Pattern Recognition in Epileptic EEG Signals via Dynamic Mode Decomposition
Abstract
1. Introduction
2. Materials and Methods
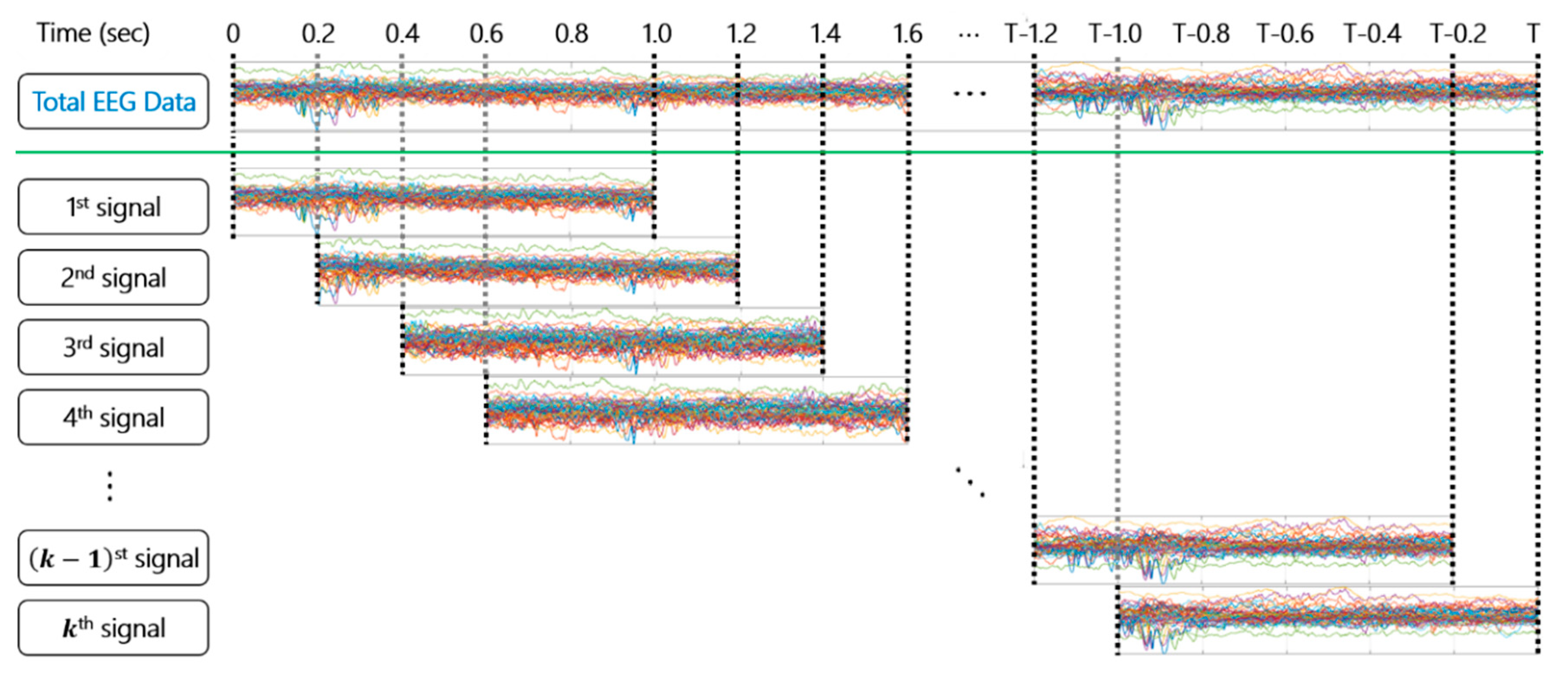
2.1. Dataset
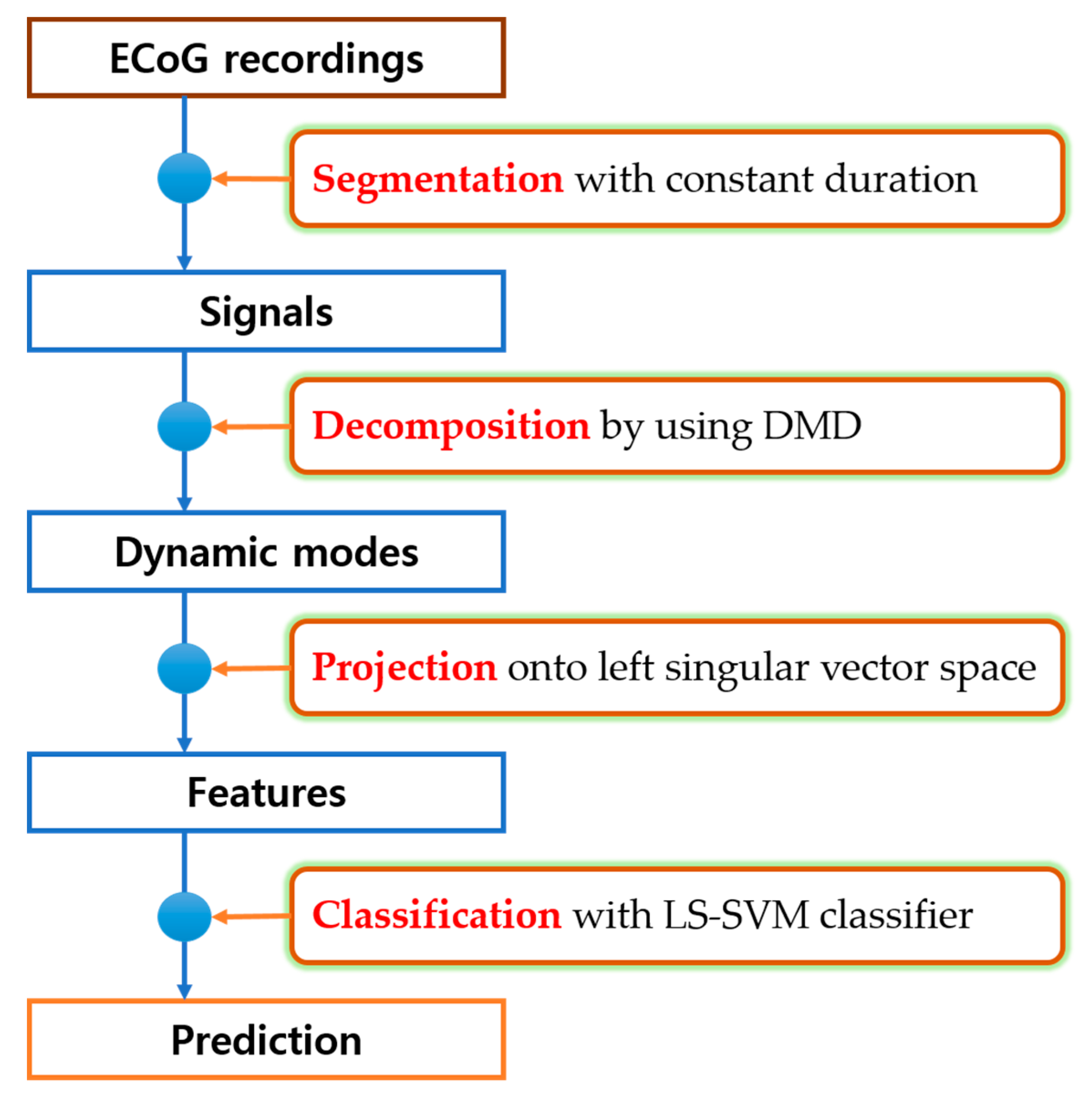
2.2. Dynamic Mode Decomposition
- (a)
- Compute the reduced and appropriately truncated SVD [19] of the data matrix in Equation (5):where the columns of and are orthonormal eigenvectors of and , respectively, the diagonal entries of are the square roots of the non-negative eigenvalues of both and , and refers to the reduced rank of the approximated matrix given in Equation (7). The columns of are called the left singular vectors of .
