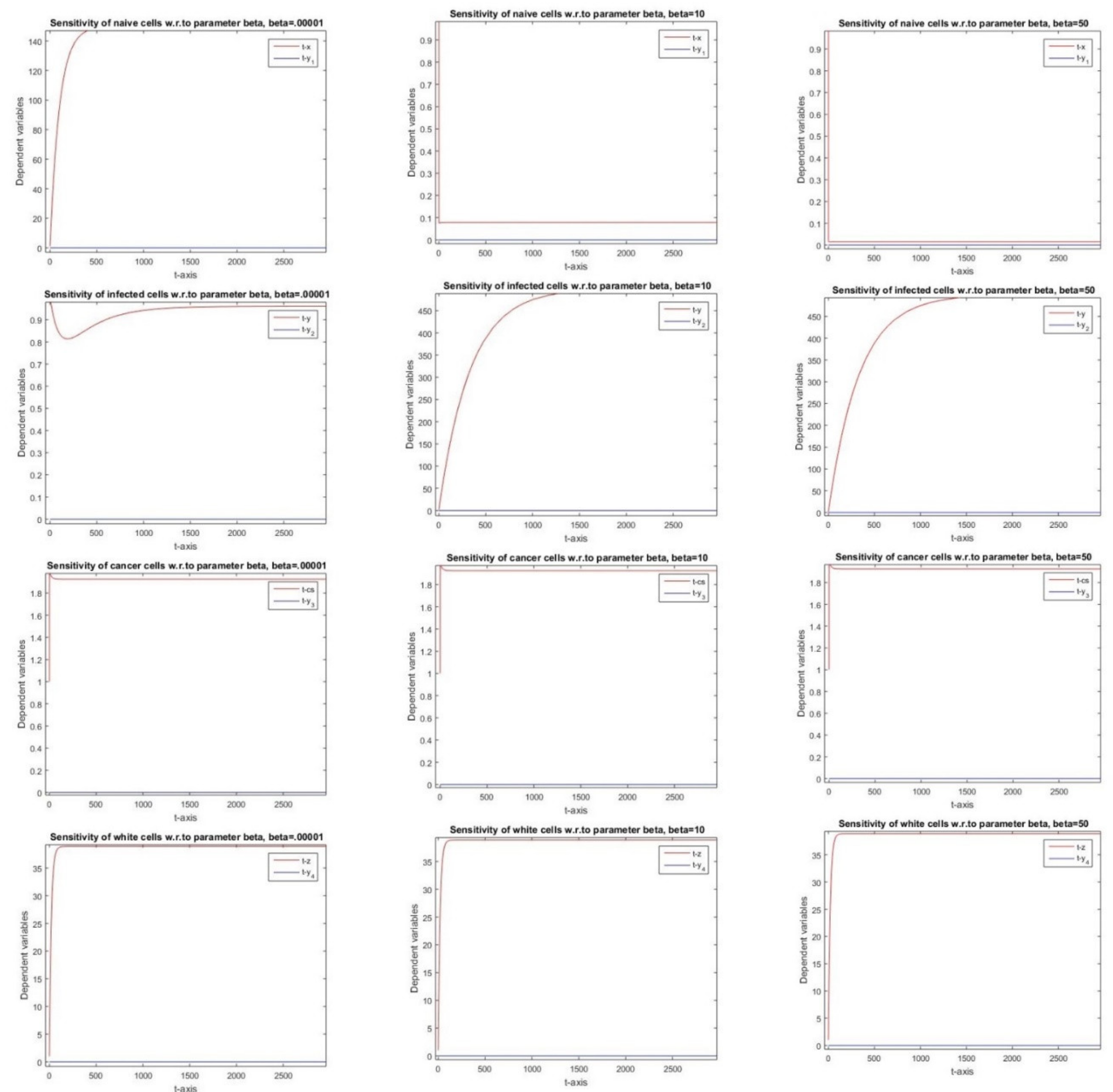
2.2.3. Sensitivity with Respect to Parameter β
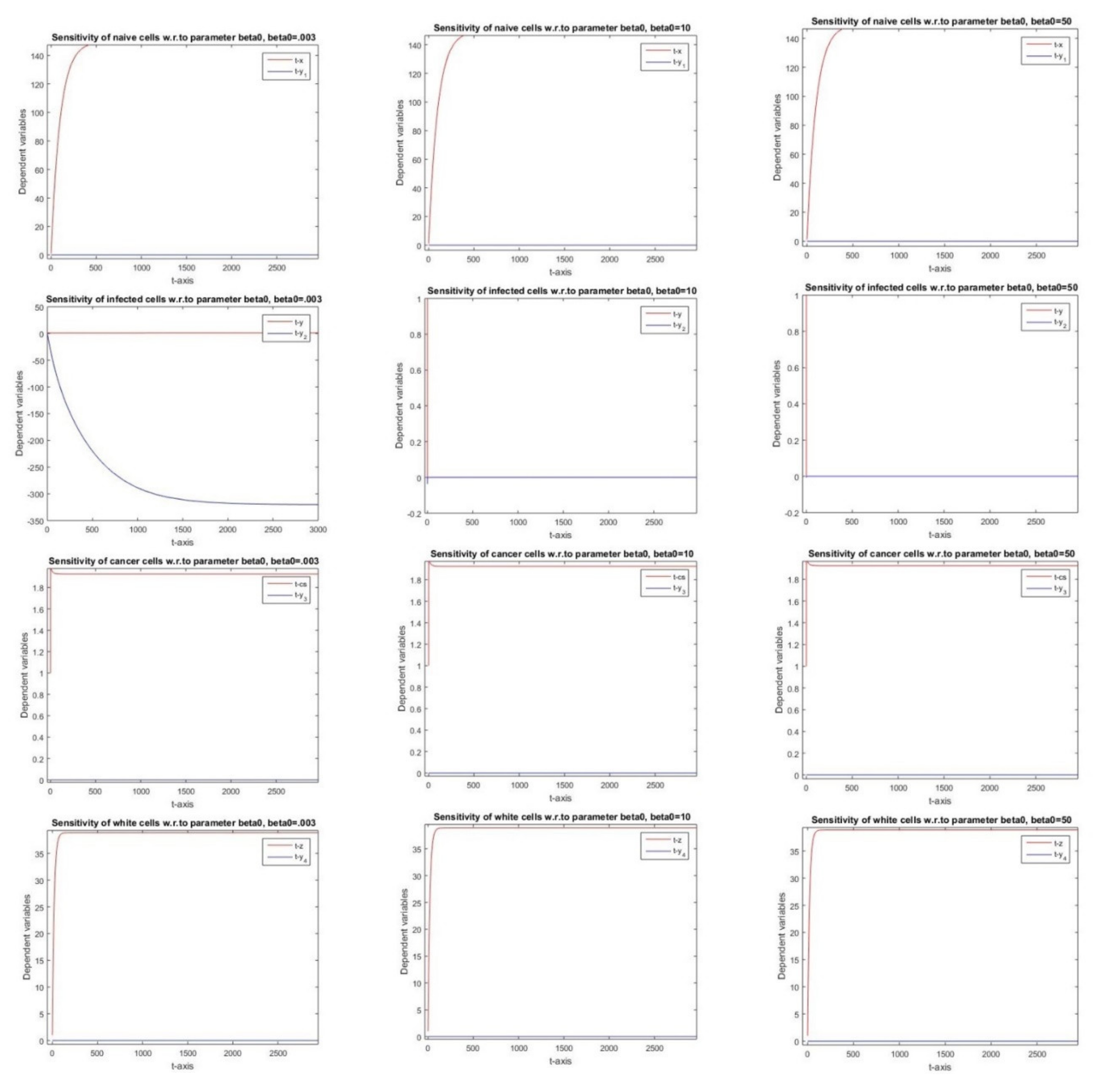
2.2.4. Sensitivity with Respect to Parameter β0
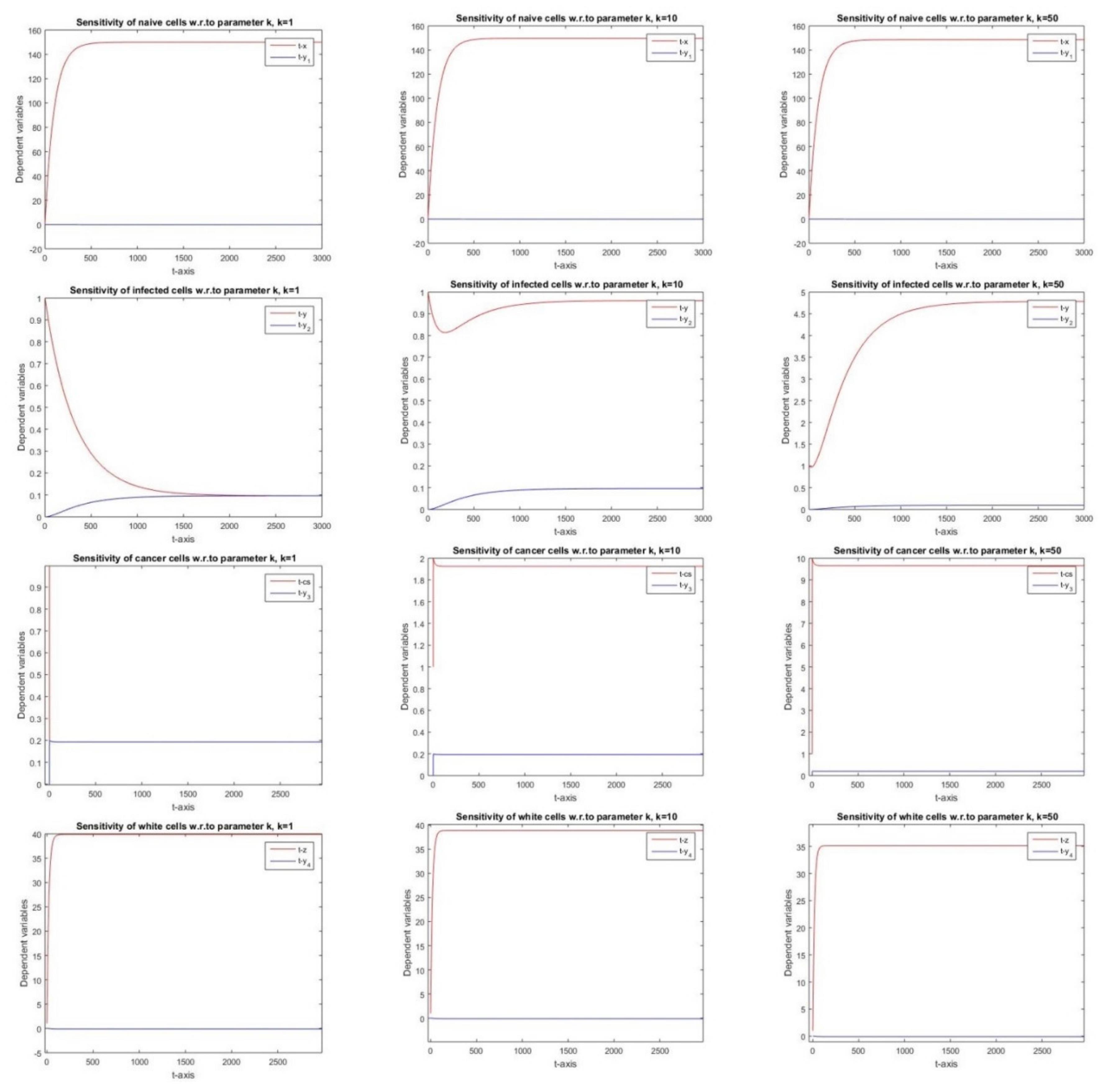
2.2.5. Sensitivity with Respect to Parameter k
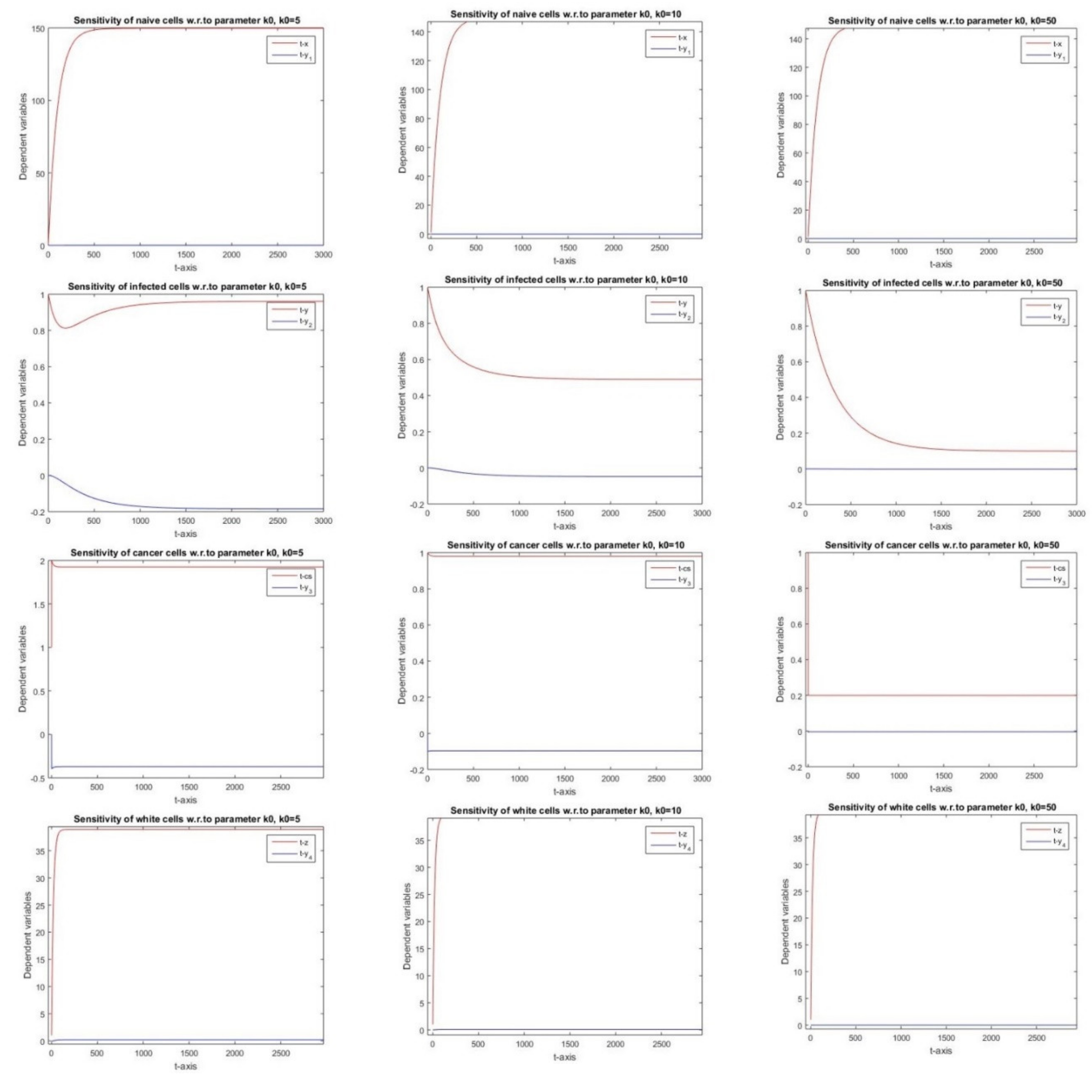
2.2.6. Sensitivity with Respect to Parameter k0
