Abstract
Traditional EEG data augmentation methods may alter the spatiotemporal characteristic distribution of brain electrical signals. This paper proposes a new method based on the Gaussian Mixture Model (GMM): First, we use the GMM to decompose data samples of the same category to obtain Gaussian coefficients and take the product of the probability coefficient and the weight matrix as the feature matrix. Then, we randomly select two EEG feature matrices and determine the similarity based on the magnitude of the correlation coefficients of their column vectors and exchange columns exceeding the threshold to obtain a new matrix. Finally, we generate new data according to the new matrix, as well as its mean and variance. Experiments on public datasets show that this method effectively retains the original data’s spatiotemporal and distribution characteristics. In classification model tests, compared with the original data without augmentation, the classification accuracy is improved by up to 29.84%. The t-SNE visualization results show that the generated data are more compact. This method can create a large number of new EEG signals similar to the original data in terms of spatiotemporal characteristics, improve classification accuracy, and enhance the performance of Brain–Computer Interface (BCI) systems.
Keywords:
EEG; Gaussian mixture model; spatiotemporal; brain–computer interface (BCI); data augmentation MSC:
68T07
1. Introduction
An electroencephalogram (EEG) is a biological signal generated by the electrical activity of neurons in the brain, which can reflect the real-time physiological state of the brain. An EEG records the spontaneous and rhythmic electrical activity of groups of neurons in the brain through scalp electrodes. It has the advantages of being non-invasive, having high temporal resolution (millisecond level), being portable, and being low-cost. EEG recordings, especially those capturing rare neurological conditions or specific events like seizures, are often scarce. Augmentation helps artificially expand the dataset, enabling better training of machine learning models for diagnosis and analysis. Specifically, augmented data help prevent overfitting in machine learning models by introducing diverse examples, ensuring they generalize well to unseen data, which is critical for accurate clinical diagnosis.Therefore, EEG is widely used in medical diagnosis (such as epilepsy detection and sleep staging) and cognitive neuroscience research (such as brain–computer interface and emotion recognition) [1]. However, EEG signals have characteristics such as nonlinearity, non-stationarity, individual differences, and low signal-to-noise ratios (usually 0.1–10 dB) [2]. In clinical practice, due to limitations in patient cooperation, data acquisition cost, and ethical issues, it is difficult to obtain high-quality EEG data. This bottleneck severely restricts the training effect of machine learning models based on EEG (such as the risk of overfitting) and their application reliability in scenarios such as early disease diagnosis and personalized treatment [3]. Therefore, developing efficient EEG data augmentation techniques to generate synthetic data with physiological rationality and diversity to expand the training set has become a key research direction to improve model generalization ability and clinical practicality [4].
Traditional EEG data augmentation methods mainly include the following:
The first method is that based on data morphology changes. Asuka Sakai et al. proposed the generation of enhanced data by moving the time axis and magnifying the measurement data potential and conducted experimental verification on a small dataset to improve the prediction accuracy [5]. Mario Michael Krell et al. proposed a rotation distortion method similar to image affine/rotation distortion to generate enhanced EEG data [6]. These methods simulate the influence of factors such as head movement and muscle tension on EEG data and generate data based on the original EEG data that can better represent time and frequency changes. However, these geometric transformations may destroy the time-domain and frequency-domain characteristics of the data, thereby affecting the data quality [7,8].
The second set of methods is based on signal segmentation and recombination. This approach segments specific time-window EEG data according to the temporal characteristics of the EEG signal and reconstructs new data by randomly selecting fragments [9,10,11,12]. These methods are intuitive and simple but may exacerbate model overfitting due to similarity after augmentation [13,14,15,16]. Another common method is to add noise. This technique involves adding random matrices from Gaussian distributions (Gaussian or salt-and-pepper noise) to the original EEG data to simulate real-world noise interference for data augmentation. This method can effectively increase dataset diversity and improve model robustness [17,18,19]. However, introducing artifacts in the original signal complicates the verification of the true psychological state response of the new EEG signal, making it challenging to validate model accuracy.
The third approach involves the use of methods based on data generation. There are two typical types. One is the Variational Autoencoder (VAE). The VAE randomly samples points from the learned latent space, then passes these samples through a decoder network, which reconstructs them into new samples. A downside of this method is that it requires a large number of data samples [20,21]. The second is Generative Adversarial Networks (GANs). GANs and their variants can generate artificial data by training a generative network and a discriminative network [22,23,24]. The generative network takes random noise from a specific distribution (e.g., Gaussian) and attempts to generate synthetic data similar to real data, while the discriminative network is trained to discriminate between real and synthetic data. These two networks are adversarial; after sufficient training, the generative network produces similar signals [4,25,26]. A common drawback of these methods is that they require a certain amount of data to support the adversarial training of the generator and discriminator [27,28,29].
A common characteristic of the above three methods for EEG data augmentation is that they do not fully consider the distribution characteristics of the original data. The newly constructed data may alter the spatiotemporal features of the original EEG, leading to data distortion. In view of this, this paper proposes an EEG data augmentation method based on the Gaussian Mixture Model (GMM). This method employs the parameters of each Gaussian submodel to ensure that the generated data retain the original EEG signals’ spatiotemporal dynamic properties.
The novelty and contributions of this study are outlined as follows:
(1) We propose an EEG data augmentation method based on Gaussian mixture model (GMM) feature reconstruction. First, we perform Gaussian mixture model decomposition on data samples of the same type to obtain the features of the sample type, utilizing the characteristics of the Gaussian submodels (probability multiplied by weight) here. Secondly, we randomly select two samples of the same type, analyze the similarity of their feature components, and exchange those with similar features to form new submodel features. Finally, we generate new data based on the features of the submodels (probability multiplied by weight), mean, and variance.
(2) By analyzing the performance of augmented electroencephalogram (EEG) samples and original EEG samples in terms of temporal and spatial characteristics, the differences in spatiotemporal characteristics among the enhanced data are demonstrated.
(3) Classification tasks are performed on EEG signals generated by traditional methods and the method proposed in this paper to compare improvements in classification performance.
The rest of this paper is organized as follows. Section 2 mainly introduces our proposed method and experimental data, including the principles of Gaussian feature reconstruction and data generation. Section 3 primarily discusses experiments and results. Section 4 discusses and analyzes experimental results. Finally, Section 5 summarizes this work.
2. Materials and Methods
2.1. The BCI Competition IV Dataset 2a
The BCI Competition IV Dataset 2a dataset, released by an authoritative competition, covers multiple subjects and experimental conditions and has a great influence in the field of brain–computer interface, attracting many top scientific research institutions and scholars to participate, which is of great significance for exploring the relationship between brain electricity and cognitive function and in verifying brain–computer interface technology.
It is an electroencephalogram (EEG) dataset based on visually induced motor imagery collected by the American Enco company [30]. The experiment was conducted with 9 participants, each completing two sessions. Each session included data from 4 categories, with 72 trials per category. Therefore, each participant contributed a total of 288 trials, comprising 72 trials for left-hand motor imagery, 72 trials for right-hand motor imagery, 72 trials for foot motor imagery, and 72 trials for tongue motor imagery.
2.2. Gaussian Mixture Model
A GMM (Gaussian mixture model) is a probabilistic model that postulates that all data points are generated from a mixture of a finite number of Gaussian distributions with unknown parameters [31]. A Gaussian mixture model is a probability density function that consists of a mixture of several Gaussian distributions, where each Gaussian distribution is referred to as a component and the number of Gaussian mixture models is K. The linear combination of these Gaussian distributions constitutes a Gaussian mixture model with the following probability distribution:
For simplicity of expression, here, x is a one-dimensional variable, but he derivation applies to multidimensional variables. is the weight coefficient of the k-th Gaussian distribution, , and ; denotes the density of the k-th Gaussian distribution, , where denotes the mean of the k-th Gaussian distribution and denotes the standard deviation of the k-th Gaussian distribution. Formula (2) represents the k-th submodel.
The optimal parameters are the peak point where gets its maximum value. . A GMM is a probability model with both observed variables and hidden variables, which cannot be directly estimated and need to be estimated by the expectation-maximization (EM) algorithm [32]. The expectation (E) step and maximization (M) step of the EM algorithm are alternately and iteratively executed. The E step creates a function to calculate the log-likelihood expectation by using the current parameter estimates, and the M step estimates the parameter that maximizes the expected logarithm. These parameter estimates are then used to determine the distribution of the latent variables in the next E step.
The observed data () are reconstructed by a Gaussian mixture model, and the probability is described as Formula (3), where . The estimation of the parameter of the Gaussian mixture model can be achieved using the EM algorithm.
A hidden variable () describes the i-th EEG trial (x)belonging to the k-th model () according to the probability () in a Gaussian mixture model; then, data () are observed according to the probability distribution of the k-th submodel. The hidden variable () is unknown, indicating whether the observed data of EEG motor imagery are derived from the k-th submodel or not:
The complete data include the EEG motor imagery observation data () and the hidden variable (); then, the complete data are .
According to the I.I.D. (Independent Identical Distribution) hyperthesis, a given set of complete data’s likelihood function is expressed as follows:
where . The log-likelihood function for complete data is expressed as follows:
Step E: The probability that the data are generated by each submodel is estimated. First, the Q function is determined, and the evaluated expectation is calculated using the current parameter estimates:
, which is denoted as , is calculated by the method shown in Formula (8):
Substituting and into Formula (8), we obtain the following:
Step M: The parameters of each Gaussian model are updated. Step M is the model parameter in the new iteration round, which is obtained by solving the maximum value of the function expressed to :
Taking the partial derivative of Equation (9) and setting the partial derivative to 0, the parameters are updated as follows:
Steps E and M are iterated until the values of the log-likelihood function converge.
2.3. Gaussian Mixture Model-Based EEG Feature
Unlike traditional EEG microstates, Gaussian mixture model (GMM)-based EEG microstates are distinguished by their use of the GMM to decompose EEG microstates into a mixture representation rather than a unique one-hot representation. It has been proven that the classification capability of the GMM-based microstate model under MI tasks is augmentation [33].
2.4. Gaussian Mixture Model-Based EEG Data Augmentation Method
As shown in Algorithm 1, taking a ten-component Gaussian mixture model (GMM) as an example, the preprocessed electroencephalogram (EEG) data () serve as the input to the model, corresponding to a three-dimensional dataset. Here, N represents the number of sensors on the EEG device, corresponding to the ‘channel’ variable in the algorithm, and T is the number of sampling points per sample, corresponding to the ‘n_samples’ variable in the algorithm. The ‘n_components’ variable indicates the number of submodels in the GMM, which is set to 10. Algorithm process for EEG Data augmentation method based on Gaussian mixture model.
In this paper, each sampling point of the original multi-channel EEG data sample is divided into a linear combination of multiple Gaussian distributions, and the duration, frequency or coverage of each Gaussian submodel in the sample period constitutes the characteristics, covering the dynamic spatial–temporal information. The points are exchanged with the same characteristics in different samples at random so as to improve the data diversity while maintaining the overall characteristics of the sample. Then, new EEG data are generated through reconstruction. The principle is shown in Figure 1.