- (b)
- Define a low-rank approximation of in Equation (6):
- (c)
- Compute the eigendecomposition of in Equation (8):where the columns of and the diagonal entries of are the eigenvectors and the eigenvalues of , respectively.
- (d)
- Since the eigenvalues in Equation (9) are also the eigenvalues of , the DMD mode (i.e., the eigenvector of ) corresponding to the DMD eigenvalue in Equation (9) is given by:where is the th column of in Equation (9).
2.3. Feature Extraction
2.4. Classification
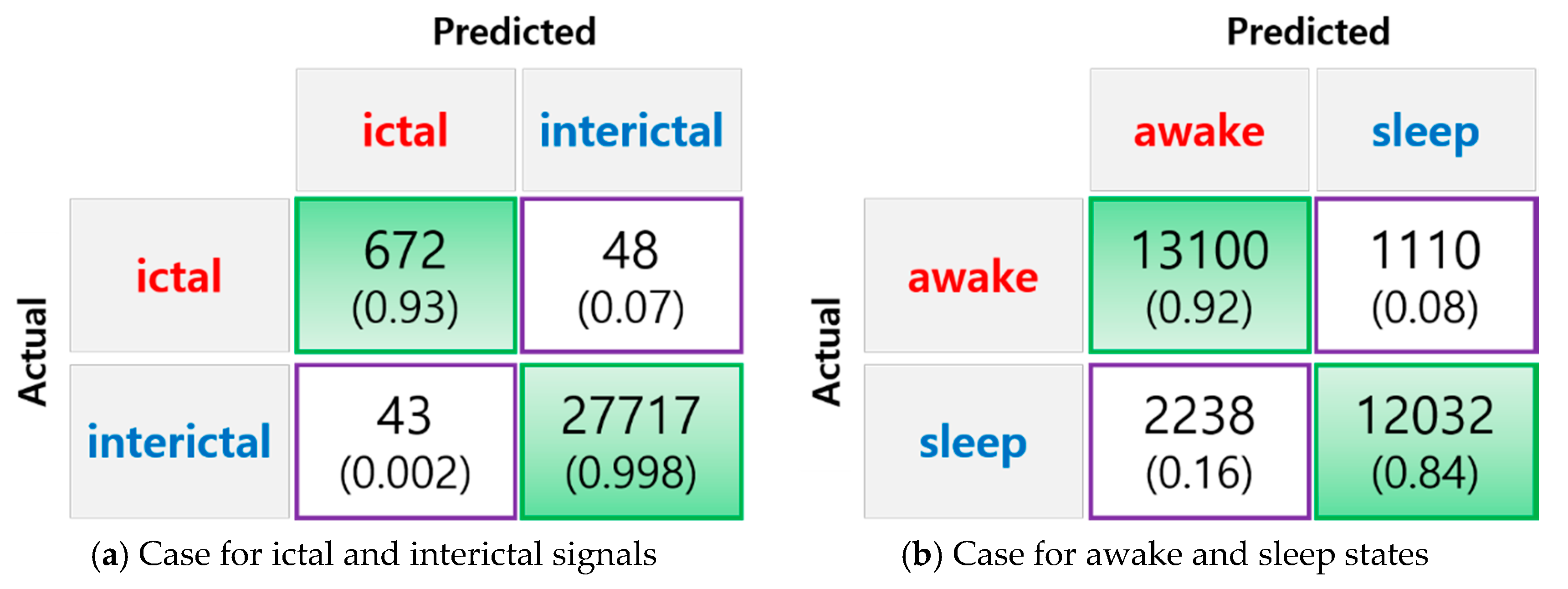
3. Results
4. Discussion
- (a)
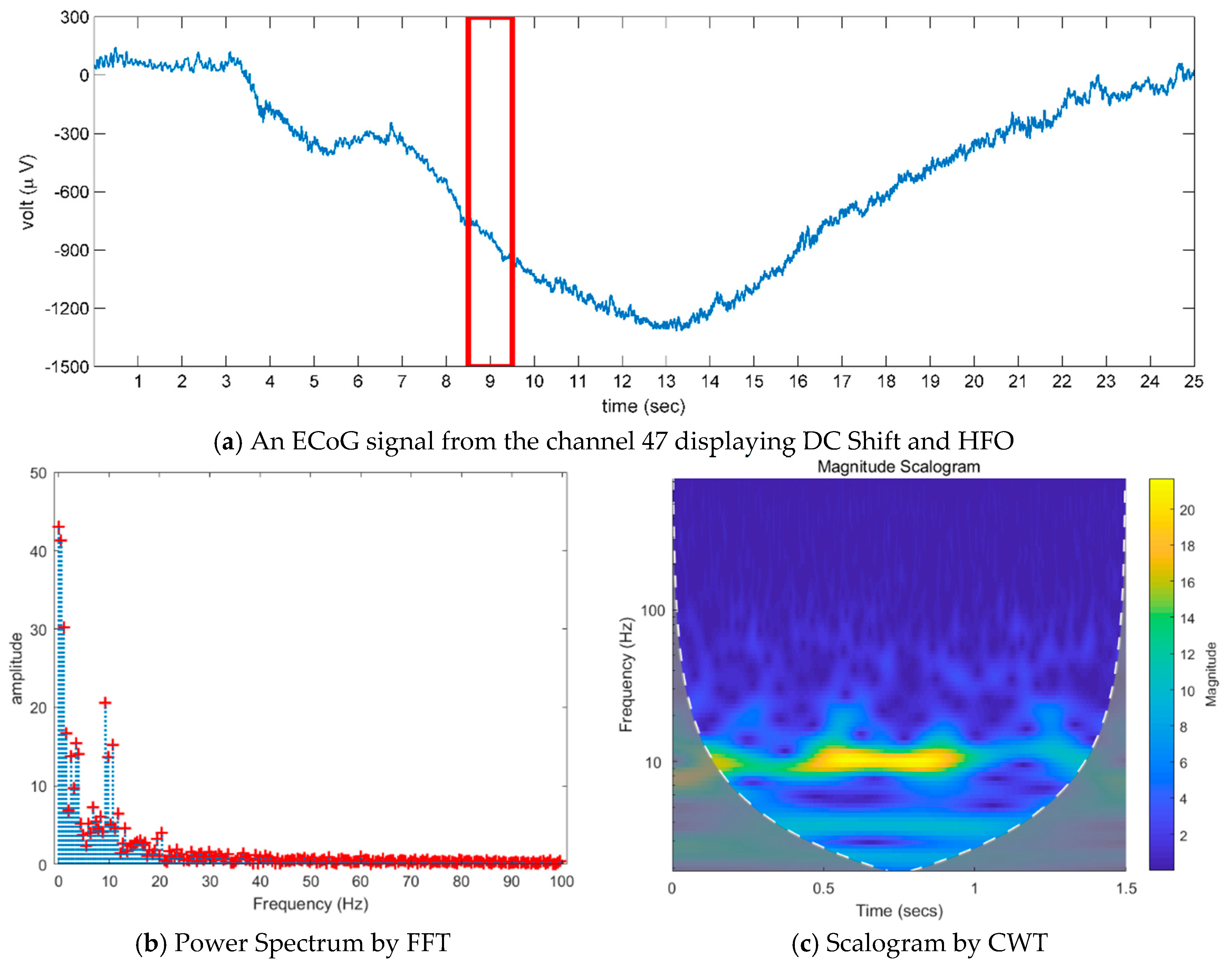
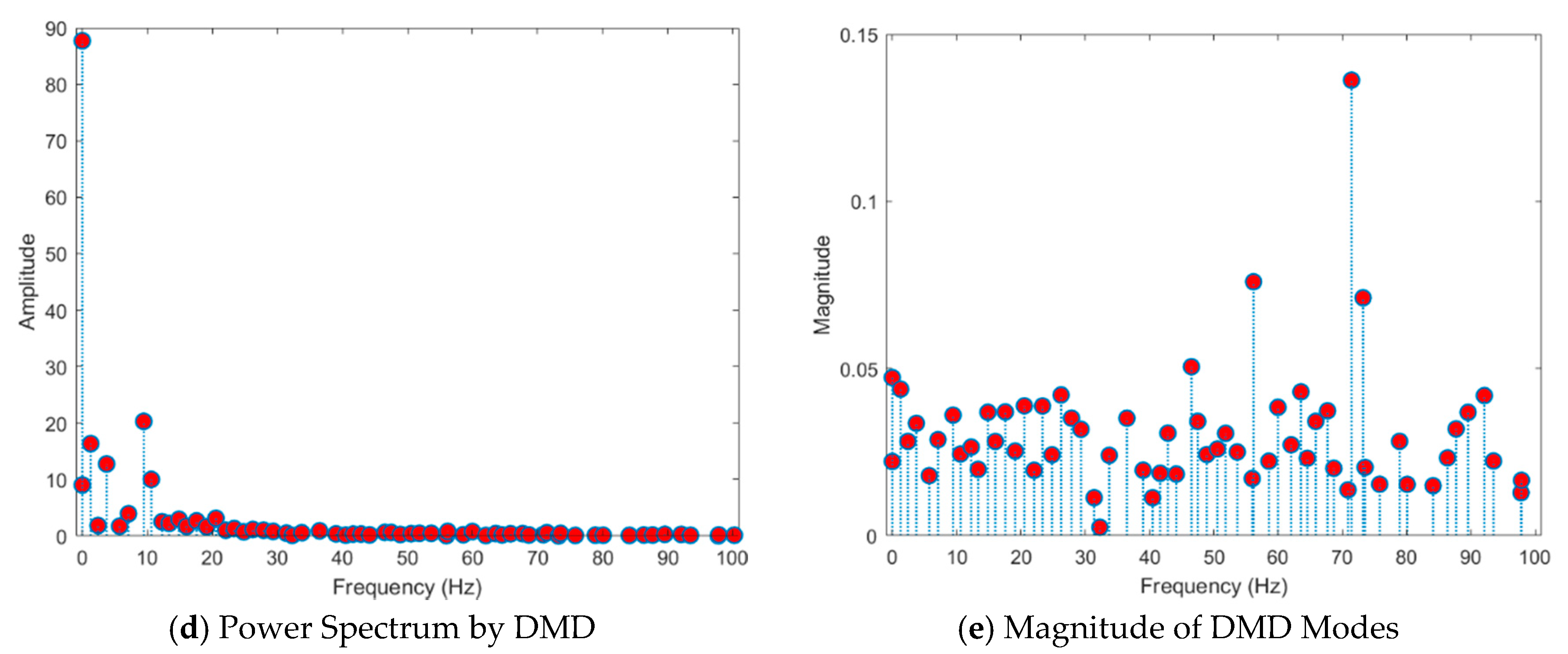
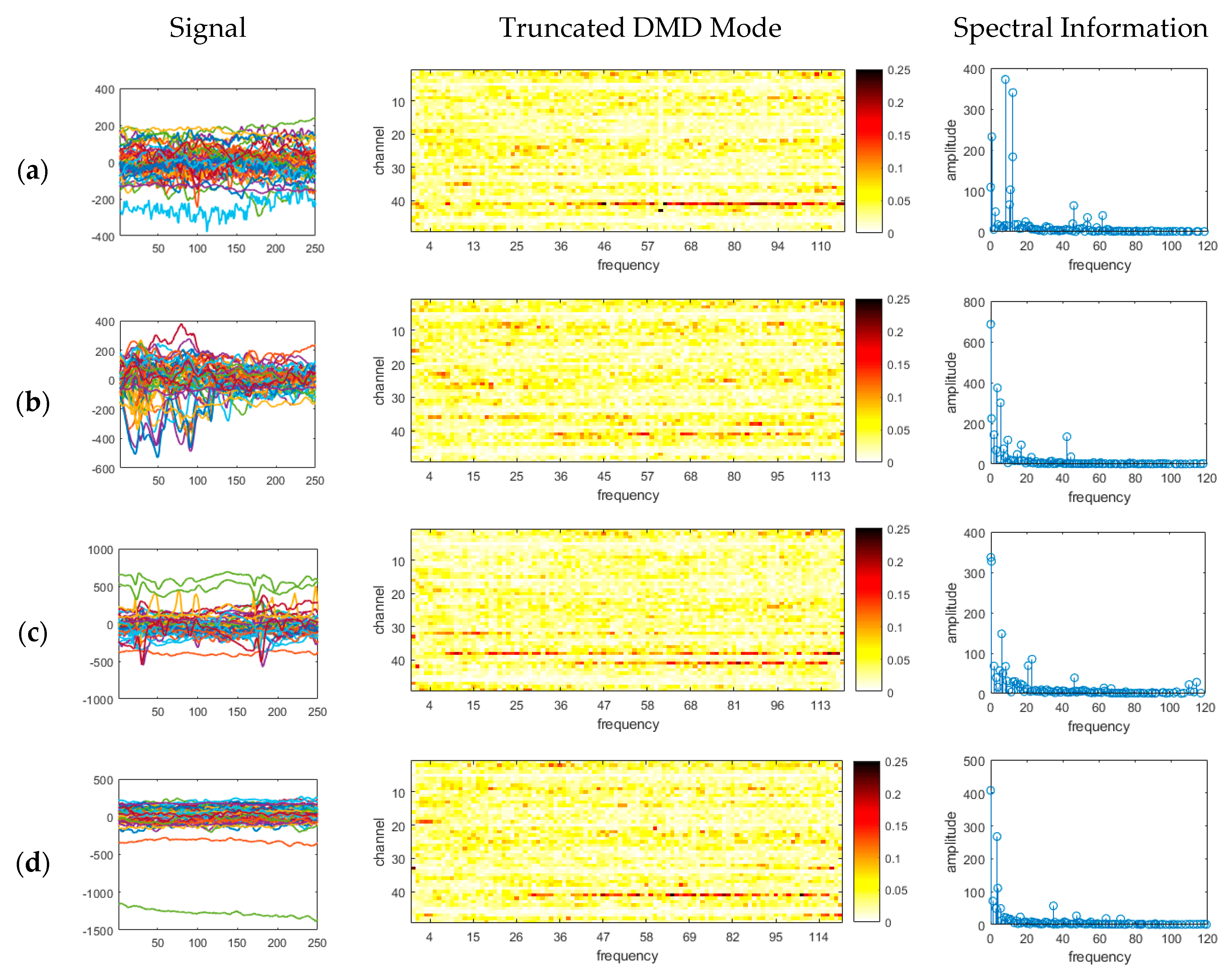
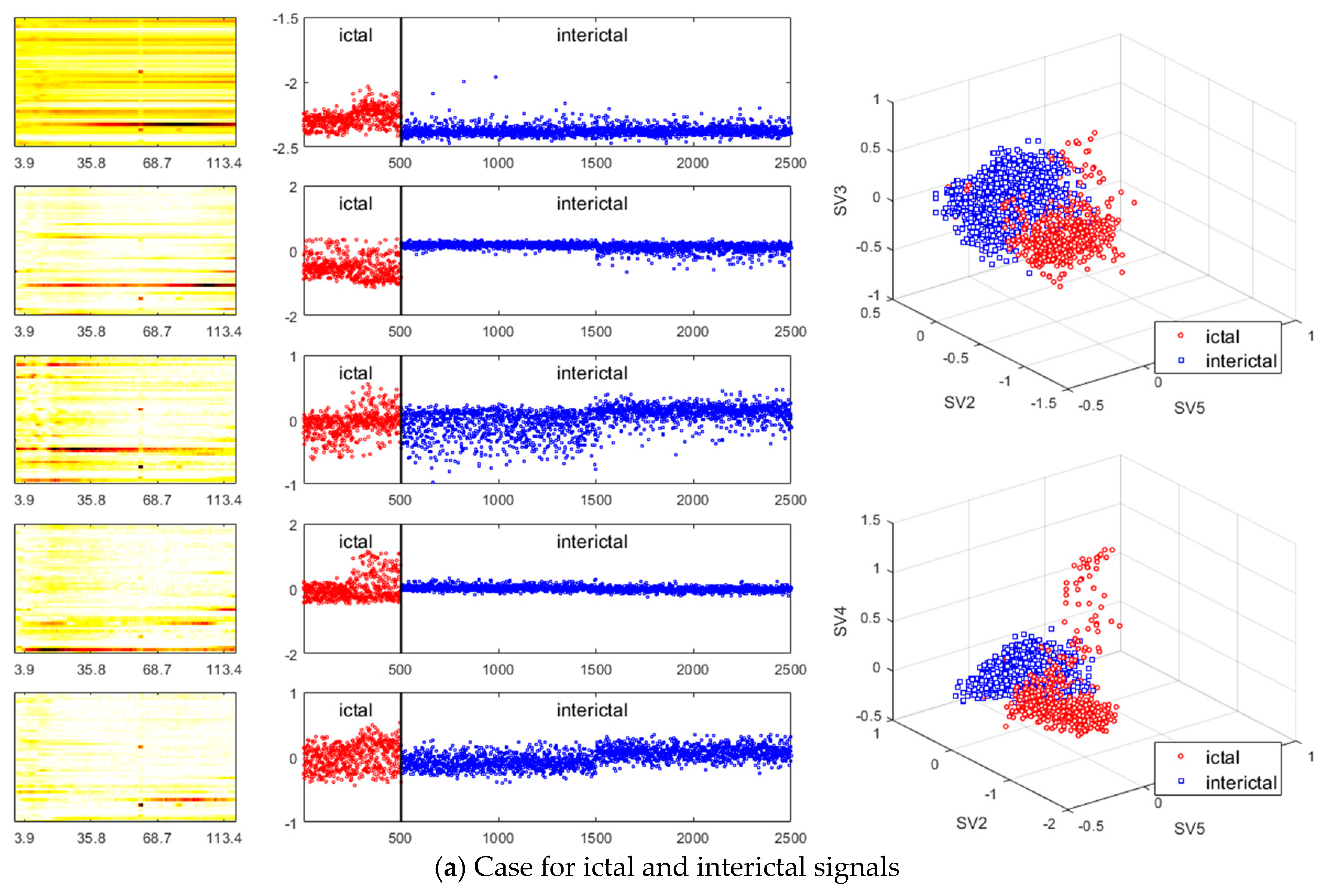
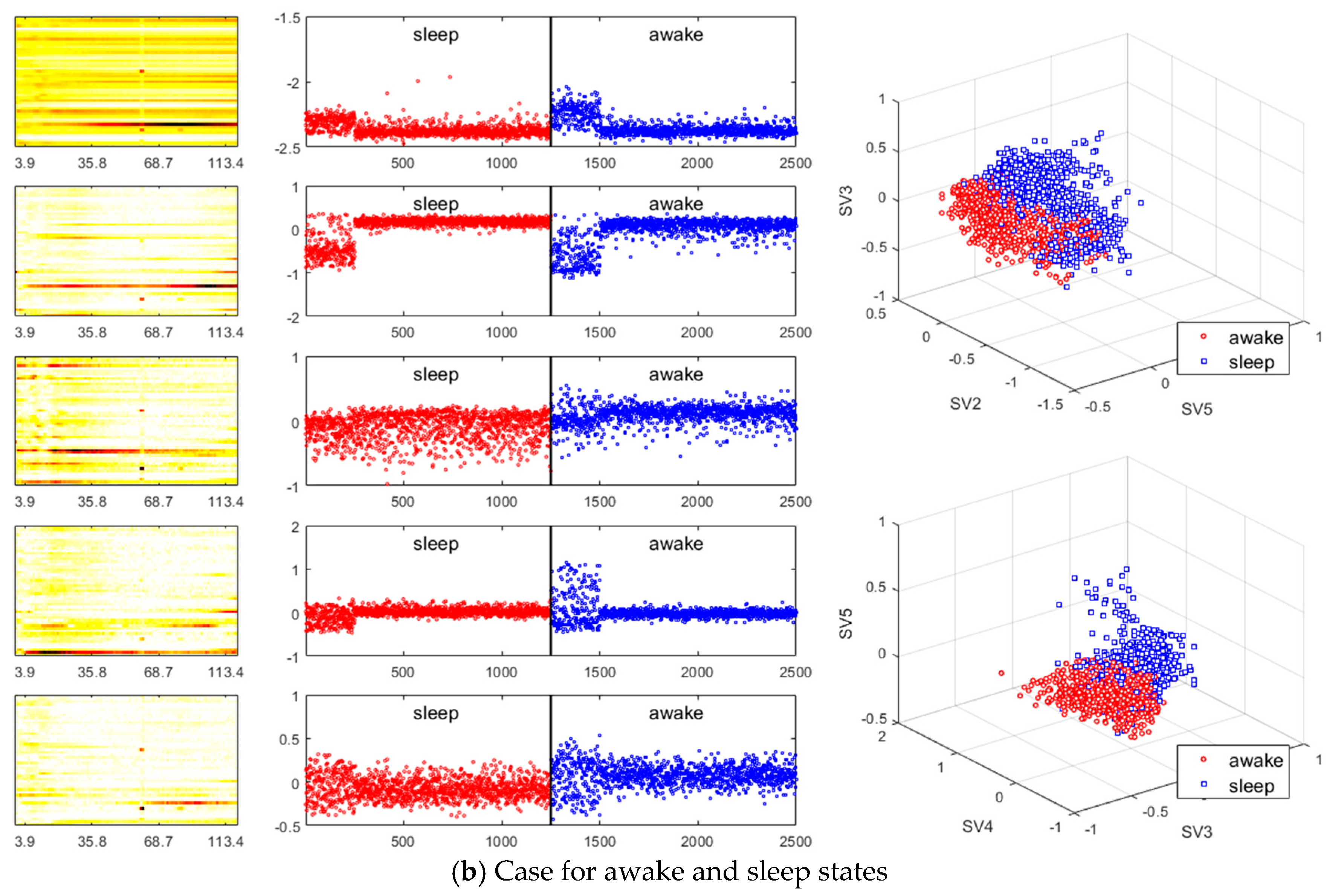
- Existing studies on the identification of the ictal onset zone mostly focus on increasing statistical accuracy by finding the proper feature method for automatic seizure detection [26,27,28,29]. In the extracted feature, it is difficult to directly understand the mechanisms and characteristics, such as the ictal slow shift and HFO, of the ictal state for the individual patient. Figure 1 and Figure 2 show the method presented in this paper directly interprets the dynamic mode of EEG signal to locate the position of the specific channel and frequency in which the slow shift phenomenon and HFO phenomenon occur and to track the state change over time of the pre-ictal and ictal state. Therefore, it is expected that the method presented in this paper can contribute significantly to further studies related to the prediction of the state of the ictal, which plays an important role in the epilepsy study.
