Abstract
Transmission electron microscopy imaging provides a unique opportunity to inspect the detailed structure of infected lung cells with SARS-CoV-2. Unlike previous studies, this novel study aims to investigate COVID-19 classification at the lung cellular level in response to SARS-CoV-2. Particularly, differentiating between healthy and infected human alveolar type II (hAT2) cells with SARS-CoV-2. Hence, we explore the feasibility of deep transfer learning (DTL) and introduce a highly accurate approach that works as follows: First, we downloaded and processed 286 images pertaining to healthy and infected hAT2 cells obtained from the electron microscopy public image archive. Second, we provided processed images to two DTL computations to induce ten DTL models. The first DTL computation employs five pre-trained models (including DenseNet201 and ResNet152V2) trained on more than one million images from the ImageNet database to extract features from hAT2 images. Then, it flattens and provides the output feature vectors to a trained, densely connected classifier with the Adam optimizer. The second DTL computation works in a similar manner, with a minor difference in that we freeze the first layers for feature extraction in pre-trained models while unfreezing and jointly training the next layers. The results using five-fold cross-validation demonstrated that TFeDenseNet201 is 12.37× faster and superior, yielding the highest average ACC of 0.993 (F1 of 0.992 and MCC of 0.986) with statistical significance ( from a t-test) compared to an average ACC of 0.937 (F1 of 0.938 and MCC of 0.877) for the counterpart (TFtDenseNet201), showing no significance results ( from a t-test).
Keywords:
SARS-CoV-2; COVID-19; human alveolar type 2 cells; transmission electron microscopy; deep transfer learning; AI applications in respiratory diseases MSC:
92B05; 68T09
1. Introduction and Related Work
SARS-CoV-2 is the virus behind a respiratory disease, named COVID-19, in which the main target organ is the human lung [1]. When SARS-CoV-2 binds to the ACE2 receptor in alveolar type II (AT2) cells of alveoli in the lungs, the ACE2 receptor becomes occupied, leading to more lung injuries and thereby impaired lung function, which is attributed to damaging the alveoli [2,3,4]. As the number of COVID-19 patients with lung infections significantly outnumbered healthcare workers, researchers developed AI-based techniques to accurately detect lung infections in COVID-19 patients using different imaging modalities. Hussein et al. [5] proposed a custom convolutional neural network (custom-CNN) to accurately classify COVID-19 patients using two previously studied chest X-ray image datasets from Kaggle and GitHub. In the first dataset, the images were categorized into three class labels, including COVID-19, normal (non-COVID-19), and viral pneumonia. For the second dataset, the images were categorized into COVID-19 and normal (non-COVID-19). Their custom-CNN consisted of a series of convolution and max pooling layers, followed by flattening the extracted feature vectors provided to a densely connected classifier composed of a stack of five dense layers interleaved with three dropout layers and two BatchNormalization layers. Custom-CNN was trained on a random split of the dataset images while testing the performance on the remaining split. The reported results on a random 20% testing split of the first dataset demonstrated that custom-CNN achieved an accuracy of 0.981, a precision of 0.976, a recall of 0.983, and an F1 of 0.973. For the second dataset, the custom-CNN achieved an accuracy of 0.998, a precision of 0.999, a recall of 0.997, and an F1 of 0.998.
To identify pulmonary diseases using X-ray and CT scan images, Abdullahi et al. [6] presented a CNN named PulmoNet, composed of 26 layers using wide residual blocks (of two convolutional layers with a dropout layer), followed by a GlobalAveragePooling layer and one dense layer with SoftMax activation to yield predictions. The Adam optimizer was used with categorical cross-entropy loss. They formulated the classification problem into a multiclass classification of pulmonary diseases and also reported the results for the binary class classification task pertaining to pulmonary diseases, described as follows: The dataset had 16,435 images: 883 images for bacterial pneumonia, 1478 images for viral pneumonia, 3749 for COVID-19, and 10,325 images for healthy cases. They divided the dataset and utilized an 85:15 training-to-testing split ratio. Then, they conducted a cross-validation based on randomly assigning examples according to the predefined split ratio and averaging the results of the testing according to five runs of the cross-validation. Particularly, for the task of testing the performance using four classes, they achieved an average accuracy of 0.940. For testing three-class classification pertaining to bacterial pneumonia, COVID-19, and healthy cases, the presented model achieved an average accuracy of 0.954. Also, they achieved an average accuracy of 0.994 for the binary class classification pertaining to discrimination between COVID-19 and healthy image cases. For the binary class classification between pneumonia and healthy image cases, their model achieved an average accuracy of 0.983.