| Algorithm 1: EEG Data Augmentation Method Based on Gaussian Mixture Model |
| Input: original_data = (trial, channel, n_samples), original_labels = (trial,) |
| Output: gmm_data |
| Step1: Set model parameters. n_components = 10; random_stata = 42; = 0.8 |
| Step2: With the same label data, calculate the GGM features |
| probs = gmm.predict_proba (original_data) |
| means = gmm.means_ |
| covariances = gmm. covariances_ |
| weights = gmm.weights_robs |
| Step3: Sampling. |
| for do |
| for do |
| for do |
| end |
| end |
| end |
| Step4: Calculate the product matrix. |
| weighted_probs_values = gmm.weights_ * probs |
| Step5: Swap similar points. |
| weighted_probs_values = swap_columns (weighted_probs_values, ) |
| Step6: Fit the data. |
| data_generate_sampel = np.matmul (weighted_probs_values, fitted_values) |
| Step7: Swap channels and reconstruct the data. |
| gmm_data = GMM_FEATURE (probability = probability, random_state=42) |
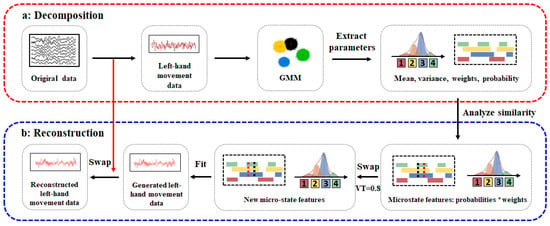
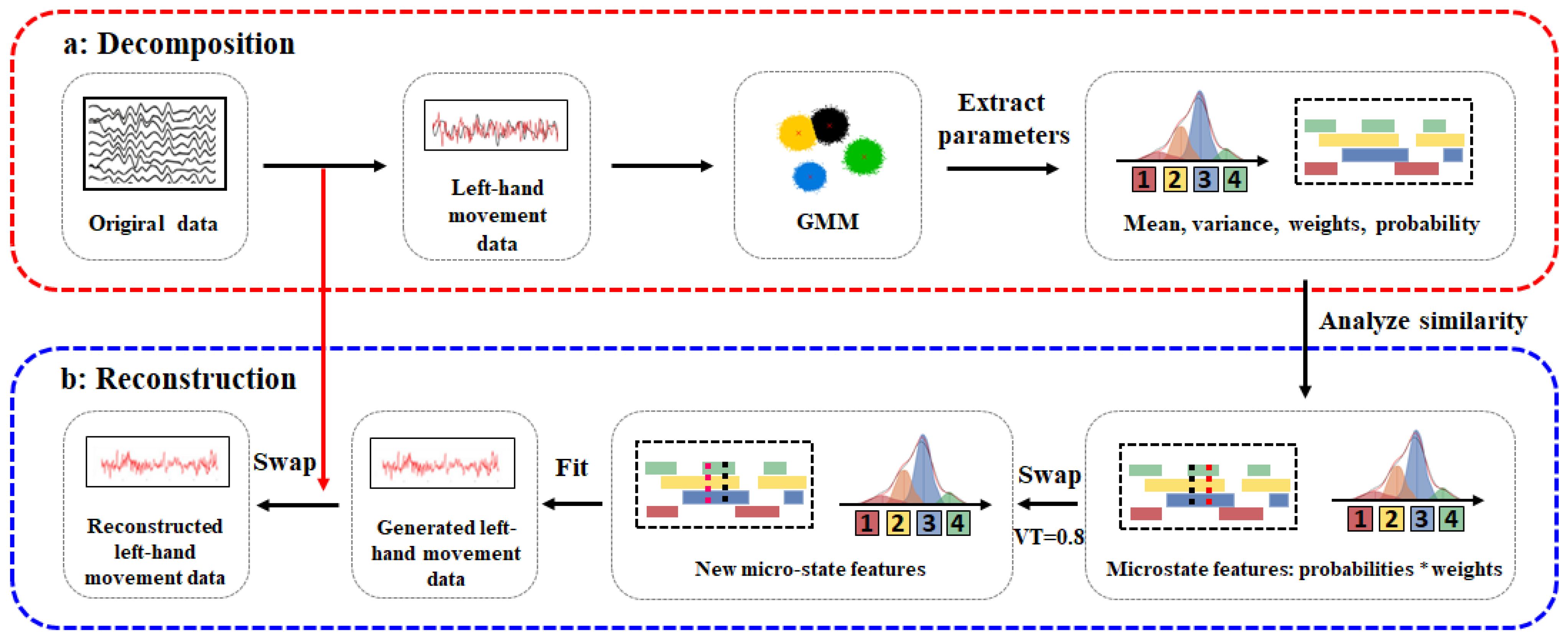
Figure 1.
Principle diagram of the EEG data augmentation method based on Gaussian mixture models. Data preprocessing mainly includes filtering (0–38 HZ), segmentation and baseline correction, removal of artifacts, and referencing. (a) Decomposition: First the original EEG data are preprocessed; then, the Gaussian mixture model (GMM) is used for decomposition. The product matrix of weights and probabilities is calculated for adjacent data points with the same label. Points with similarity coefficients greater than a threshold are swapped to obtain a new feature matrix. (b) Reconstruction: First, a random seed is set to ensure the reproducibility of results. For each sample, a channel is randomly selected, and channels are swapped between the original data and the fitted data. Finally, new EEG data are generated.
2.4.1. Decomposition
As shown in Figure 1a, during the decomposition process, we first preprocess the original EEG data, then apply the Gaussian mixture model to calculate the product matrix of weights and probabilities for adjacent data points with the same label. Points with a similarity coefficient greater than a threshold are exchanged to obtain a new feature matrix. The steps are outlined as follows:
Step 1: Preprocess the original data with filtering, denoising, and other operations.
Step 2: Conduct Gaussian decomposition on the data based on labels to obtain the probability, weight, mean, and variance for each category.
Step 3: Multiply the weight of each sample by the probability matrix to obtain the feature matrix.
Step 4: Calculate the similarity coefficients of the feature matrix points for adjacent data with the same label. For points with a similarity coefficient greater than 0.8, exchange their positions to generate a new feature matrix. The purpose is to reduce overfitting in the generated data.
2.4.2. Reconstruction
As shown in Figure 1b, during the reconstruction process, we set a random seed to ensure the reproducibility of the results. For each sample, a channel is randomly selected, and the original data and fitted data are exchanged for that channel, ultimately generating new EEG data. The steps are outlined as follows:
Step 1: Combine the feature matrix with the mean and variance to refit the EEG data.
Step 2: Set a random seed to ensure the reproducibility of the results.
Step 3: Randomly select a channel for each sample according to a certain probability.
Step 4: Exchange the original data and fitted data for the corresponding channel.
Step 5: Reconstruct to obtain new EEG data.
2.5. Evaluation and Criteria
2.5.1. Pearson Correlation Coefficient
The Pearson correlation coefficient is a statistical indicator used to measure the linear correlation between two variables (X and Y). The specific formula is expressed as follows:
where = ; E denotes the mathematical expectation; and and are the population means of X and Y, respectively.
2.5.2. Classification Accuracy