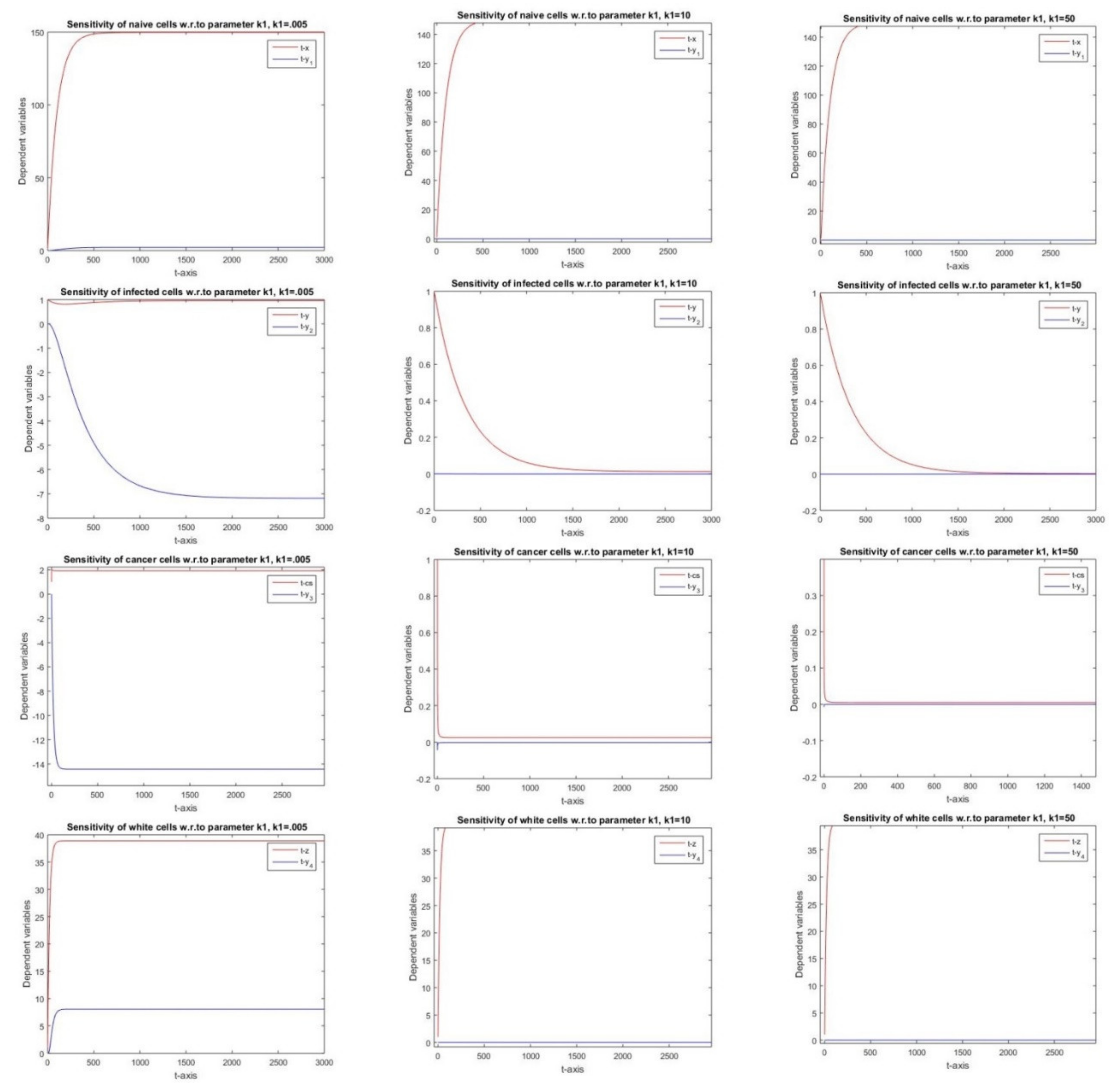
2.2.7. Sensitivity with Respect to Parameter k1
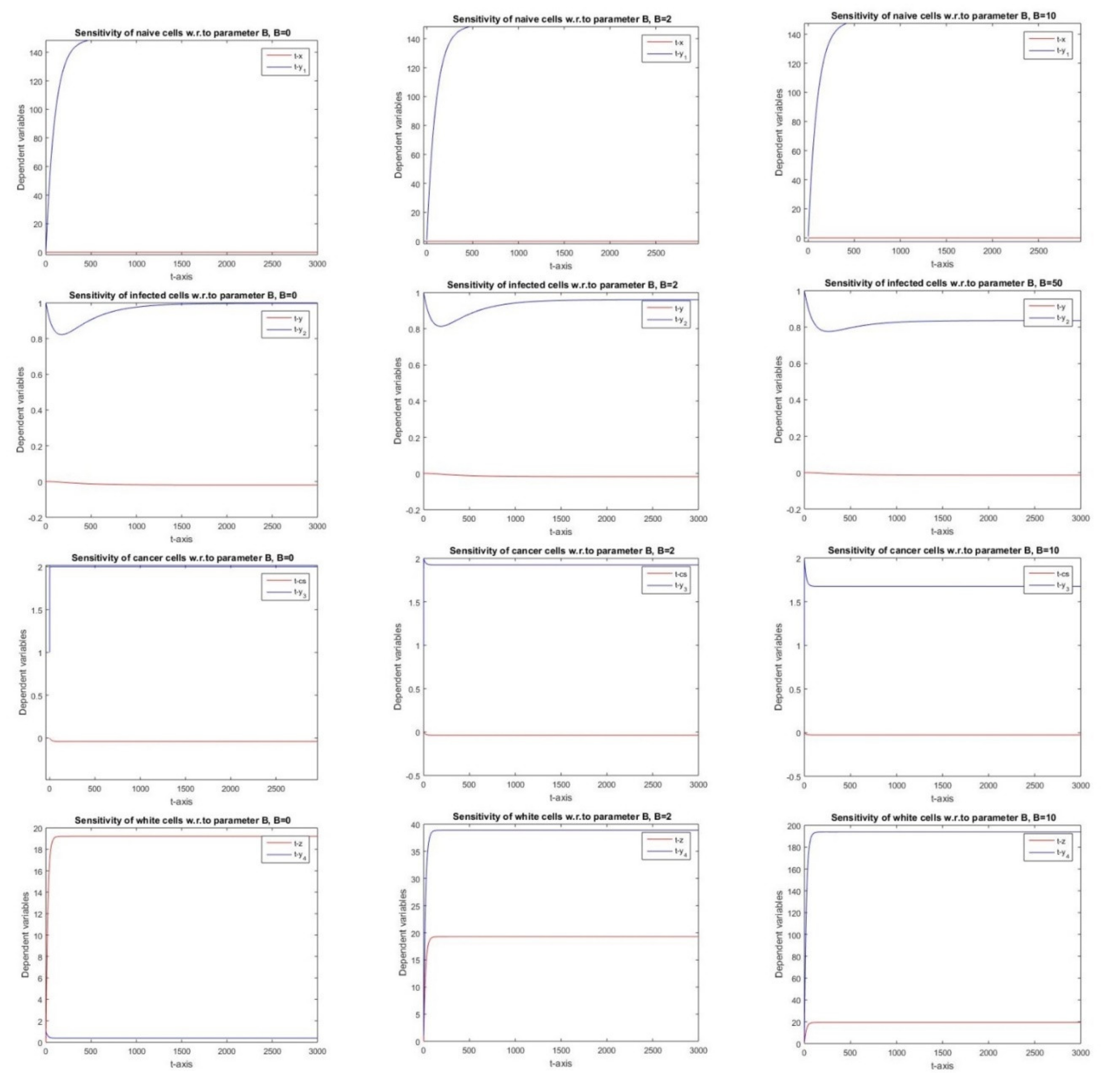
2.2.8. Sensitivity with Respect to Parameter B
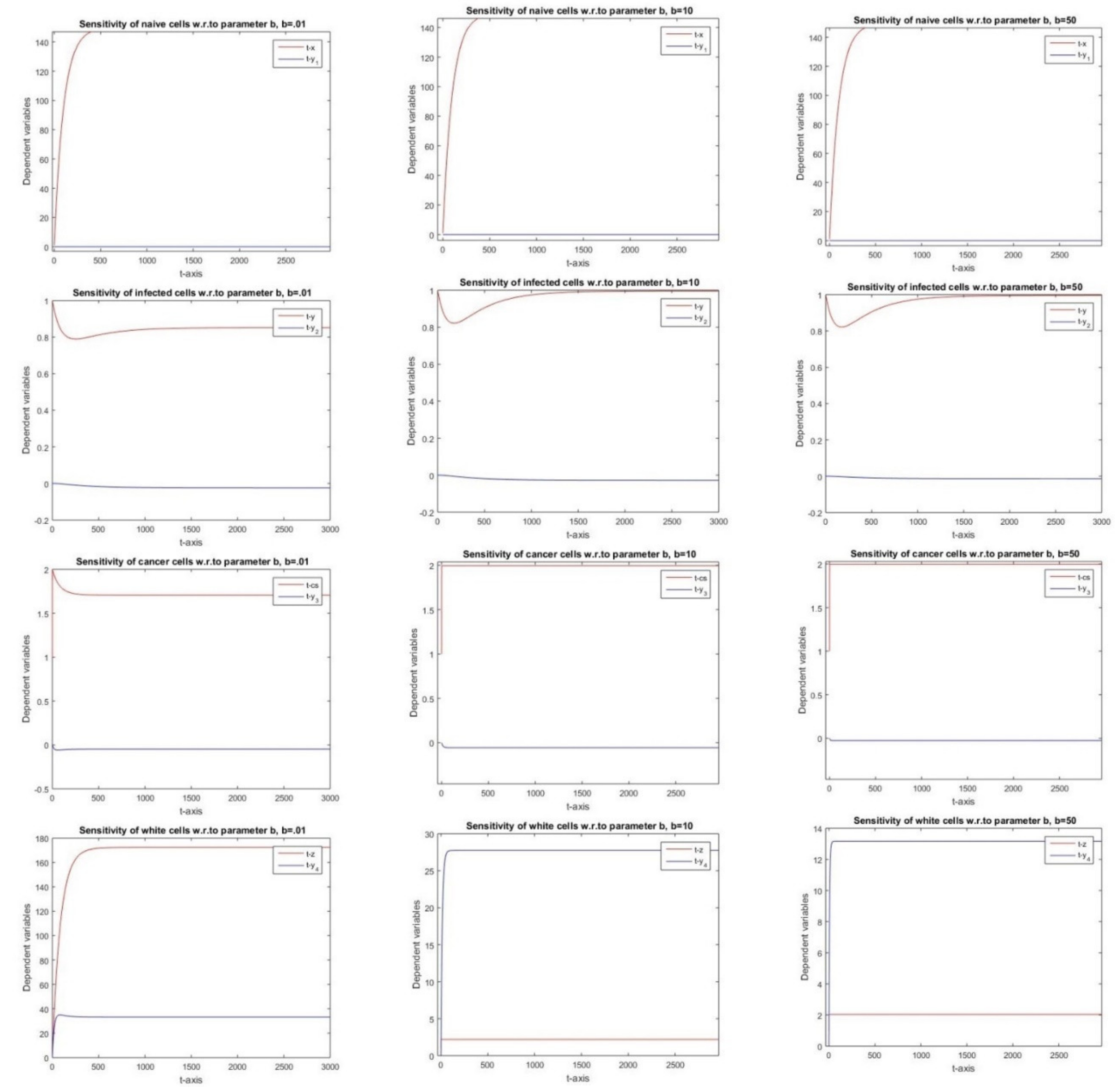
2.2.9. Sensitivity with Respect to Parameter b
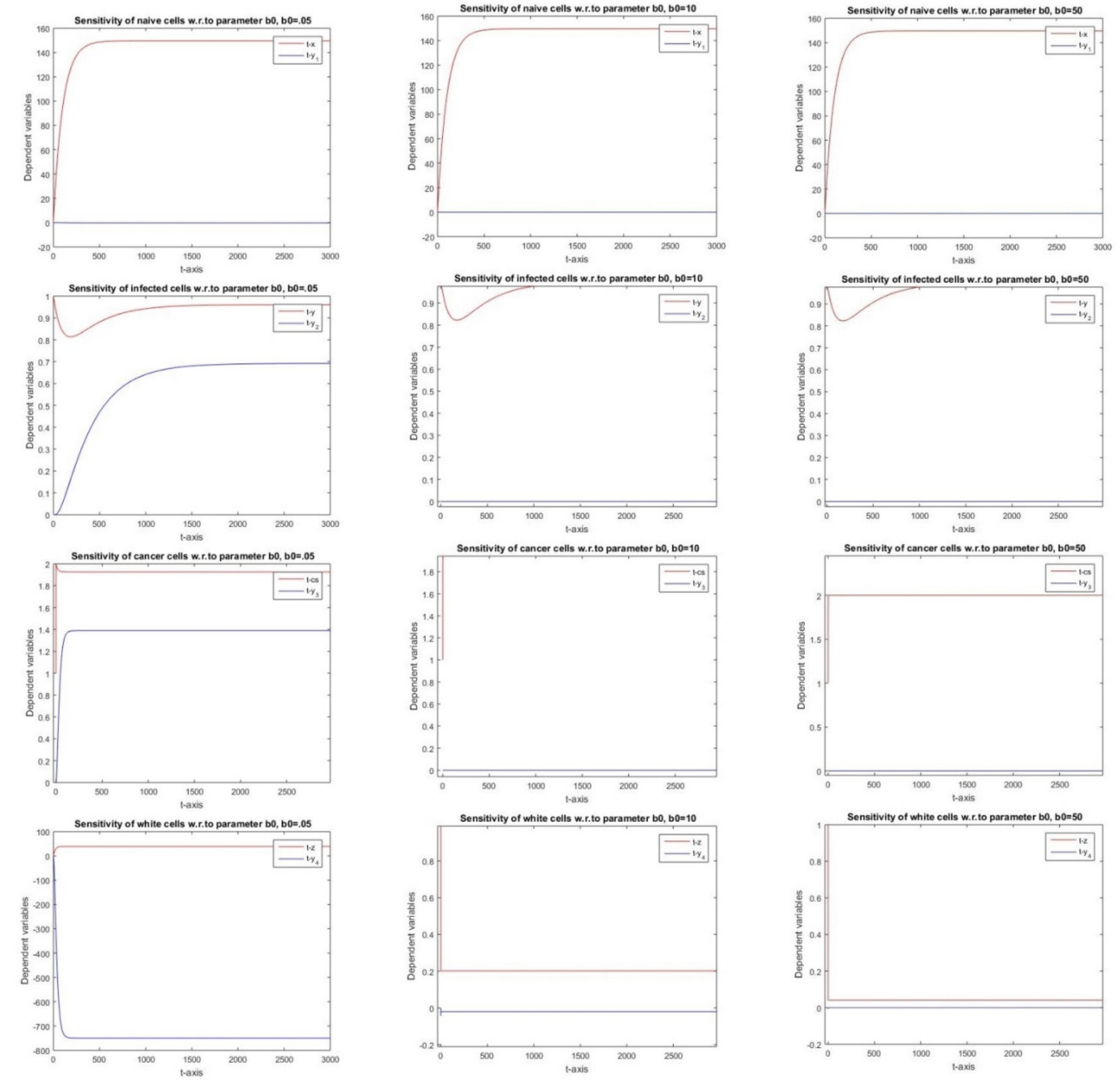
2.2.10. Sensitivity Analysis w.r.ro Parameter b0