Talukder et al. [7] presented a deep transfer learning (DTL) approach to detecting COVID-19 using X-ray images, working as follows: First, they used two datasets, where the first had 2000 COVID-19 X-ray images pertaining to COVID-19 and normal cases, and the second had 4352 chest X-ray images pertaining to these four classes: COVID-19, normal, lung opacity, and viral pneumonia. They divided the two datasets and utilized an 80:10 training-to-validation split ratio, while the remaining was used for testing. Image augmentation was performed through operation, such as shears, rotation, flipping, and zooming. For DTL, six pre-trained models (Xception, InceptionResNetV2, ResNet50, ResNet50V2, EfficientNetB0, and EfficientNetB4) were used. Then, they unfroze the first layers in the feature extraction part of pre-trained models and unfroze the next layers, including the GlobalAveragePool layer, two BatchNormalization layers, and two dense layers for prediction. The results demonstrated that EfficientNetB4, when applied to 208 testing images from the first dataset, achieved the highest accuracy of 1.00. Moreover, EfficientNetB4, when applied to 480 testing images from the second dataset, achieved an accuracy of 0.9917 and an F1 of 0.9914.
Abdullah et al. [8] presented a hybrid DTL approach to detect COVID-19 using chest X-ray images. First, they downloaded the images from the COVID-19 radiography database at Kaggle, selecting 2413 images pertaining to COVID-19 and 6807 images related to normal cases. They divided the dataset and utilized a 70:30 training (including validation)-to-testing split ratio. To induce the hybrid model, they utilized VGG16 and VGG19 pre-trained models, in which they applied the feature extraction part of these two pre-trained models to extract features from the image training set. Then, they concatenated the output of the two flattened feature vectors provided as inputs to train densely connected classifiers of two dense layers and one dropout layer. Also, the input feature vectors were provided to train machine learning algorithms including random forest, naive Bayes, KNN, neural network (NN), and support vector machines (SVMs) with several kernels, including linear, sigmoid, and radial. Applying the experimental results to the 30% testing data (i.e., 2766 images out of 9220) demonstrates the superiority of the hybrid DTL approach when coupled with NN, achieving the highest MCC of 0.814, followed by SVM with a linear kernel, which yielded an MCC of 0.805 when compared to existing pre-trained models of densely connected classifiers in the binary classification task. Others have proposed deep learning approaches to detect COVID-19 using X-ray and CT images [9,10,11,12,13,14]. Table 1 provides an overview of the existing works compared to our proposed work.
Although these recent studies aimed to detect lungs infected with SARS-CoV-2 in COVID-19 patients, the novelty of our study is attributed to the summarized contributions as follows:
- (1)
- Unlike the existing studies that aim to detect lungs infected with SARS-CoV-2 using different imaging modalities (e.g., CT and X-rays), to the best of our knowledge, this is the first study to investigate the feasibility of DTL in lungs infected with SARS-CoV-2 at the cellular level within alveoli in human lungs using transmission electron microscopy (TEM) images.
- (2)
- The images generated via TEM unveiled the detailed structures of hAT2 cells and other characteristics of SARS-CoV-2, including viral particles dispersed inside the cell cytoplasm. We downloaded and processed 286 images pertaining to infected and healthy (control) human alveolar type II (hAT2) cells in alveoli from the electron microscopy public image archive (EMPIAR) at https://www.ebi.ac.uk/empiar/EMPIAR-10533/ (accessed on 13 March 2023).
- (3)
- We formulated the problem as a binary class classification problem and induced ten DTL models using two DTL computations [15], where in the first DTL computation, we applied five pre-trained models (DenseNet201 [16], NasNetMobile [17], ResNet152V2 [18], VGG19 [19], and Xception [20]) to extract features from hAT2 images, followed by flattening the extracted features into feature vectors provided as inputs to train a densely connected classifier of three layers, including two dense layers and one dropout layer. We refer to the induced models via the first DTL computation as TFeDenseNet201, TFeNasNetMobile, TFeResNet152V2, TFeVGG19, and TFeXception. For the second DTL computation, we froze the first layers in the pre-trained models while unfreezing and jointly training the next layers, including a densely connected classifier. The induced models via such DTL computation are referred to as TFtDenseNet201, TFtNasNetMobile, TFtResNet152V2, TFtVGG19, and TFtXception.