The classification accuracy rate refers to the proportion of the total number of samples that the model correctly predicts, reflecting the model’s ability to classify the overall data. It is one of the important indicators for evaluating the performance of the model in classification problems and is widely selected and used because of its intuitiveness, universality, applicability, and significant role in the preliminary screening of models. The calculation formula is expressed as follows:
refers to the number of samples that are actually positive and correctly predicted as positive by the model, refers to the number of samples that are actually negative and correctly predicted as negative by the model, refers to the number of samples that are actually negative but incorrectly predicted as positive by the model, and refers to the number of samples that are actually positive but incorrectly predicted as negative by the model.
3. Experiment and Results
3.1. Data Preprocessing
Each sample data point in the experiment is a segment of 2–6 s from each trial. Preprocessing is carried out using Python 3.8.8 and the MNE 0.23.4 toolkit [34]. The baseline correction, principal component analysis (PCA), and artifact rejection preprocessing steps are detailed in an article by Liao et al. [33].
3.2. Comparison of Data Characteristics Generated by Different Augmentation Methods
To compare the differences in spatiotemporal characteristics between traditional augmentation methods and the method proposed in this paper, we carefully selected four time-domain augmentation methods—noise addition [18,19], time reverse [19], time masking [19], and signal flip [19]. Meanwhile, three frequency-domain augmentation methods, namely Frequency shift [19], Fourier transform surrogate [13], and bandpass filter [19], as well as two spatial-domain augmentation methods, namely channel symmetry and channel shuffle [15,16], are chosen as the basis for the experiment. The augmented data used for comparison are generated by the augmentation functions in the Braindecode library [35], while the modeling of the Gaussian mixture model (GMM) is carried out using relevant classes in the sklearn.mixture module [36]. The parameter settings are as follows: the probability parameter is set to 0.5; the frequency-shifting parameter (sfreq_1) is set to 2.0; the bandpass filtering parameter (sfreq_2) is set to 100; the random seed parameter (random_state) is set to 21; the mask parameter (mask_len_samples) is set to 100; the noise parameter (std)is set to 0.16; and is set to 0.8, which is a user-defined value based on multiple experiments.
3.2.1. Waveform Comparison
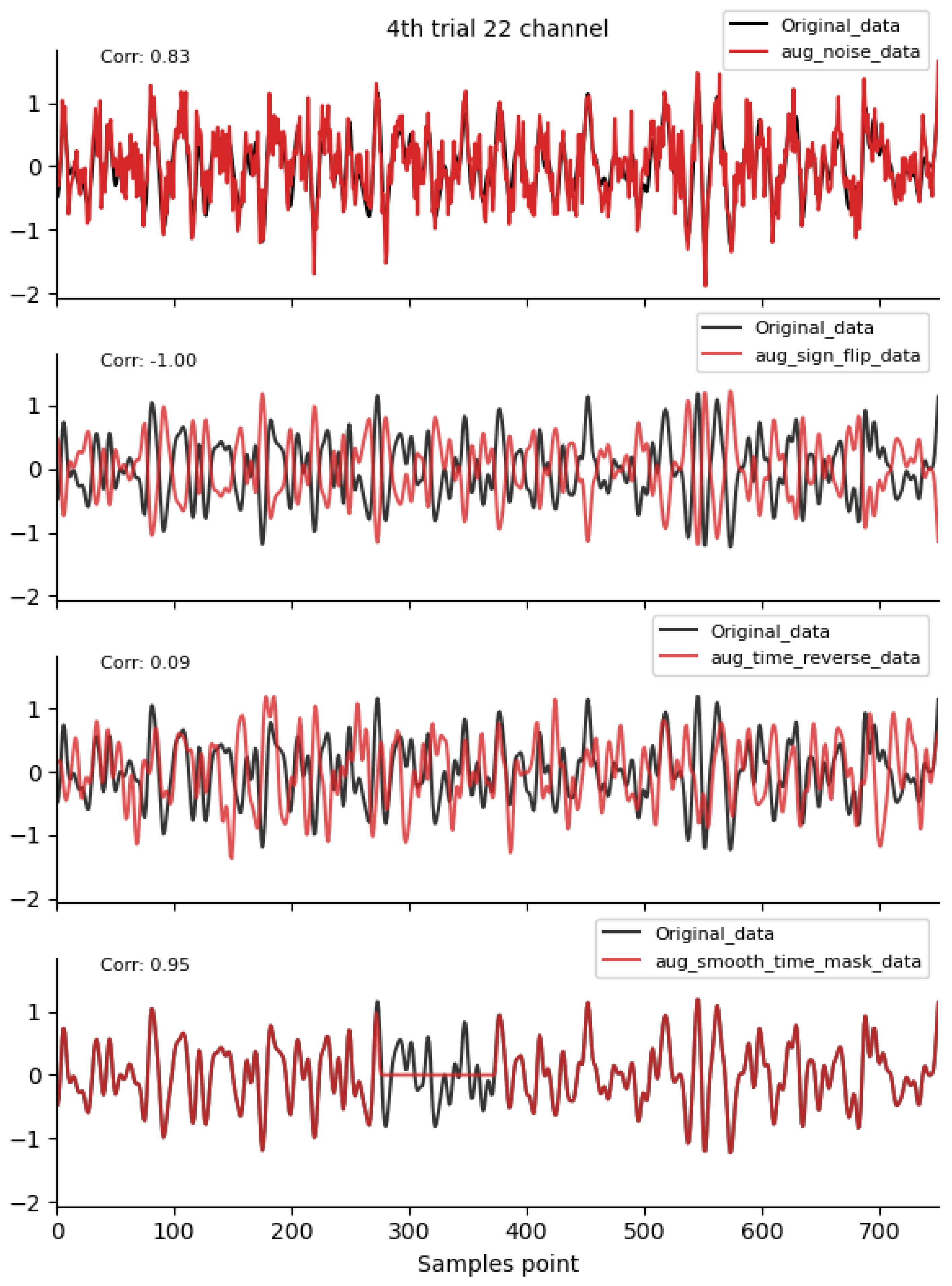
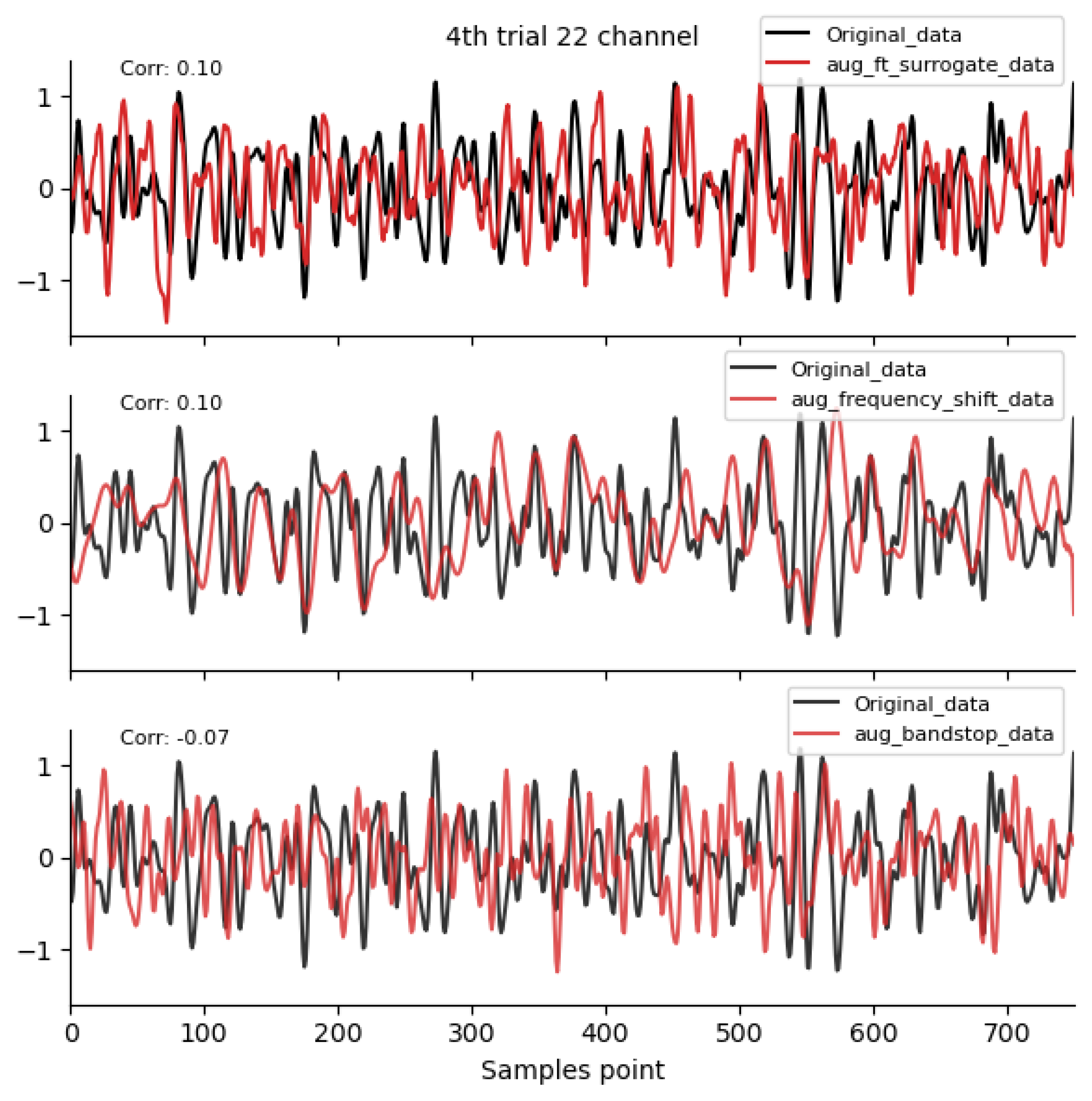
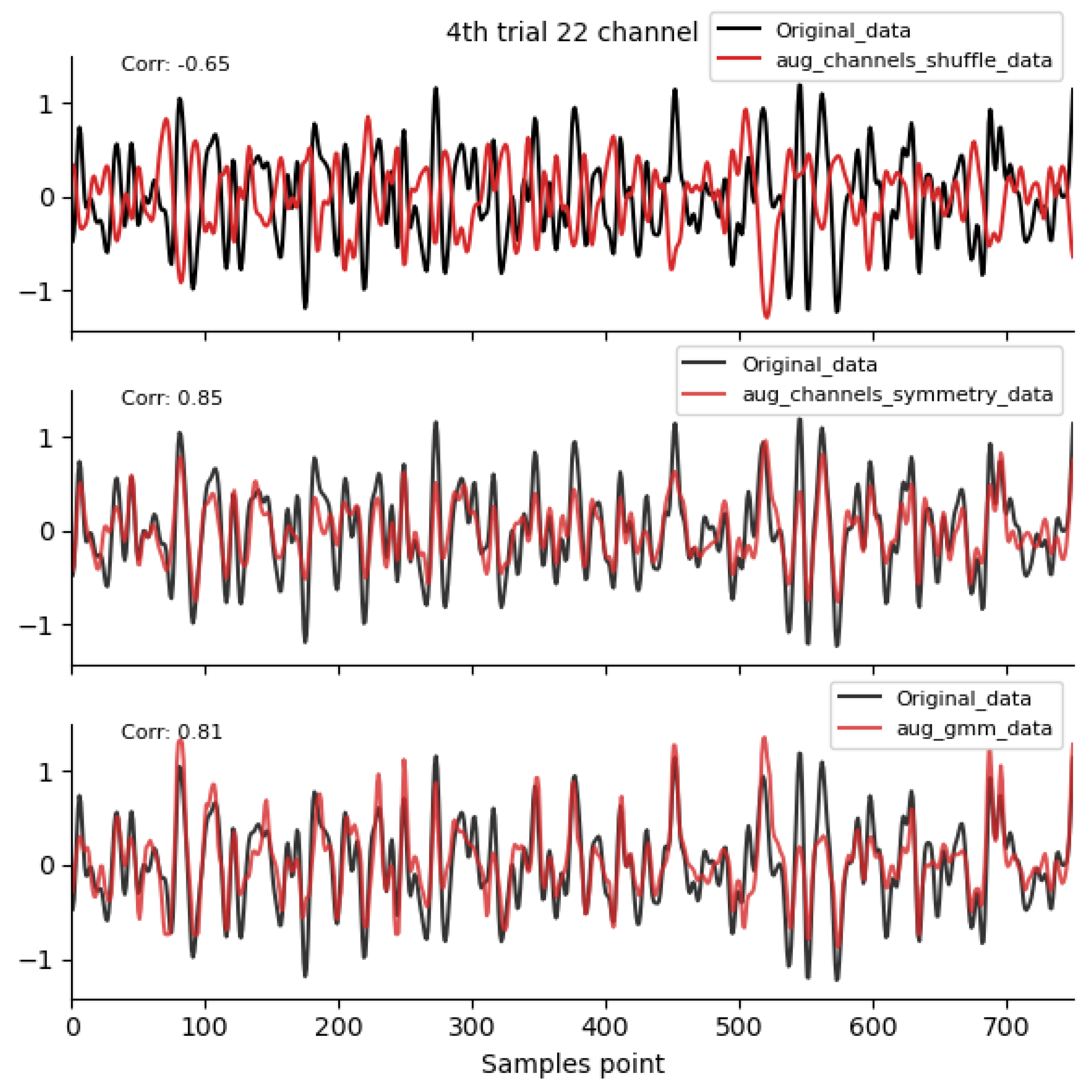
Figure 2, Figure 3 and Figure 4 shows the time-domain waveforms of the first participant, the fourth trial, imagined left-hand movement, and channel 22, respectively, for data generated by ten EEG data augmentation methods.
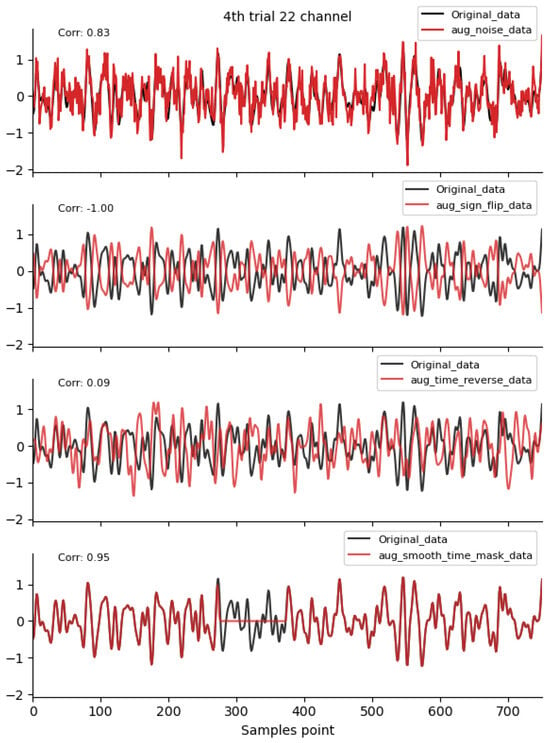
Figure 2.
Time-domain augmentation method waveform diagram. Data sources: subject 1, trial 4, left-hand motor imagery, and channel 22.
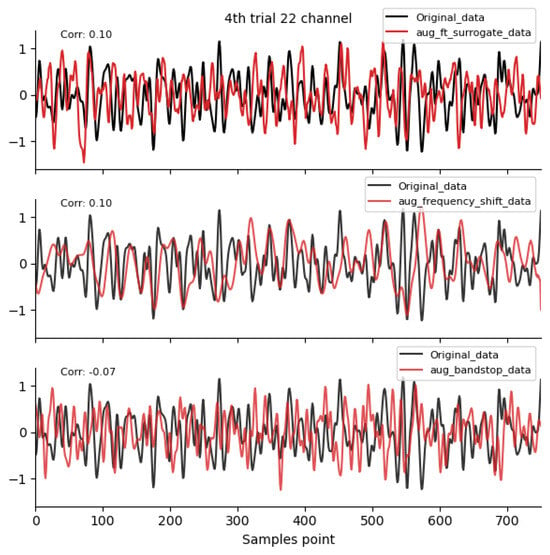
Figure 3.
Frequency-domain augmentation method waveform diagram. Data sources: subject 1, trial 4, left-hand motor imagery, and channel 22.
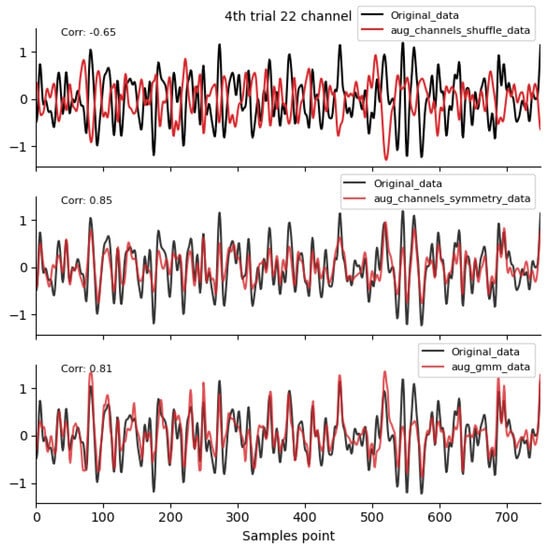
Figure 4.
Spatial-domain and GMM augmentation method waveform diagram. Data sources: subject 1, trial 4, left-hand motor imagery, and channel 22.
By comparing the data correlation of augmentation data and original data in the same channel, it can be observed from the figure that the similarity coefficients of data generated by operations such as noise addition, time masking, signal flip, channel symmetry, channel shuffle, and the method proposed in this paper are all above 0.6. This indicates that the proposed method can basically preserve the time-domain characteristics of the original data.