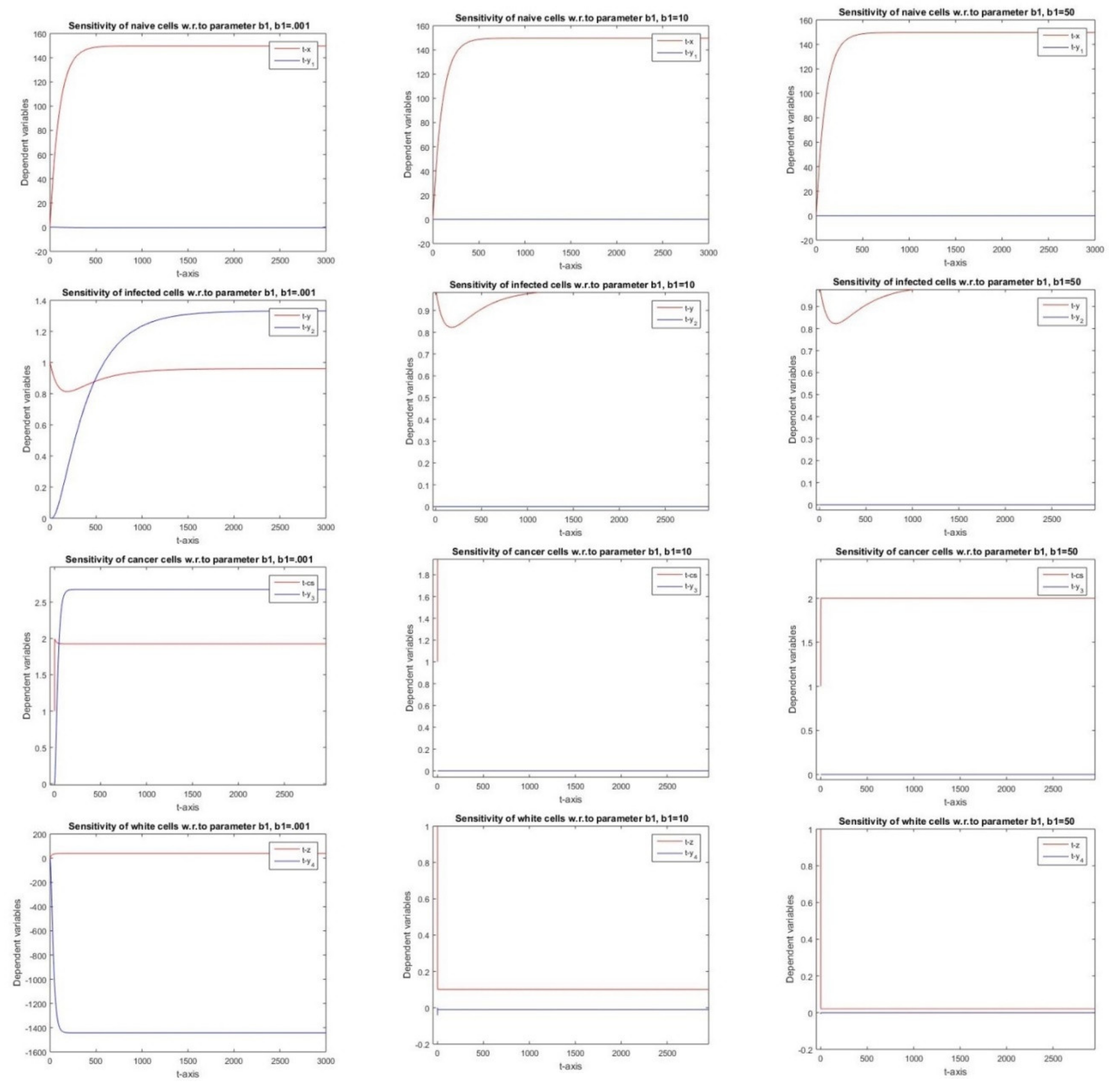
2.2.11. Sensitivity Analysis w.r.ro Parameter b1
3. Results
3.1. Parameter A
3.2. Parameter a0
3.3. Parameter β
3.4. Parameter β0
3.5. Parameter k
3.6. Parameter k0
3.7. Parameter k1
3.8. Parameter B
3.9. Parameter b
3.10. Parameter b0
3.11. Parameter b1
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gross, L.J. Points of View: The Interface of Mathematics and Biology. Cell Biol. Educ. 2004, 3, 85–87. [Google Scholar] [CrossRef] [PubMed]
- Elango, P. The Role of Mathematics in Biology; South Eastern University of Sri Lanka: Oluvil, Sri Lanka, 2015. [Google Scholar]
- Dreij, K.; Chaudhry, Q.A.; Jernström, B.; Morgenstern, R.; Hanke, M. A method for efficient calculation of diffusion and reactions of lipophilic compounds in complex cell geometry. PLoS ONE 2011, 6, e23128. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.; Chaudhry, Q.A.; Siddiqui, S. Mathematical modeling and optimization of complex structures: A review. Complex Adapt. Syst. Modeling 2016, 4, 1–3. [Google Scholar] [CrossRef]
- Khalid, S.; Chaudhry, Q.A. Quantitative analysis of cancer risk assessment in a mammalian cell with the inclusion of mitochondria. Comput. Math. Appl. 2019. [Google Scholar] [CrossRef]
- Chaudhry, Q.A.; Hanke, M.; Morgenstern, R.; Dreij, K. Surface reactions on the cytoplasmatic membranes—Mathematical modeling of reaction and diffusion systems in a cell. J. Comput. Appl. Math. 2014, 262, 244–260. [Google Scholar] [CrossRef]
- Dreij, K.; Chaudhry, Q.A.; Jernström, B.; Hanke, M.; Morgenstern, R. In silico modeling of intracellular diffusion and reaction of benzo. Pyrene Diol Epoxide 2012, 211, S60–S61. [Google Scholar]
- Felman, A. What to Know about Leukemia. Available online: https://www.medicalnewstoday.com/articles/142595 (accessed on 10 January 2020).