- (b)
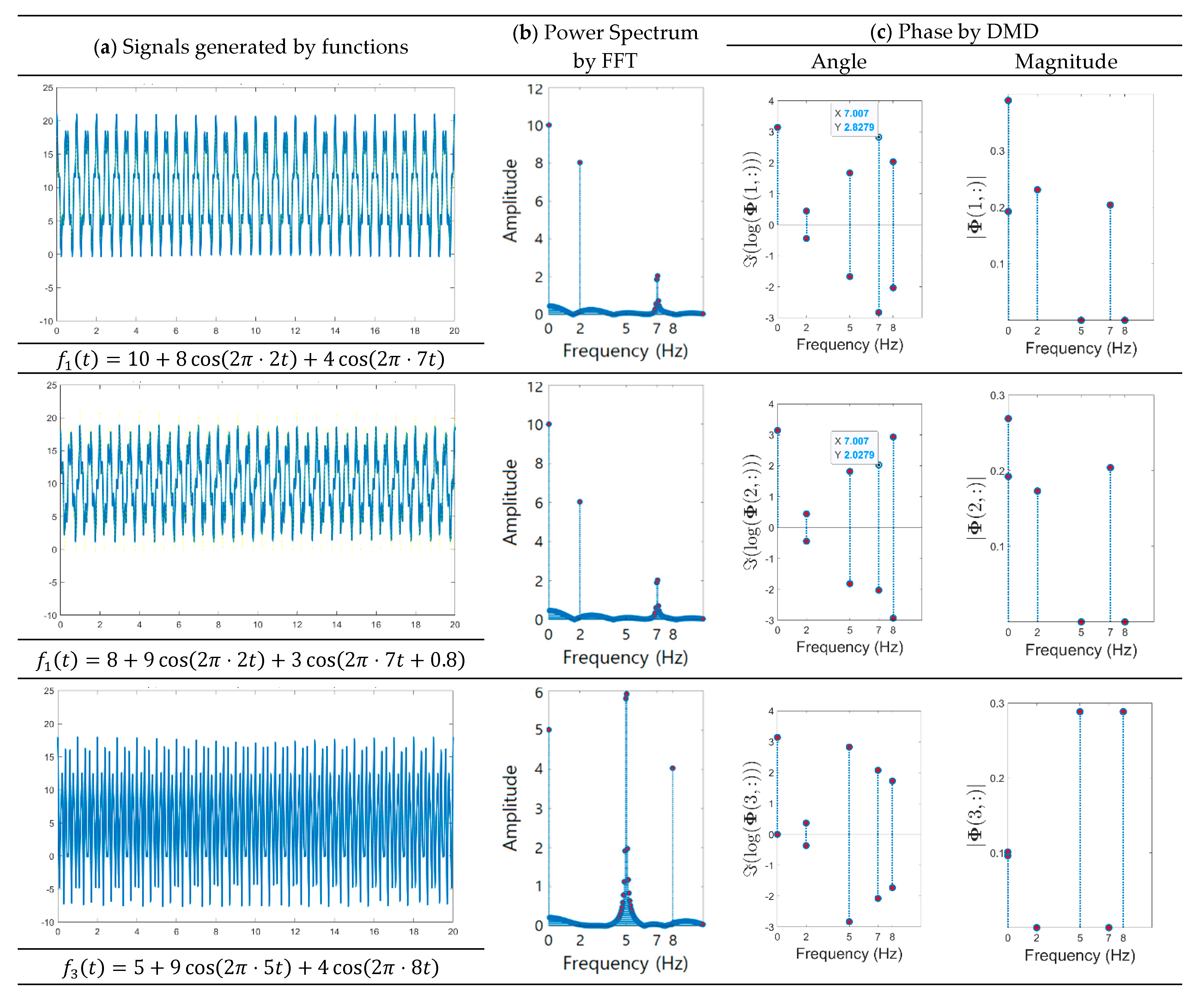
- A method of investigating the correlation between channels was recently proposed in [30] as another way of studying the mechanism of ictal phenomena for the prediction of epilepsy. In the work, it was shown that the information about changes in the network correlation provides insight for epileptic brain behavior, demonstrating that other locations of the brain are involved in the seizure other than the focus, and that there might be early indications for the seizure. The method that we present is to obtain left singular vectors that are clustered into correlated channels through the dimension reduction process for the libraries of the dynamic mode with SVD applied. Each of the left singular vectors is divided into channels that are simultaneously involved in the ictal and interictal and channels that only respond to the ictal state (for example, the figures in the third row of Figure 6a show the components of the channels and frequencies that react in both ictal and interictal states, whereas the figures in its second row identify the components that react in the ictal state only). In other words, the components of the dynamic mode contain information on both the channel and frequency, so a more sophisticated analysis can be made.