3.2.2. Topographic Brain Map Comparison
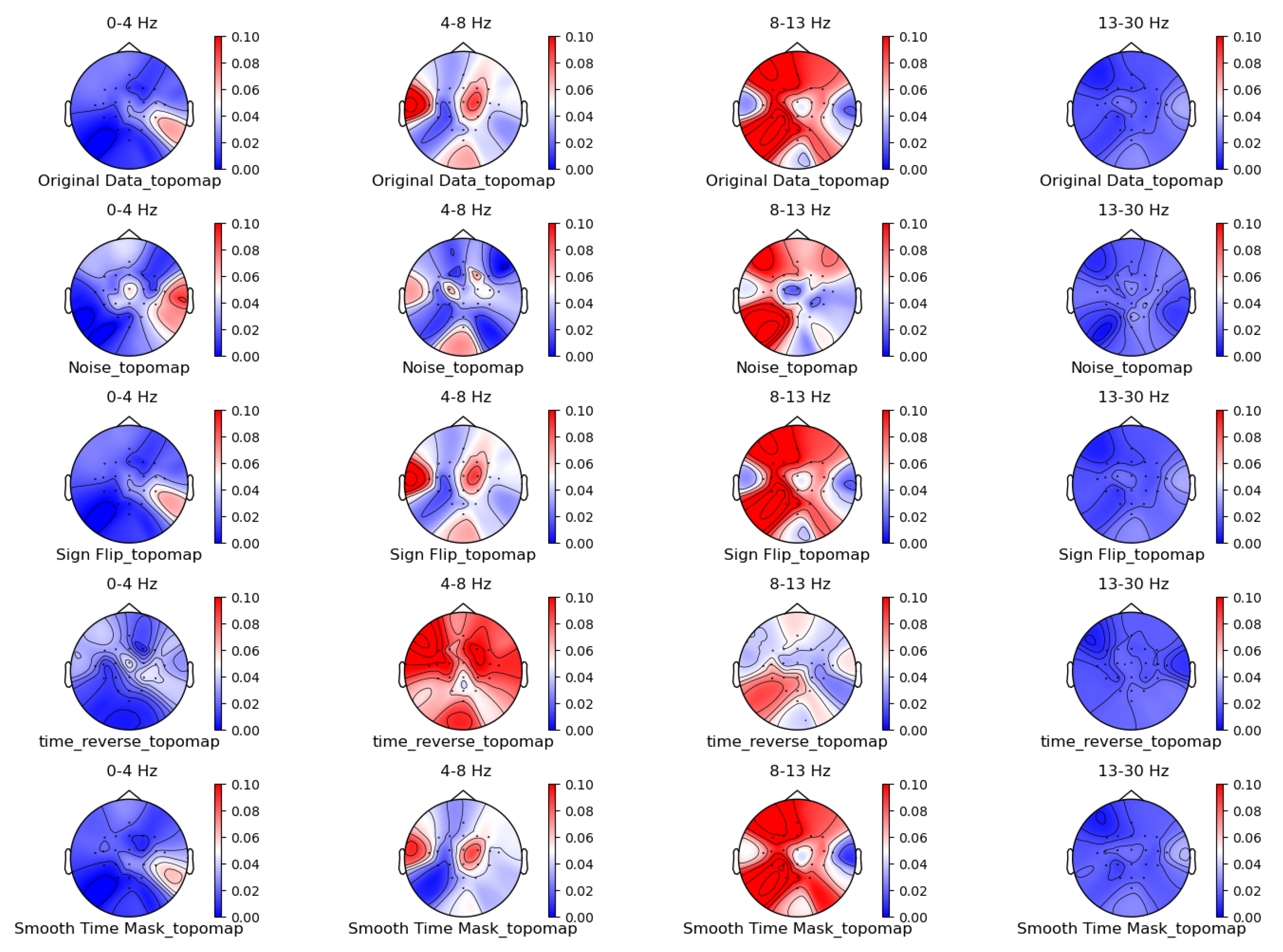
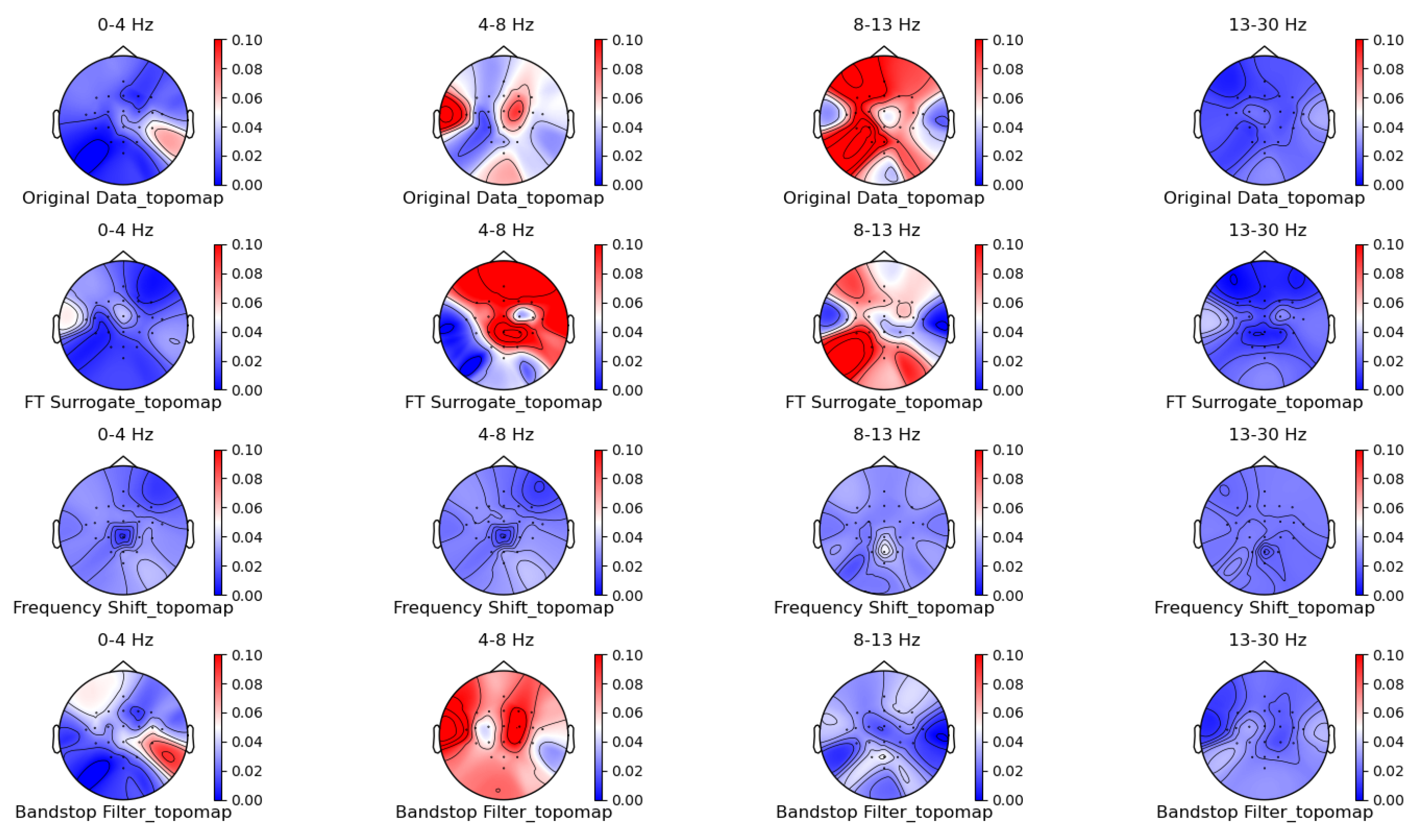
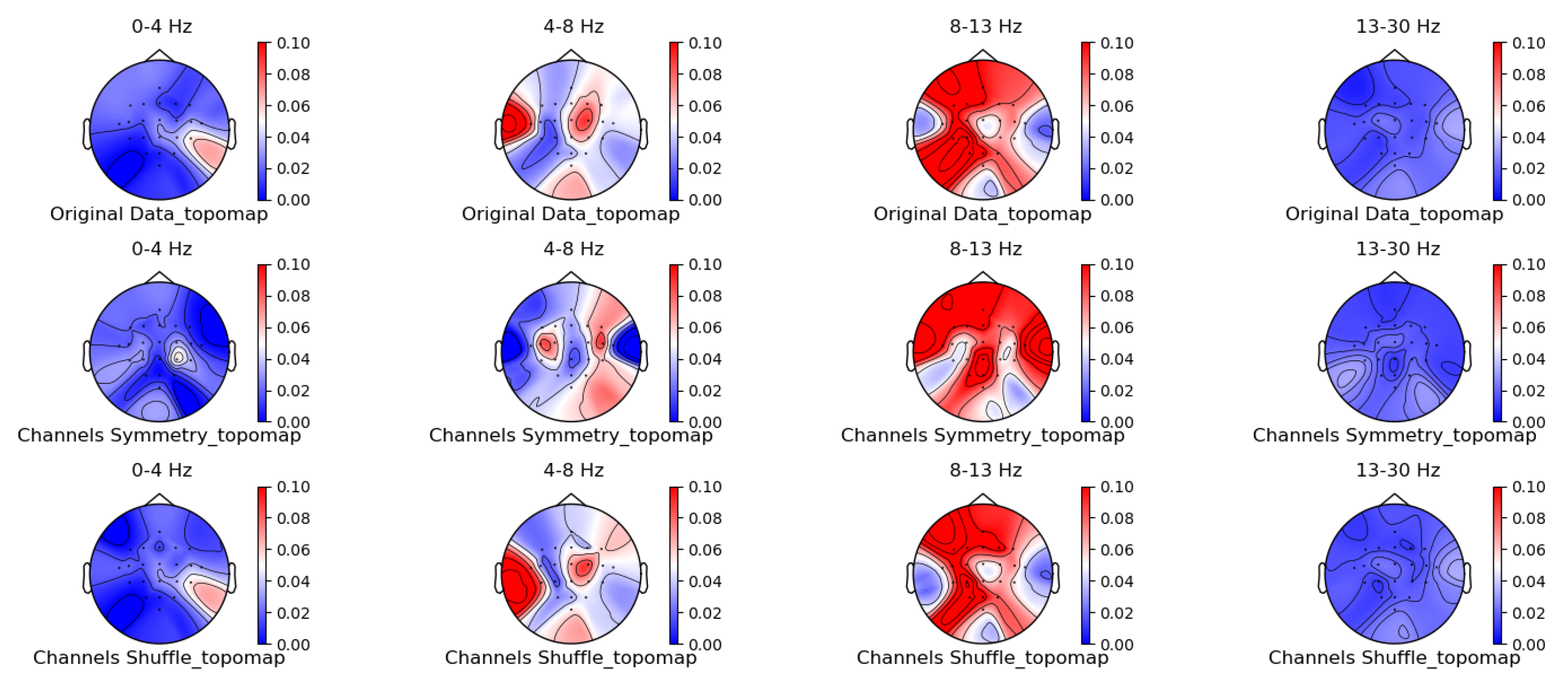
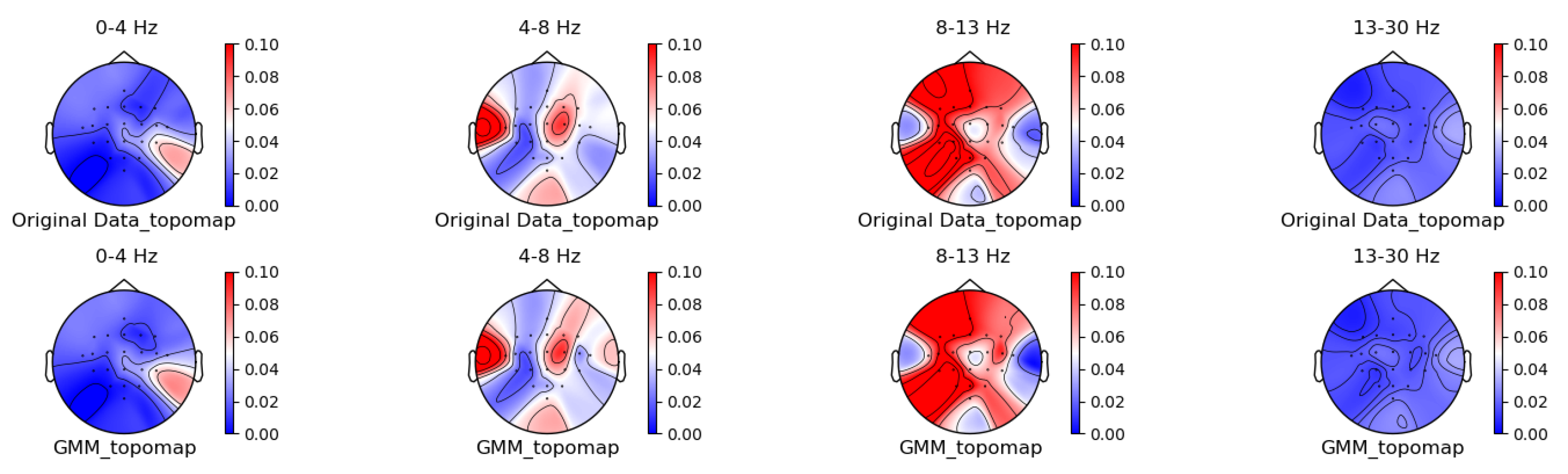
Figure 5, Figure 6, Figure 7 and Figure 8 shows the topographic brain maps of the first subject; the fourth trial; imagined left-hand movement; and the 0–4 Hz (Delta), 4–8 Hz (Theta), 8–13 Hz (Alpha), and 13–30 Hz (Beta) frequency bands, respectively, for data generated by the same ten EEG data augmentation methods.
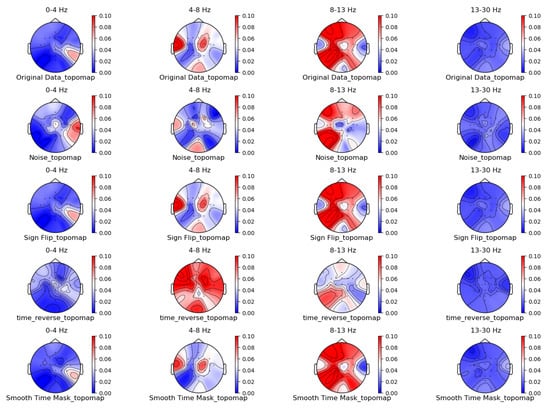
Figure 5.
Time-domain augmentation method for topographic brain maps using left-hand motor imagery. Data source: subject 1, trial 4.
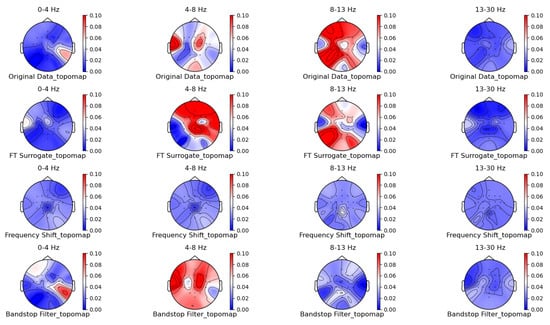
Figure 6.
Frequency-domain augmentation method for topographic brain maps using left-hand motor imagery. Data source: subject 1, trial 4.