- Marieb, E.N.; Hoehn, K.N. Human Anatomy & Physiology; Pearson Education: London, UK, 2015. [Google Scholar]
- Martini, F.H.; Nath, J.L.; Bartholomew, E.F. Fundamentals of Anatomy & Physiology; Global Edition; Pearson: London, UK, 2018. [Google Scholar]
- MacGill, M. What Does the Lymphatic System Do? Available online: https://www.medicalnewstoday.com/articles/303087 (accessed on 10 January 2020).
- Gray, H.; Lewis, W.H. Anatomy of the Human Body; Lea & Febiger: Philadelphia, PA, USA, 1918. [Google Scholar]
- Lim, H.; Yeo, K.; Angeli, V. Lipid biology and lymphatic function: A dynamic interplay with important physiological and pathological consequences. J. Clin. Cell Immunol. 2014, 5, 2. [Google Scholar]
- Bertuccio, P.; Bosetti, C.; Malvezzi, M.; Levi, F.; Chatenoud, L.; Negri, E.; Vecchia, C.L. Trends in mortality from leukemia in Europe: An update to 2009 and a projection to 2012. Int. J. Cancer 2013, 132, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; Dandona, L. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar] [PubMed]
- Combest, A.J.; Danford, R.C.; Andrews, E.R.; Simmons, A.; Mcatee, P.; Reitsma, D.J. Overview of the Current Treatment Paradigm in Chronic Lymphocytic Leukemia, Part 2. J. Hematol. Oncol. Pharm. 2016, 6, 89–94. [Google Scholar]
- Hus, I.; Roliński, J. Current concepts in diagnosis and treatment of chronic lymphocytic leukemia. Contemp. Oncol. 2015, 19, 361. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wang, X. Therapeutic advancement of chronic lymphocytic leukemia. J. Hematol. Oncol. 2012, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Scheithauer, H.; Belka, C.; Lauber, K.; Gaipl, U.S. Immunological Aspects of Radiotherapy; Springer: Berlin, Germany, 2014. [Google Scholar]
- Watanabe, Y.; Dahlman, E.L.; Leder, K.Z.; Hui, S.K. A mathematical model of tumor growth and its response to single irradiation. Theor. Biol. Med. Model. 2016, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Gatza, E.; Wahl, D.R.; Opipari, A.W.; Sundberg, T.B.; Reddy, P.; Liu, C.; Glick, G.D.; Ferrara, J.L. Manipulating the bioenergetics of alloreactive T cells causes their selective apoptosis and arrests graft-versus-host disease. Sci. Transl. Med. 2011, 3, ra67–ra68. [Google Scholar] [CrossRef] [PubMed]
- Agur, Z.; Vuk-Pavlović, S. Mathematical modeling in immunotherapy of cancer: Personalizing clinical trials. Mol. Ther. 2012, 20, 1–2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Agarwal, M.; Bhadauria, A.S. Mathematical Modeling and Analysis of Leukemia: Effect of External Engineered T Cells Infusion. Appl. Appl. Math. 2015, 10, 249–266. [Google Scholar]
- Saltelli, A. Sensitivity Analysis. 2017. Available online: http://www.andreasaltelli.eu/file/repository/Bergen_Andrea_Thursday_SA.pdf (accessed on 10 January 2020).
- Zi, Z. Sensitivity analysis approaches applied to systems biology models. IET Syst. Biol. 2011, 5, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q. Sensitivity Analysis in Biological Modelling. 2012. Available online: http://www.math.ualberta.ca/~hwang/sensitivity (accessed on 10 January 2020).
- Davis, T.A. MATLAB Primer, 8th ed.; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Bolboacă, S.D. Medical Diagnostic Tests: A Review of Test Anatomy, Phases, and Statistical Treatment of Data. Comput. Math. Methods Med. 2019. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anjum, A.U.R.; Chaudhry, Q.A.; Almatroud, A.O. Sensitivity Analysis of Mathematical Model to Study the Effect of T Cells Infusion in Treatment of CLL. Mathematics 2020, 8, 564. https://doi.org/10.3390/math8040564
Anjum AUR, Chaudhry QA, Almatroud AO. Sensitivity Analysis of Mathematical Model to Study the Effect of T Cells Infusion in Treatment of CLL. Mathematics. 2020; 8(4):564. https://doi.org/10.3390/math8040564
Chicago/Turabian StyleAnjum, Asad Ur Rehman, Qasim Ali Chaudhry, and A. Othman Almatroud. 2020. "Sensitivity Analysis of Mathematical Model to Study the Effect of T Cells Infusion in Treatment of CLL" Mathematics 8, no. 4: 564. https://doi.org/10.3390/math8040564
APA StyleAnjum, A. U. R., Chaudhry, Q. A., & Almatroud, A. O. (2020). Sensitivity Analysis of Mathematical Model to Study the Effect of T Cells Infusion in Treatment of CLL. Mathematics, 8(4), 564. https://doi.org/10.3390/math8040564