- (c)
- The outcome of machine learning algorithms, such as an artificial neural network (ANN) and support vector machine (SVM) algorithms, can be effectively used as physiological feedback to advanced implantable medical devices (IMDs), which will then expectantly operate in a closed-loop fashion. As a result, the efficacy of the treatment will be improved by delivering intense targeted stimulation [31]. However, due to the inherent uncertainty of the brain signals and because EEG and ECoG vary a lot depending on age, environments, drug intake, etc., it is extremely challenging [32]. In addition, in the case of epilepsy, seizure patterns for each patient not only are unique but also vary with time [33]. As a result, a generic machine learning algorithm will not efficiently work for the same patient as well as for large patient groups [34]. A more common way to approach this problem is to collect continuous EEG and ECoG data, which captures unique ictal patterns and interictal patterns for individual patients. The collected data is then used to train machine learning algorithms and to develop a patient-specific seizure prediction algorithm. However, this approach is not scalable to large patient groups as developing patient-specific machine learning models requires enormous efforts in collecting, labeling, and training the machine learning algorithms. A number of studies are underway on the development of an online seizure advisory system that automates this process [35]. However, due to the risk and complexity involved in ECoG, it is strictly restricted to clinical use. There is no ECoG data obtained from the measurement of healthy individuals’ epilepsy [30]. For this reason, from a clinical point of view, tools for prediction and identification based on ECoG data should be able to produce accurate results for subsequent observational data with the number of measurement data from a patient being minimized. Moreover, the tools should train the ECoG dataset obtained from an individual patient, considering the inherent specificities of the patient. The methodology proposed in this work meets those requirements above. In order to avoid possible errors in the derived results, these tools implement an intuitive configuration and fast calculation speed so that the features created by the tools enable us to instantly interpret the attributes associated with known ictal states and to judge whether and when the epileptic seizure occurs by monitoring the changes of magnitudes (i.e., whether the value is meaningful or not) and imaginary (i.e., phase angle) regarding the modes between channels in some special banded frequencies. Moreover, the methodology allows real-time visual monitoring of the modes.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fisher, R.S.; Boas, W.E.; Blume, W.; Elger, C.; Genton, P.; Lee, P.; Engel, J. Epileptic seizures and epilepsy: Definitions proposed by the International League Against Epilepsy (ILAE) and the International Bureau for Epilepsy (IBE). Epilepsia 2005, 46, 470–472. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Inouchi, M.; Matsuhashi, M.; Matsumoto, R.; Hitomi, T.; Daifu-Kobayashi, M.; Kobayashi, K.; Nakatani, M.; Kanazawa, K.; Shimotake, A.; et al. Interictal Slow and High-Frequency Oscillations: Is it an Epileptic Slow or Red Slow? J. Clin. Neurophysiol. 2019, 36, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Abduhay, E.; Alafeef, M.; Abdelhay, A.; Al-Bashir, A. Classification of Normal, Ictal and Inter-ictal EEG via Direct Quadrature and Random Forest Tree. J. Med. Biol. Eng. 2017, 37, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.F.; Wang, H.C.; Chang, W.L. Combination of EEG Complexity and Spectral Analysis for Epilepsy Diagnosis and Seizure Detection. EURASIP J. Adv. Signal Process. 2010, 2010, 853434. [Google Scholar] [CrossRef]
- Redelico, F.O.; Traversaro, F.; Garcia, M.C.; Silva, W.; Rosso, O.A.; Risk, M. Classification of normal and pre-ictal EEG signals using permutation entropies and a generalized linear model as a classifier. Entropy 2017, 19, 72. [Google Scholar] [CrossRef]
- Kanazawa, K.; Matsumoto, R.; Imamura, H.; Matsuhashi, M.; Kikuchi, T.; Kunieda, T.; Mikuni, N.; Miyamoto, S.; Takahashi, R.; Ikeda, A. Intracranially recorded ictal direct current shifts may precede high frequency oscillations in human epilepsy. Clin. Neurophysiol. 2015, 126, 47–59. [Google Scholar] [CrossRef]
- Ochoa, J.G.; Rusyniak, W.G. Description of Ictal HFO Mapping in Patients with Both Temporal and Extratemporal Seizure Focus. Neurol. Res. Int. 2016, 5, 1–4. [Google Scholar] [CrossRef]
- Parvez, M.Z.; Paul, M. Classification of Ictal and Interictal EEG signals. In Proceedings of the 10th IASTED Conference on Biomedical Engineering, Innsbruck, Austria, 13–15 February 2013; ACTA Press: Alberta, AB, Canada, 2013; pp. 113–141. [Google Scholar]
- Rilling, G.; Flandrin, P.; Gonçalves, P. On empirical mode decomposition and its algorithms. In Proceedings of the IEEE-EURASIP Workshop on Nonlinear Signal and Image Processing, Grado, Italy, 8–11 June 2003; pp. 8–11. [Google Scholar]
- Kutz, J.N. Data-Driven Modeling & Scientific Computation: Methods for Complex Systems & Big Data; Oxford University Press: Oxford, UK, 2013; pp. 354–357. [Google Scholar]
- Ódor, G.; Kelling, J. Critical synchronization dynamics of the Kuramoto model on connectome and small world graphs. Sci. Rep. 2019, 9, 19621. [Google Scholar] [CrossRef]
- Lobier, M.; Siebenhühner, F.; Palva, S.; Palva, J.M. Phase transfer entropy: A novel phase-based measure for directed connectivity in networks coupled by oscillatory interactions. Neuroimage 2014, 85, 853–872. [Google Scholar] [CrossRef]
- Kutz, J.N.; Brunton, S.L.; Brunton, B.W.; Proctor, J.L. Dynamic Mode Decomposition: Data-Driven Modeling of Complex Systems; SIAM: Philadelphia, PA, USA, 2016; pp. 119–128. [Google Scholar]
- Solaija, M.S.J.; Saleem, S.; Khurshid, K.; Hassan, S.A.; Kamboh, A.M. Dynamic Mode Decomposition Based Epileptic Seizure Detection from Scalp EEG. IEEE Access. 2018, 6, 38683–38692. [Google Scholar] [CrossRef]
- Tu, J.H.; Rowley, C.W.; Luchtenburg, D.M.; Brunton, S.L.; Kutz, J.N. On dynamic mode decomposition: Theory and applications. J. Comput. Dyn. 2014, 1, 391–421. [Google Scholar] [CrossRef]
- Murai, T.; Hitomi, T.; Matsuhashi, M.; Matsumoto, R.; Kawamura, Y.; Kanda, M.; Takahashi, R.; Ikeda, A. Scalp-EEG could record both ictal DC shift and HFO together even with time constant 2 sec. J. Clin. Neurophysiol. 2019, in press. [Google Scholar]
- Ikeda, A.; Taki, W.; Kunieda, T.; Terada, K.; Mikuni, N.; Nagamine, T.; Yazawa, S.; Ohara, S.; Hori, T.; Kaji, R.; et al. Focal ictal DC shifts in human epilepsy as studied by subdural and scalp recording. Brain 1999, 122, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.J. Dynamic mode decomposition of numerical and experimental data. J. Fluid Mech. 2010, 656, 5–28. [Google Scholar] [CrossRef]
- Antoulas, A.C. Approximation of Large-Scale Dynamical System; SIAM: Philadelphia, PA, USA, 2005; pp. 31–57. [Google Scholar]
- Brunton, L.A.; Johnson, J.G.; Ojemann, J.G.; Kutz, J.N. Extracting spatial–temporal coherent patterns in large-scale neural recordings using dynamic mode decomposition. J. Neurosci. Methods 2016, 258, 1–15. [Google Scholar] [CrossRef]
- Gavish, M.; Donoho, D.L. The optimal hard threshold for singular values is . IEEE Trans. Inf. Theory 2014, 60, 5040–5053. [Google Scholar] [CrossRef]
- Kecman, V.; Huang, T.-M.; Vogt, M. Iterative Single Data Algorithm for Training Kernel Machines from Huge Data Sets: Theory and Performance. In Support Vector Machines: Theory and Applications; Wang, L., Ed.; Springer: Berlin, Germany, 2005; pp. 255–274. [Google Scholar]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Focus on Epilepsy. Available online: https://www.nature.com/articles/nn.3964.pdf (accessed on 19 May 2019).