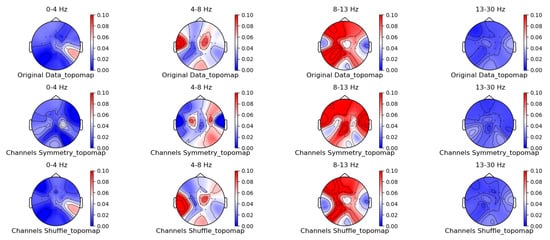
Figure 7.
Spatial-domain augmentation method for topographic brain maps using left-hand motor imagery. Data source: subject 1, trial 4.
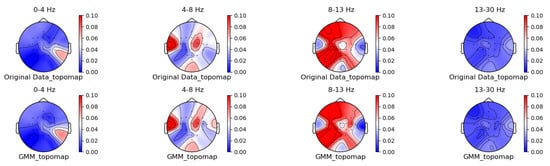
Figure 8.
GMM augmentation method for topographic brain maps using left-hand motor imagery. Data source: subject 1, trial 4, left-hand motor imagery.
From the original topographic brain map data, it is evident that different frequency bands exhibit significant variations in activity across various brain regions. Specifically, the -wave band (8–13 Hz) shows the most pronounced activity in the frontal and occipital lobes. The activation of these areas is closely related to the planning, simulation, and execution of imagined left-hand movements. In contrast, other frequency bands, such as waves, waves, and waves, show relatively weaker activity during imagined left-hand movement tasks and have less distinct associations with specific brain regions compared to the -wave band.
Comparing the topographic brain maps generated by each data augmentation method, techniques such as signal flipping, time masking, channel shuffling, and the Gaussian mixture model (GMM) generally preserve characteristics similar to those of the original topographic brain maps. Among these, the topographic brain map produced by the method proposed in this paper and the time masking method show the highest similarity. In contrast, other data augmentation methods exhibit greater deviations.
3.3. Comparison of the Effectiveness of Data Generated by Different Augmentation Methods on Classification Models
3.3.1. Dataset Partitioning and Model Parameters
To accurately evaluate the performance of each data augmentation method on different models, we use a five-fold cross-validation approach. Specifically, for each participant, the original dataset is divided into five subsets. Each time, one subset is used as the test set, and the remaining subsets are used as the training set. The augmented dataset is formed by concatenating the original data with the augmented data and is also randomly divided into five subsets. Similarly, each time, one subset is used as the test set, and the remaining subsets are used as the training set. This process is repeated five times, and the average classification accuracy for each participant under the model is calculated. Finally, the average classification accuracy across nine participants is computed as the performance metric.
The parameter design and code of each model can be found at https://github.com/liaoliao3450/EEG-Data-Augmentation-Method (accessed on 20 December 2024) (EEG Data Augmentation Method Based on the Gaussian Mixture Model). Introductions to the five models are presented as follows:
(1) FBCSP (Filter Bank Common Spatial Pattern): FBCSP is a method commonly used in electroencephalogram (EEG) signal processing [37]. It combines filter banks and Common Spatial Pattern (CSP) features to effectively extract features from multi-channel EEG signals.
(2) LSTM (Long Short-Term Memory Network): The long short-term memory network is a type of Recurrent Neural Network (RNN) suitable for sequence prediction and other tasks involving time-series data [38]. LSTM can learn long-term dependencies and has been widely applied in various fields, such as natural language processing and time-series prediction.
(3) EEGNet: EEGNet is a lightweight Convolutional Neural Network (CNN) specifically designed for EEG signal classification [39]. It uses depth-wise separable convolution to reduce computational complexity while maintaining high accuracy.
(4) ShallowNet: ShallowNet is a simplified CNN architecture designed specifically for EEG data [40]. It focuses on efficient computation and ease of implementation while still providing competitive performance.
(5) Deep4Net: Deep4Net is a deep learning framework for Brain–Computer Interface (BCI) based on EEG [41]. It combines various types of convolutional layers and temporal convolutional networks to capture the spatial and temporal features of EEG signals. and parameter settings.
3.3.2. Classification Result
For each model, we record the average classification accuracy of each data augmentation method across nine participants, as shown in Table 1.

Table 1.
Comparison of average classification accuracy in K-fold cross-validation between data generated by different augmentation methods and the original mean data accuracy on dataset .
Through the analysis of the average classification accuracies of different augmentation methods across five models, we obtained the following results:
GMM augmentation achieved the highest average accuracy of 80.64%, with a standard deviation of 2.11, indicating that among all augmentation methods, GMM augmentation provided the best performance improvement. It delivered the highest average classification accuracy for all models, particularly excelling on Deep4Net.
For FT surrogate, the average accuracy was 78.90%, with a standard deviation of 3.55, making it the second-best enhancement method, closely approaching the performance of GMM augmentation. This method significantly improved the accuracy on FBCSP and Deep4Net but had a minimal impact on EEGNet.
For time reverse, the average accuracy was 78.72%, with a standard deviation of 1.45. This method performed well across all models and also showed relatively good stability in terms of data consistency, although it had a narrow range and limited variation.
For sign flip, the average accuracy was 78.89%, with a standard deviation of 4.01, similar to channel symmetry, demonstrating good consistency. While it performed well on ShallowNet and Deep4Net, its improvements on FBCSP and EEGNet were minimal.
Under noise addition, the average accuracy was 78.53%, with a standard deviation of 4.10, performing well overall and significantly enhancing the accuracy on FBCSP and EEGNet but having almost no effect on ShallowNet, while showing significant improvement on Deep4Net.
For time masking, the average accuracy was 78.39%, with a standard deviation of 2.67. Although ranked middling overall, it improved performance on all models, especially on ShallowNet and Deep4Net.
Under bandstop filter, the average accuracy was 78.20%, with a standard deviation of 1.85. This method significantly augmentedthe accuracy on Deep4Net and maintained relatively stable performance.
Under channel symmetry, the average accuracy was 77.75%, with a standard deviation of 3.00, indicating consistent and stable performance across all models.
For channel shuffle, the average accuracy was 75.66%, with a standard deviation of 4.52, showing the most variability among all methods. Its performance varied across models, providing some improvement on FBCSP and Deep4Net.
Under frequency shift, the average accuracy was 74.73%, with a standard deviation of 4.72, similar to noise addition, performing well but with larger fluctuations. It performed best on FBCSP but offered limited improvements on other models.
Using the original data as a baseline, the average accuracy was 52.76%, with a standard deviation of 8.74—the lowest among all methods.
From these analyses, it is evident that GMM augmentation and FT surrogate are the two most effective data augmentation methods, significantly improving the model’s classification accuracy. Time reverse and Bandstop filter also provided relatively stable performance improvements. In contrast, the original data (without augmentation) performed the worst, indicating that employing appropriate data augmentation techniques can significantly enhance the classification performance of models.
3.3.3. ROC Curve Comparison
After completing the five-fold cross-validation, we began to plot the ROC curves. For each category, we first collected the FPR (false-positive rate) and TPR (true-positive rate) data from all five folds. Then, in order to make these discrete data points better reflect the performance of the model over the entire range, we interpolated them. By establishing a continuous functional relationship between FPR and TPR, we obtained smooth ROC curves. Such curves can more accurately reflect the overall performance of the model in multiple experiments, reducing the impact of randomness and contingency in individual experiments.
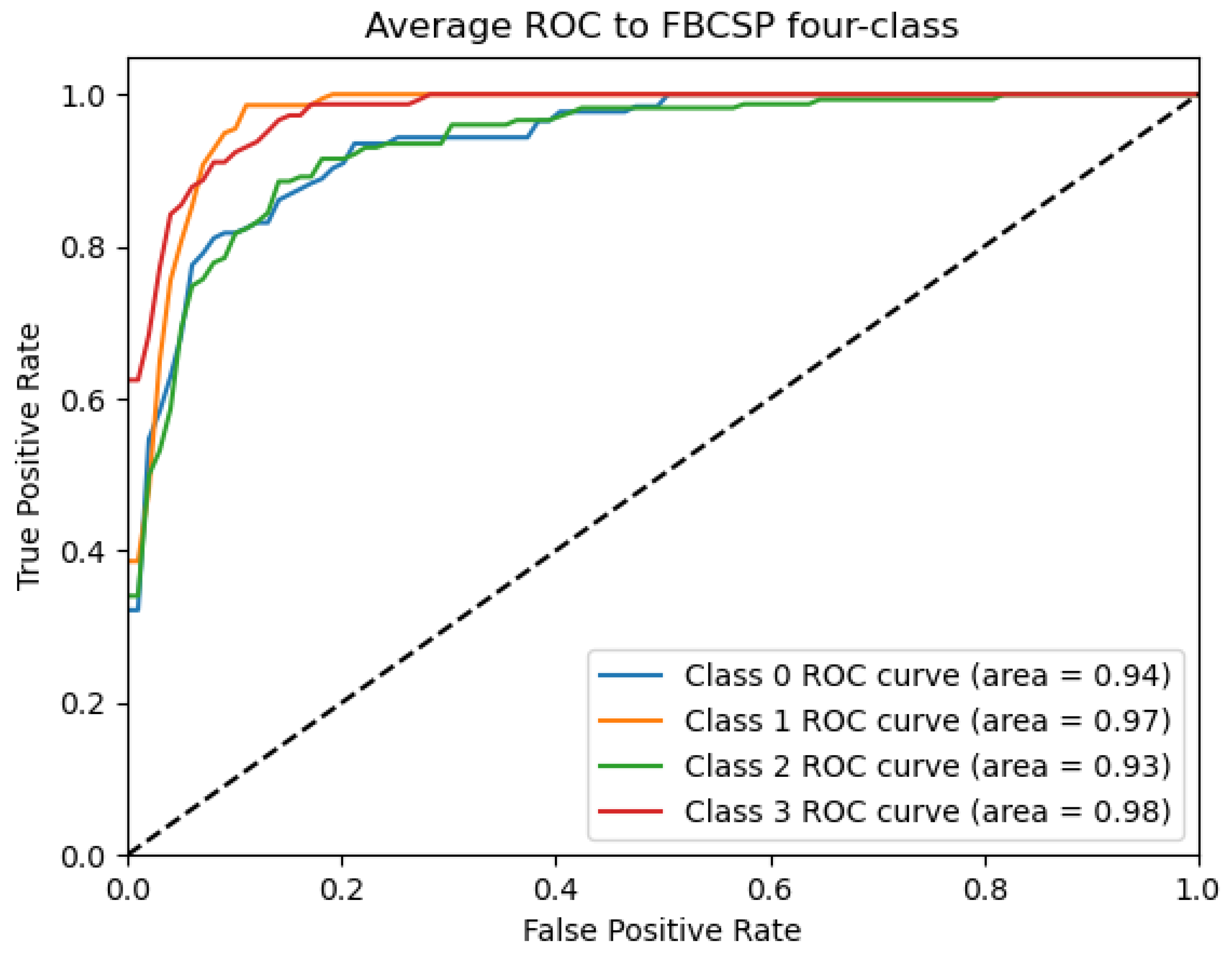
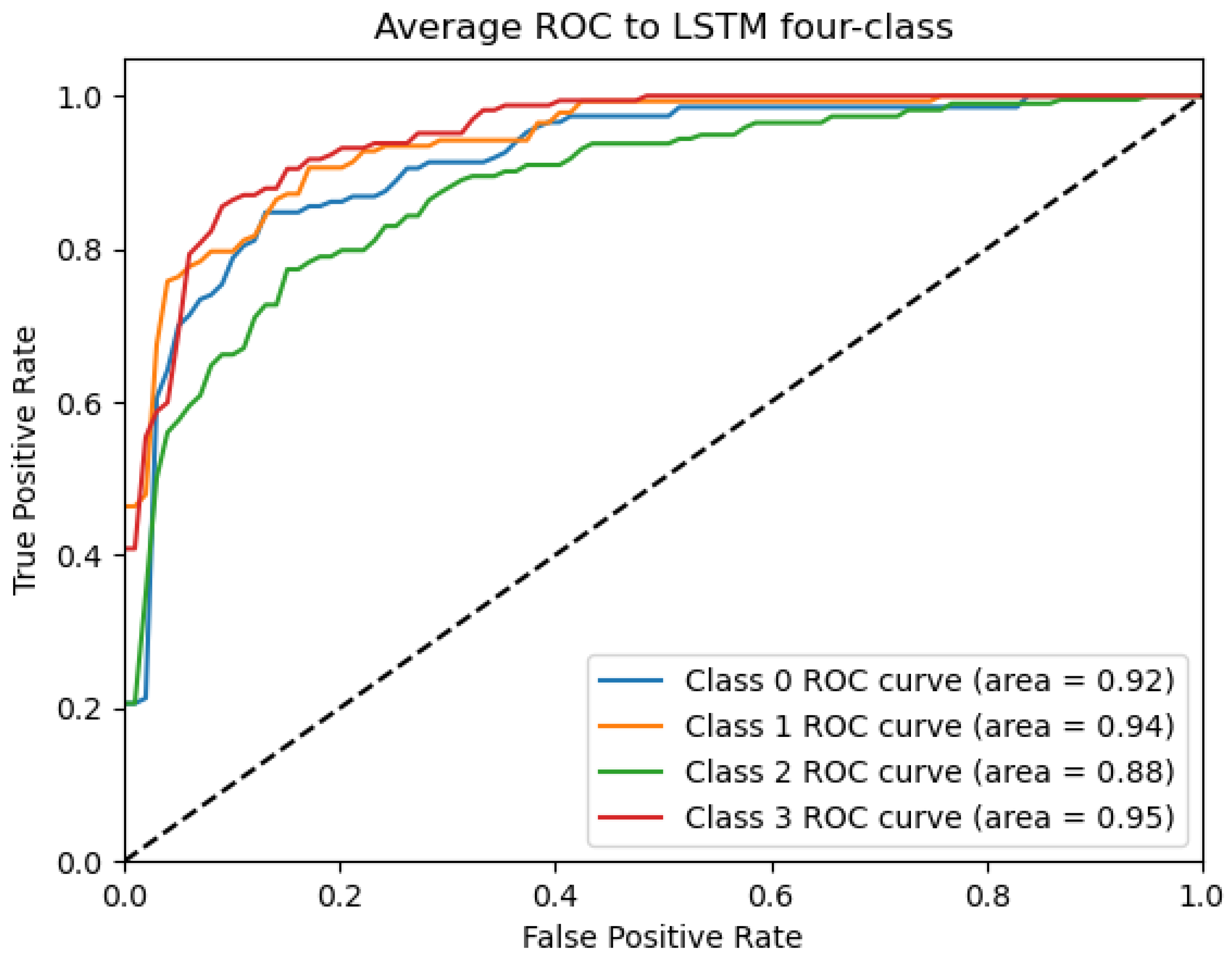
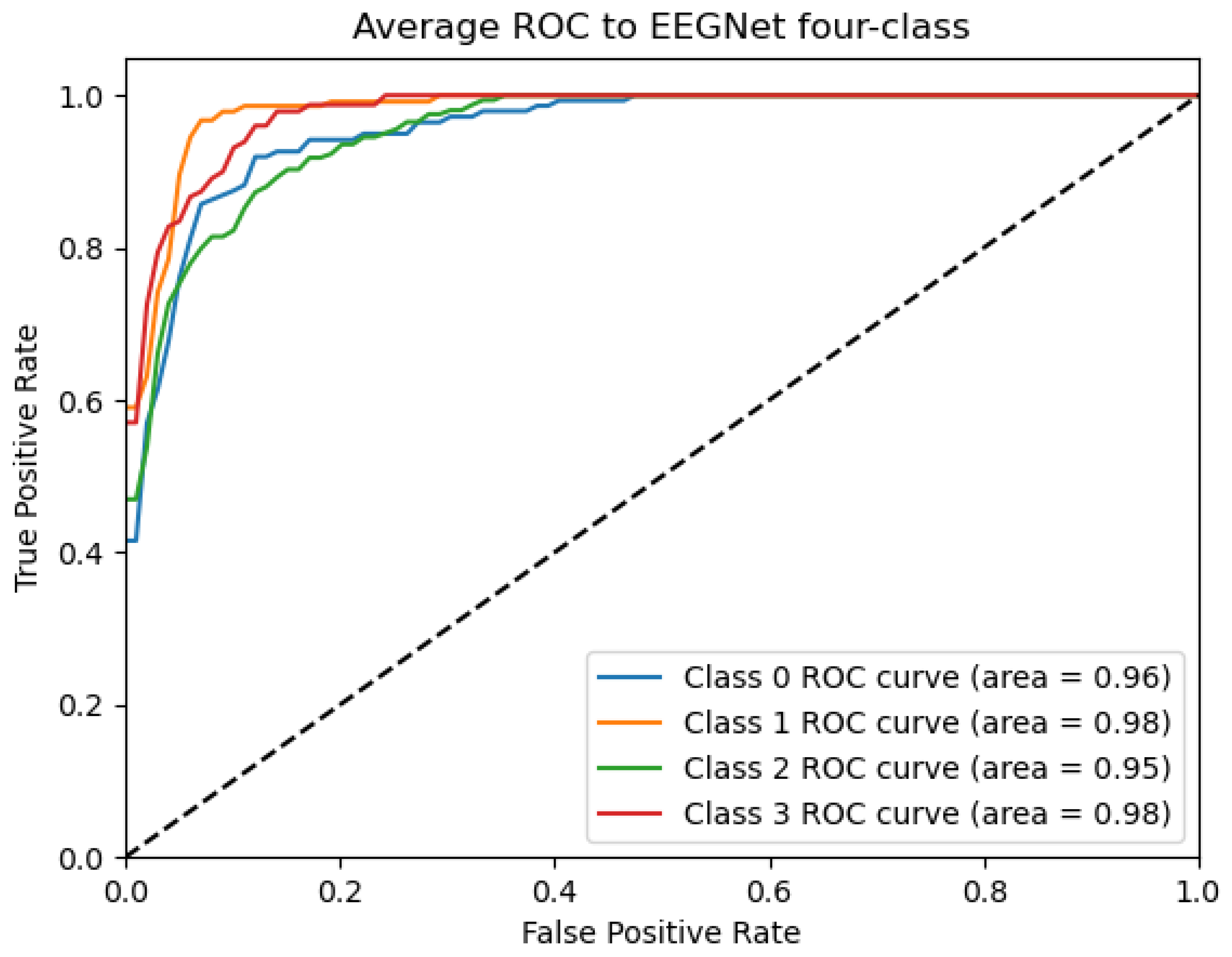
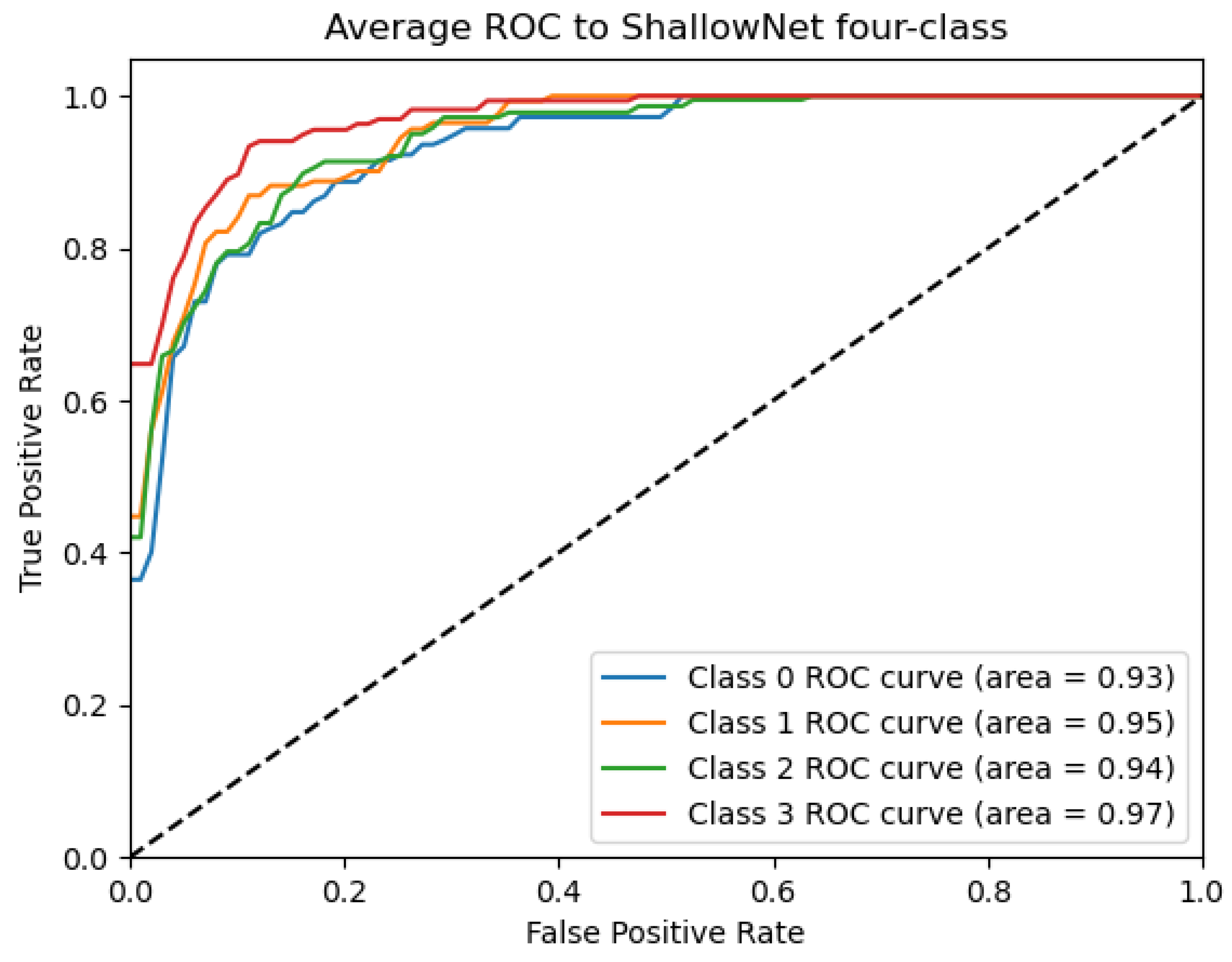
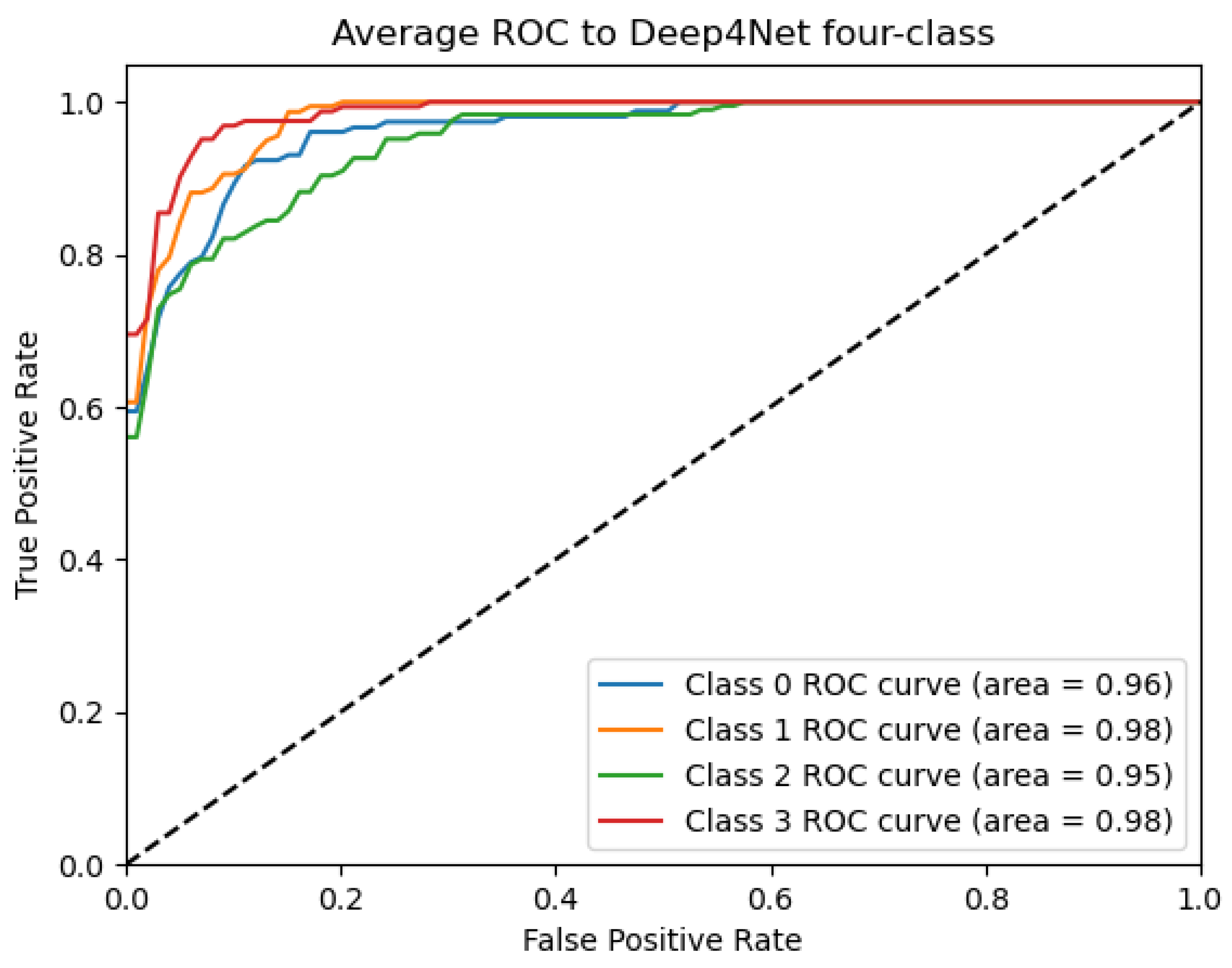
Figure 9, Figure 10, Figure 11, Figure 12 and Figure 13 show that there are significant differences in the working performance of the five models on the augmentation data. These models have a larger area under the ROC curve, indicating that they have a strong ability to distinguish between positive and negative samples and can better capture the potential patterns and rules in the data. Among them, the Deep4Net model has the highest sensitivity and can more accurately identify positive classes. This reflects the fact that the model has a stronger ability to distinguish between positive and negative samples, especially in capturing positive classes, in which case it is more sensitive and effective.