- Imamura, H.; Matsumoto, R.; Inouchi, M.; Matsuhashi, M.; Mikuni, N.; Takahashi, R.; Ikeda, A. Ictal wideband ECoG: Direct comparison between ictal slow shifts and high frequency oscillations. Clin. Neurophysiol. 2011, 122, 1500–1504. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, L.; Shen, Y.; Wang, J.; Zhao, R.; Dai, J. A new algorithm for classification of ictal and pre-ictal epilepsy ECoG using MI and SVM. In Proceedings of the 2017 International Conference on Signals and Systems(ICSigSys), Sanur, Indonesia, 16–18 May 2017; pp. 212–216. [Google Scholar]
- Lu, Y.; Ma, Y.; Chen, C.; Wang, Y. Classification of single-channel EEG signals for epileptic seizures detection based on hybrid features. Technol. Health Care 2018, 26, 337–346. [Google Scholar] [CrossRef]
- Nandy, A.; Alahe, M.A.; Nasim Uddin, S.M.; Alam, S.; Nahid, A.; Awal, M.A. Feature Extraction and Classification of EEG Signals for Seizure Detection. In Proceedings of the 2019 International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), Dhaka, Bangladesh, 10–12 January 2019; pp. 480–485. [Google Scholar]
- Zhou, M.; Tian, C.; Cao, R.; Wang, B.; Niu, Y.; Hu, T.; Guo, H.; Xiang, J. Epileptic Seizure Detection Based on EEG Signals and CNN. Front. Aging Neurosci. 2018, 12. [Google Scholar] [CrossRef]
- Yekutieli, Z.; Ben-Jacob, E. ECoG Correlation Variation for Epilepsy Research. Epilepsy J. 2018, 4, 1–6. [Google Scholar] [CrossRef]
- Ramgopal, S.; Thome-Souza, S.; Jackson, M.; Kadish, N.E.; Fernández, I.S.; Klehm, J.; Bosl, W.; Reinsberger, C.; Schachter, S.; Loddenkemper, T. Seizure detection, seizure prediction, and closed-loop warning systems in epilepsy. Epilepsy Behav. 2014, 37, 291–307. [Google Scholar] [CrossRef] [PubMed]
- Andrzejak, R.G.; Chicharro, D.; Elger, C.E.; Mormann, F. Seizure prediction: Any better than chance? Clin. Neurophys. 2009, 120, 1465–1478. [Google Scholar] [CrossRef]
- Freestone, D.R.; Karoly, P.J.; Cook, M.J. A forward-looking review of seizure prediction. Curr. Opin. Neurol. 2017, 30, 167–173. [Google Scholar] [CrossRef]
- Cook, M.J.; O’Brien, T.J.; Berkovic, S.F.; Murphy, M.; Morokoff, A.; Fabinyi, G.; D’Souza, W.; Yerra, R.; Archer, J.; Litewka, L.; et al. Prediction of seizure likelihood with a long-term, implanted seizure advisory system in patients with drug-resistant epilepsy: A first-in-man study. Lancet Neurol. 2013, 12, 563–571. [Google Scholar] [CrossRef]
- Karuppiah Ramachandran, V.R.; Alblas, H.J.; Le, D.V.; Meratnia, N. Towards an Online Seizure Advisory System—An Adaptive Seizure Prediction Framework Using Active Learning Heuristics. Sensors 2018, 18, 1698. [Google Scholar] [CrossRef]
| Record Name | State | No. of Snapshots (Total/Ictal) | Time (sec) (Total/Ictal) | No. of Signals (Total/Ictal) | Type |
|---|---|---|---|---|---|
| Pt1_ictal1 | Awake | 692,000/13,201 | 2768/52.8 | 11,835/264 | Training |
| Pt1_ictal2 | Sleep | 735,000/16,234 | 2940/64.9 | 14,690/325 | Training |
| Pt1_ictal3 | Sleep | 737,750/24,407 | 2951/97.6 | 14,750/489 | Testing |
| Pt1_ictal4 | Awake | 734,750/11,545 | 2939/46.2 | 14,690/231 | Testing |
| Statistics | Measurement | ||
|---|---|---|---|
| Accuracy | Sensitivity | Specificity | |
| Ictal vs. Interictal | 99.68% | 93.99% | 99.83% |
| Awake vs. Sleep | 88.24% | 85.41% | 91.55% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, J.-H.; Tsuda, I.; Lee, Y.J.; Ikeda, A.; Matsuhashi, M.; Matsumoto, R.; Kikuchi, T.; Kang, H. Pattern Recognition in Epileptic EEG Signals via Dynamic Mode Decomposition. Mathematics 2020, 8, 481. https://doi.org/10.3390/math8040481
Seo J-H, Tsuda I, Lee YJ, Ikeda A, Matsuhashi M, Matsumoto R, Kikuchi T, Kang H. Pattern Recognition in Epileptic EEG Signals via Dynamic Mode Decomposition. Mathematics. 2020; 8(4):481. https://doi.org/10.3390/math8040481
Chicago/Turabian StyleSeo, Jong-Hyeon, Ichiro Tsuda, Young Ju Lee, Akio Ikeda, Masao Matsuhashi, Riki Matsumoto, Takayuki Kikuchi, and Hunseok Kang. 2020. "Pattern Recognition in Epileptic EEG Signals via Dynamic Mode Decomposition" Mathematics 8, no. 4: 481. https://doi.org/10.3390/math8040481
APA StyleSeo, J.-H., Tsuda, I., Lee, Y. J., Ikeda, A., Matsuhashi, M., Matsumoto, R., Kikuchi, T., & Kang, H. (2020). Pattern Recognition in Epileptic EEG Signals via Dynamic Mode Decomposition. Mathematics, 8(4), 481. https://doi.org/10.3390/math8040481