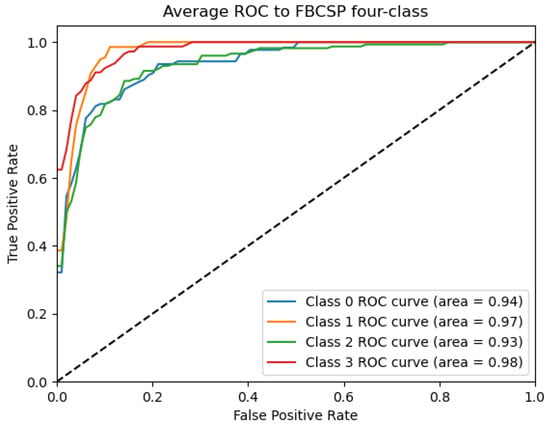
Figure 9.
ROC_FBCSP curve: data from subject 1 augmented by the GMM method.
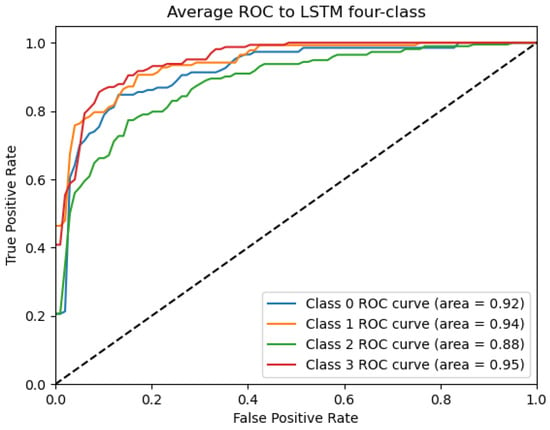
Figure 10.
ROC_LSTM Curve: Data from subject 1 augmented by the GMM method.
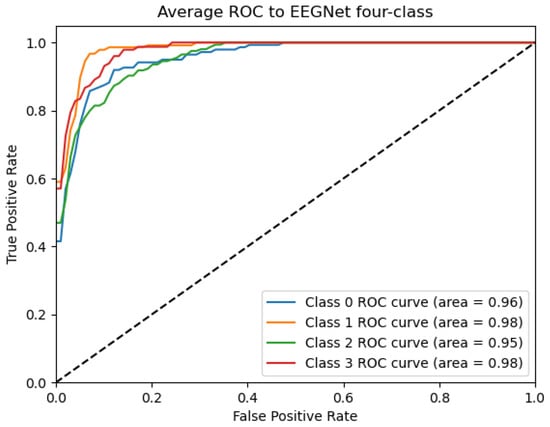
Figure 11.
ROC_EEGNET Curve: Data from subject 1 augmented by the GMM method.
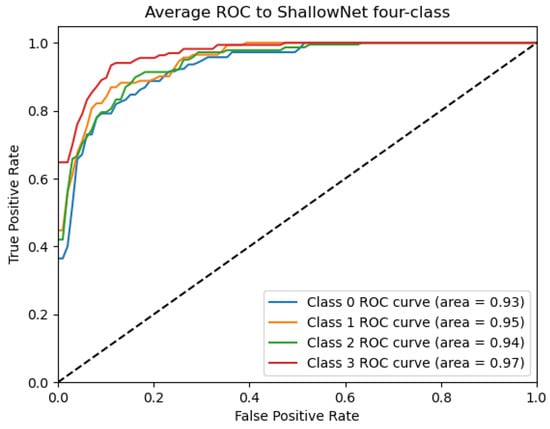
Figure 12.
ROC_ShallowNet curve: Data from subject 1 augmented by the GMM method.
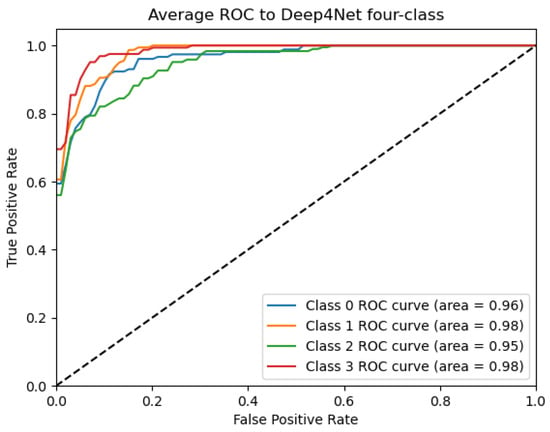
Figure 13.
ROC_Deep4Net curve: data from subject 1 augmented by the GMM method.
3.4. Comparison of Visualization Results Between Original Data and the Data Augmented Using the Gaussian Mixture Model (GMM)
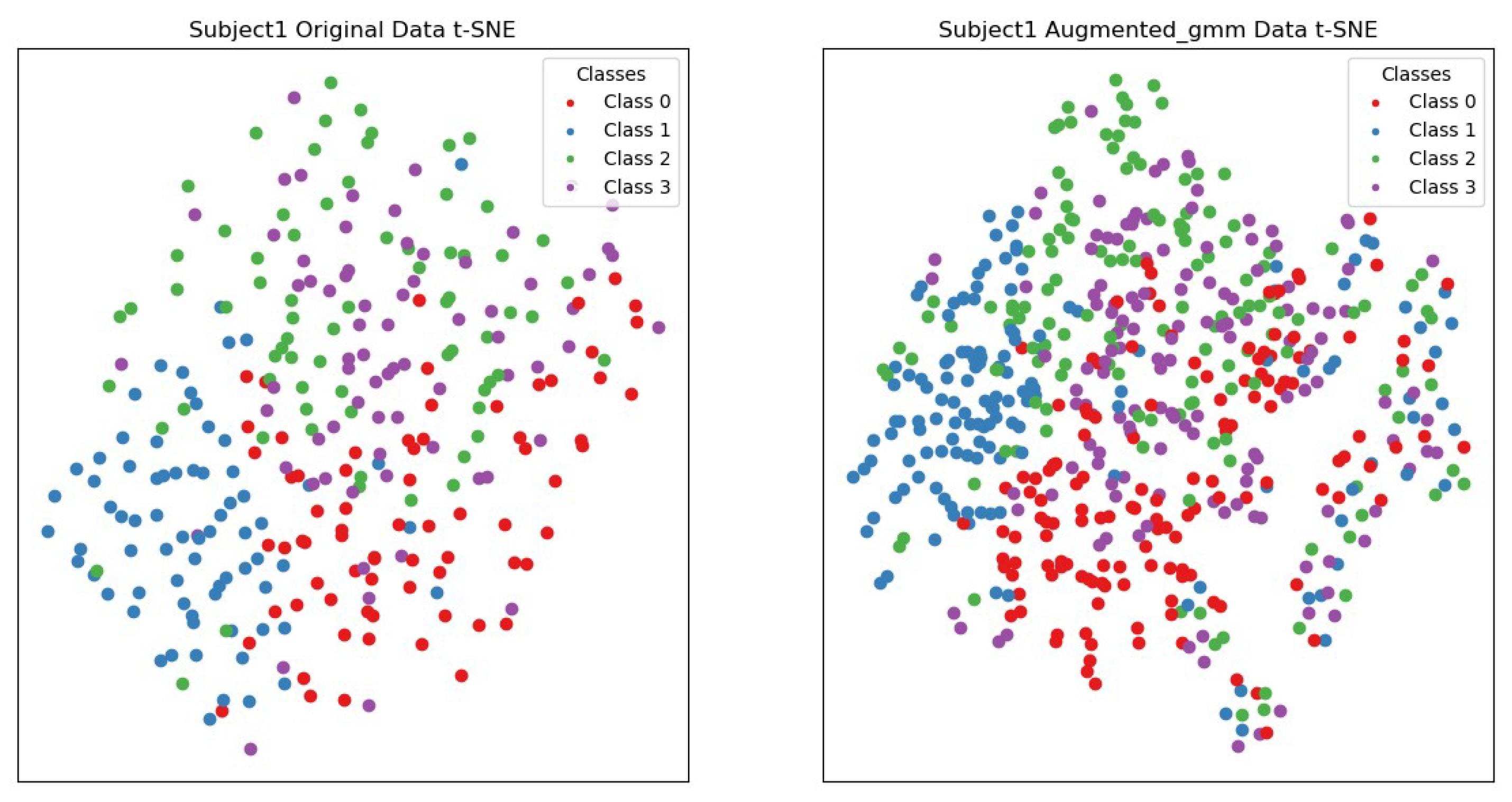
As shown in Figure 14, by comparing the t-SNE visualization of the original data and the data generated by the augmentation method for the first participant, it was found that the original data had unclear boundaries among points from different categories, while the augmented data showed clearer cluster structures. Points from each category were relatively gathered together, with distinct boundaries, improving the distinguishability between different categories. Meanwhile, the original data had dispersed points within the same category, and the augmented data exhibited more compact local structures. Points from the same category were closely gathered together, enhancing intra-class consistency.
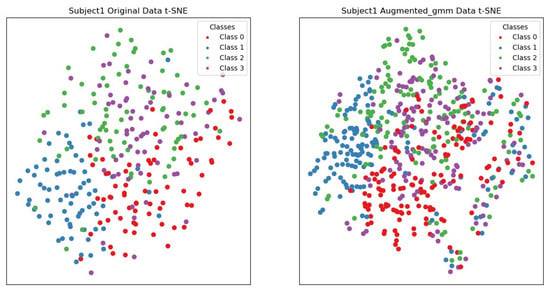
Figure 14.
t-SNE visualization of subject 1’s original data and the data augmented using the Gaussian mixture model (GMM). Red represents the data for left-hand motor imagery, blue represents the data for right-hand motor imagery, green represents the data for motor imagery of both feet, and purple represents the data for tongue motor imagery.
In summary, the augmentation methods investigated in this paper improve the visualization of the data, making points from different classes more clearly clustered together and increasing the separation between different classes. This improvement helps enhance the performance of subsequent machine learning models, as clearer class boundaries and higher local structure compactness aid the model in learning and classifying data more accurately.
4. Discussion
Data augmentation plays a significant role in improving model generalization, reducing overfitting risks, and enhancing performance on unseen data. Its core aim is to increase data diversity, allowing the model to better learn data patterns. After comparing the impacts of various data augmentation methods, we found the following:
(1) The method based on the Gaussian mixture model (GMM) deeply analyzes the characteristics of EEG signals and accurately models them using the GMM. Compared with traditional methods, it can more finely capture the true distribution, complex structures, and patterns in EEG signals, thereby extracting more representative and discriminative features.
(2) The GMM is a probabilistic model and can accurately model the distribution of EEG signals. By reasonably setting the model parameters, it is possible to better fit real data, thereby improving the accuracy and effectiveness of data augmentation. During the data augmentation process, new samples that are more in line with actual observations are generated based on the probability distribution learned by the model. These new samples not only exhibit diversity but also maintain similarity with the original data, providing a more comprehensive learning material for the model. Experiments show that on the t-SNE distribution map, the original data and the augmentation data have similar and more compact cluster distributions, whereas on the brain topography map, the original data and the augmentation data have exhibit similar performance in terms of electrical brain activities in specific areas, while on the model, they achieve higher evaluation scores.
(3) EEG signals vary among different individuals. The method based on the GMM can adapt to the data characteristics of different individuals by adjusting model parameters, especially the threshold of the feature similarity coefficient. This personalized data augmentation strategy can better meet the needs of the model for different data, making the model more accurate and stable in the learning and classification process.
(4) This method can solve the challenges of traditional EEG data augmentation methods, such as insufficient data diversity, signal distortion, and so on, providing an effective and reliable data augmentation solution for EEG signal processing and analysis. However, it bases the Gaussian mixture decomposition on similar data, requiring an increase in the number of submodels for complex, multidimensional, and highly variable data, which leads to higher computational complexity compared to geometric transformations and results in limited real-time performance.
(5) Experiments were conducted using only one dataset, which may limit the universality of the research results to a certain extent. However, the data augmentation method based on the Gaussian mixture model (GMM) proposed in this paper is designed based on the modeling of the data distribution. This method can flexibly capture and adapt to the complex structures and patterns in EEG data from different tasks; therefore, it has strong potential for generalization. The research results have potential positive implications for cognitive neuroscience and clinical practice, contributing to a deeper understanding of brain signal processing mechanisms, aiding in the diagnosis and treatment of brain diseases, and promoting the development of personalized medicine.
5. Conclusions
In this paper, we innovatively propose a Gaussian feature reconstruction-based electroencephalogram (EEG) data augmentation scheme, aiming to address existing challenges such as the disruption of spatiotemporal dynamic characteristics and the reliance on a large number of real samples. By applying Gaussian mixture model decomposition to similar samples, we successfully extracted mixed features and performed exchanges and reconstructions based on the probability similarities of these features within the same sample category, thereby creating new Gaussian mixture features. This refined process depends on accurate sampling based on probability, mean, and variance, ensuring that the newly generated sample data are highly realistic. After rigorous dataset testing and evaluation, our method achieved significant improvements in motor imagery recognition accuracy, effectively enhancing the generalization capability of models, in addition to verifying the effectiveness and innovation of this method in the field of EEG data processing. More importantly, our work reveals another perspective for EEG data processing with respect to how to maintain the distribution characteristics of data to ensure the authenticity of generated data, providing new ways of thinking for future research. Although the current method has certain limitations in terms of computational resource requirements and real-time performance, its core concept of ensuring the authenticity of generated data by maintaining data distribution characteristics provides new ideas and directions for subsequent research. In the future, we plan to extend the application of the GMM method to other types of neural data analysis fields and actively explore the possibility of integrating it with advanced technologies such as diffusion models in order to further optimize the EEG data processing and analysis process and broaden its application scope and depth in the brain signal research field.
Author Contributions
Conceptualization, J.Z. and C.L.; methodology, C.L. and S.Z.; software, C.L.; validation, S.Z., C.L. and X.W. (Xiangcun Wang); formal analysis, Y.L.; investigation, S.Z.; resources, J.Z.; data curation, S.Z.; writing—original draft preparation, C.L.; writing—review and editing, C.L.; visualization, C.L. and X.W. (Xiangcun Wang); supervision, J.Z., X.W. (Xiangcun Wang) and X.W. (Xia Wu); project administration, J.Z.; funding acquisition, J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Major Research Instruments Project (62227801) of the Natural Science Foundation of China.
Data Availability Statement
To substantiate the uniqueness of my research, I have made the preliminary data and code snippets (if applicable) from this manuscript, along with my article published in September, publicly available on the respective websites https://github.com/liaoliao3450/EEG-Data-Augmentation-Method: EEG Data Augmentation Method Based on the Gaussian Mixture Model (accessed on 20 December 2024).
Conflicts of Interest
Author Shiyu Zhao was employed by the Tianyi Security Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| EEG | Electroencephalogram |
| GANs | Generative Adversarial Networks |
| VAEs | Variational Autoencoders |
| SNR | Signal-to-noise Ratio |
| GMM | Gaussian Mixture Model |
References
- Abdul Hussain, A.; Singh, A.; Guesgen, H.; Lal, S. A Comprehensive Review on Critical Issues and Possible Solutions of Motor Imagery Based Electroencephalography Brain-Computer Interface. Sensors 2021, 21, 2173. [Google Scholar] [CrossRef] [PubMed]
- Roy, Y.; Banville, H.; Albuquerque, I.; Gramfort, A.; Falk, T.H.; Faubert, J. Deep learning-based electroencephalography analysis: A systematic review. J. Neural Eng. 2019, 16, 051001. [Google Scholar] [CrossRef]
- Craik, A.; He, Y.; Contreras-Vidal, J.L. Deep learning for electroencephalogram (EEG) classification tasks: A review. J. Neural Eng. 2019, 16, 031001. [Google Scholar] [CrossRef]
- Luo, Y.; Lu, B.L. EEG Data Augmentation for Emotion Recognition Using a Conditional Wasserstein GAN. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 2535–2538. [Google Scholar] [CrossRef]
- Sakai, A.; Minoda, Y.; Morikawa, K. Data augmentation methods for machine-learning-based classification of bio-signals. In Proceedings of the 2017 10th Biomedical Engineering International Conference (BMEiCON), Hokkaido, Japan, 31 August–2 September 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Krell, M.M.; Kim, S.K. Rotational data augmentation for electroencephalographic data. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju Island, Republic of Korea, 11–15 July 2017; pp. 471–474. [Google Scholar] [CrossRef]
- Lashgari, E.; Liang, D.; Maoz, U. Data augmentation for deep-learning-based electroencephalography. J. Neurosci. Methods 2020, 346, 108885. [Google Scholar] [CrossRef] [PubMed]
- Um, T.T.; Pfister, F.M.J.; Pichler, D.; Endo, S.; Lang, M.; Hirche, S.; Fietzek, U.; Kulić, D. Data augmentation of wearable sensor data for parkinson’s disease monitoring using convolutional neural networks. In Proceedings of the 19th ACM International Conference on Multimodal Interaction, New York, NY, USA, 6–9 June 2017; ICMI ’17. pp. 216–220. [Google Scholar] [CrossRef]
- Lotte, F. Signal Processing Approaches to Minimize or Suppress Calibration Time in Oscillatory Activity-Based Brain–Computer Interfaces. Proc. IEEE 2015, 103, 871–890. [Google Scholar] [CrossRef]
- Pei, Y.; Luo, Z.; Yan, Y.; Yan, H.; Jiang, J.; Li, W.; Xie, L.; Yin, E. Data Augmentation: Using Channel-Level Recombination to Improve Classification Performance for Motor Imagery EEG. Proc. IEEE 2021, 15, 645–952. [Google Scholar] [CrossRef]
- Kim, S.J.; Lee, D.H.; Choi, Y.W. CropCat: Data Augmentation for Smoothing the Feature Distribution of EEG Signals. In Proceedings of the 2023 11th International Winter Conference on Brain-Computer Interface (BCI), Gangwon, Republic of Korea, 20–22 February 2023; pp. 1–4. [Google Scholar] [CrossRef]
- Luo, Y.; Zhu, L.Z.; Wan, Z.Y.; Lu, B.L. Data augmentation for enhancing EEG-based emotion recognition with deep generative models. J. Neural Eng. 2020, 17, 056021. [Google Scholar] [CrossRef] [PubMed]
- Schwabedal, J.T.C.; Snyder, J.C.; Cakmak, A.; Nemati, S.; Clifford, G.D. Addressing Class Imbalance in Classification Problems of Noisy Signals by using Fourier Transform Surrogates. arXiv 2018. [Google Scholar] [CrossRef]
- Rommel, C.; Moreau, T.; Gramfort, A. CADDA: Class-wise Automatic Differentiable Data Augmentation for EEG Signals. arXiv 2021, arXiv:2106.13695. [Google Scholar]
- Deiss, O.; Biswal, S.; Jing, J.; Sun, H.; Westover, M.B.; Sun, J. HAMLET: Interpretable Human And Machine co-LEarning Technique. arXiv 2018. [Google Scholar] [CrossRef]
- Saeed, A.; Grangier, D.; Pietquin, O.; Zeghidour, N. Learning From Heterogeneous Eeg Signals with Differentiable Channel Reordering. In Proceedings of the ICASSP 2021-2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1255–1259. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Int. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Wang, F.; Zhong, S.H.; Peng, J.; Jiang, J.; Liu, Y. Data Augmentation for EEG-Based Emotion Recognition with Deep Convolutional Neural Networks. In Proceedings of the MultiMedia Modeling, Bangkok, Thailand, 5–7 February 2018; pp. 82–93. [Google Scholar] [CrossRef]
- Mohsenvand, M.N.; Izadi, M.R.; Maes, P. Contrastive Representation Learning for Electroencephalogram Classification. In Proceedings of the Machine Learning for Health NeurIPS Workshop, Virtual, 11 December 2020; Alsentzer, E., McDermott, M.B.A., Falck, F., Sarkar, S.K., Roy, S., Hyland, S.L., Eds.; PMLR. Volume 136, pp. 238–253. [Google Scholar]
- Li, Y.; Huang, J.; Zhou, H.; Zhong, N. Human Emotion Recognition with Electroencephalographic Multidimensional Features by Hybrid Deep Neural Networks. Appl. Sci. 2017, 7, 1060. [Google Scholar] [CrossRef]
- Fu, R.; Wang, Y.; Jia, C. A new data augmentation method for EEG features based on the hybrid model of broad-deep networks. Expert Syst. Appl. 2022, 202, 117386. [Google Scholar] [CrossRef]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein generative adversarial networks. In Proceedings of the 34th International Conference on Machine Learning-Volume 70, Sydney, Australia, 6–11 August 2017; ICML’17. pp. 214–223. [Google Scholar]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A. Improved training of wasserstein GANs. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Red Hook, NY, USA, 4–9 December 2017; NIPS’17. pp. 5769–5779. [Google Scholar]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.Y.; Wang, Z.; Smolley, S.P. Least Squares Generative Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2813–2821. [Google Scholar] [CrossRef]
- Hartmann, K.G.; Schirrmeister, R.T.; Ball, T. EEG-GAN: Generative adversarial networks for electroencephalograhic (EEG) brain signals. arXiv 2018. [Google Scholar] [CrossRef]
- Fahimi, F.; Dosen, S.; Ang, K.K.; Mrachacz-Kersting, N.; Guan, C. Generative Adversarial Networks-Based Data Augmentation for Brain–Computer Interface. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4039–4051. [Google Scholar] [CrossRef] [PubMed]
- Ramponi, G.; Protopapas, P.; Brambilla, M.; Janssen, R. T-CGAN: Conditional Generative Adversarial Network for Data Augmentationin Noisy Time Series with Irregular Sampling. arXiv 2018, arXiv:1811.08295. [Google Scholar]
- Zhu, F.; Ye, F.; Fu, Y.; Liu, Q.; Shen, B. Electrocardiogram generation with a bidirectional LSTM-CNN generative adversarial network. Sci. Rep. 2019, 9, 6734. [Google Scholar] [CrossRef] [PubMed]
- Bao, G.; Yan, B.; Tong, L.; Shu, J.; Wang, L.; Yang, K.; Zeng, Y. Data Augmentation for EEG-Based Emotion Recognition Using Generative Adversarial Networks. Front. Comput. Neurosci. 2021, 15, 723843. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, G.; Blankertz, B. BCI Competition 2003–Data set IIa: Spatial patterns of self-controlled brain rhythm modulations. IEEE Trans. Biomed. Eng. 2004, 51, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- De Lucia, M.; Michel, C.M.; Clarke, S.; Murray, M.M. Single-trial topographic analysis of human EEG: A new ‘image’ of event-related potentials. In Proceedings of the 2007 6th International Special Topic Conference on Information Technology Applications in Biomedicine, Tokyo, Japan, 8–11 November 2007; pp. 95–98. [Google Scholar] [CrossRef]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum Likelihood from Incomplete Data Via the EM Algorithm. J. R. Stat. Soc. Ser. B 1977, 39, 1–22. [Google Scholar] [CrossRef]
- Liao, C.; Zhao, S.; Zhang, J. Motor Imagery Recognition Based on GMM-JCSFE Model. IEEE Trans. Neural Syst. Rehabil. Eng. 2024, 32, 3348–3357. [Google Scholar] [CrossRef]
- Gramfort, A.; Luessi, M.; Larson, E.; Engemann, D.; Strohmeier, D.; Brodbeck, C.; Goj, R.; Jas, M.; Brooks, T.; Parkkonen, L.; et al. MEG and EEG data analysis with MNE-Python. Front. Neurosci. 2013, 7, 267. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ang, K.K.; Chin, Z.Y.; Wang, C.; Guan, C.; Zhang, H. Filter Bank Common Spatial Pattern Algorithm on BCI Competition IV Datasets 2a and 2b. Front. Neurosci. 2012, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain–computer interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wu, D.; Li, H.; Jiang, F.; Lu, H. ShallowNet: An Efficient Lightweight Text Detection Network Based on Instance Count-Aware Supervision Information. In Proceedings of the Neural Information Processing, Cham, Switzerland, 8–12 December 2021; pp. 633–644. [Google Scholar]
- Schirrmeister, R.; Gemein, L.; Eggensperger, K.; Hutter, F.; Ball, T. Deep learning with convolutional neural networks for decoding and visualization of EEG pathology. In Proceedings of the 2017 IEEE Signal Processing in Medicine and Biology Symposium (SPMB), Philadelphia, PA, USA, 2 December 2017; pp. 1–7. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).