- (4)
- For fairness of performance comparisons among the ten studied DTL models, we evaluated the performance on the whole dataset of 286 images using five-fold cross-validation, in which we provided the same training and testing images in each run to each model. Then, we reported the average performance results of the five runs on testing folds and reported the standard deviation.
- (5)
- Our experimental study demonstrated that TFeDenseNet201 achieved the highest average ACC of 0.993, the highest F1 of 0.992, and the highest MCC of 0.986 when utilizing five-fold cross-validation. Moreover, these performance results were statistically significant (, obtained from a t-test), demonstrating the generalization ability of TFeDenseNet201. In terms of measuring the training running time, TFeDenseNet201 was 12.37× faster than its peer, TFtDenseNet201, induced via the second DTL method. These high-performance results demonstrate that our DTL models can act as (1) an assisting AI tool in diagnostic pathology and (2) a reliable AI tool for automatic annotation when used with TEM. We provide details about the experimental study, including the processed datasets, in the Supplementary Materials.

Table 1.
A summary of the literature studies against our proposed work. VP is viral pneumonia. BP is bacterial pneumonia. LO is lung opacity. AT2 is alveolar type II. CT is computed tomography. TEM is transmission electron microscopy. ACC is accuracy; MCC is the Matthews correlation coefficient. CV is cross-validation.
Table 1.
A summary of the literature studies against our proposed work. VP is viral pneumonia. BP is bacterial pneumonia. LO is lung opacity. AT2 is alveolar type II. CT is computed tomography. TEM is transmission electron microscopy. ACC is accuracy; MCC is the Matthews correlation coefficient. CV is cross-validation.
| Year | Study | Scanning | Cell Type | Best Architecture | Class | Evaluation Using | Results |
|---|---|---|---|---|---|---|---|
| 2022 | Oğuz et al. [21] | CT | - | ResNet-50+SVM | 2-class (COVID-19, normal) | Test set | ACC (0.96); F1 (0.95); AUC (0.98) |
| 2022 | Haghanifar et al. [22] | X-ray | - | COVID-CXNet | 3-class (COVID-19, normal, pneumonia) | Test set | ACC (0.8788); F1 (0.97) |
| 2022 | Bhattacharyya et al. [23] | X-ray | - | VGG19-BRISK-RF | 3-class (COVID-19, normal, pneumonia) | Test set | ACC (0.966) |
| 2022 | Chouat et al. [24] | X-ray; CT | - | VGGNet-19 | 2-class (COVID-19, normal) | Test set | ACC (0.905) |
| 2022 | Asif et al. [25] | X-ray | - | Shallow CNN | 2-class (COVID-19, normal) | Test set | ACC (0.9968) |
| 2022 | Ullah et al. [26] | X-ray | - | CovidDetNet | 3-class (COVID-19, normal, pneumonia) | Test set | ACC (0.9840) |
| 2022 | Zouch et al. [27] | X-ray; CT | - | VGG19 | 2-class (COVID-19, normal) | Test set | ACC (0.9935) |
| 2023 | Haennah et al. [9] | CT | - | DETS-ResNet101 | 2-class (COVID-19, normal) | Five-fold CV | ACC (0.967); F1 (0.969) |
| 2023 | Salama et al. [13] | CT | - | Hybrid DL+ML | 2-class (COVID-19, normal) | Three test sets | Average ACC (0.9869) |
| 2023 | Ayalew et al. [28] | X-ray | - | CNN+SVM | 2-class (COVID-19, normal) | Test set | ACC (0.991) |
| 2023 | Constantinou et al. [29] | X-ray | - | ResNet101 | 3-class (COVID-19, non-COVID-19, normal) | Test set | ACC (0.96) |
| 2023 | Patro et al. [30] | X-ray | - | SCovNet | 2-class (COVID-19, normal) | Test set | ACC (0.9867) |
| 2023 | Zhu et al. [31] | X-ray | - | CovC-ReDRNet | 3-class (COVID-19, normal, pneumonia) | Five-fold CV | ACC (0.9756); F1 (0.9584) |
| 2023 | Chakraborty et al. [32] | X-ray | - | WAE | 3-class (COVID-19, normal, VP) | Test set | ACC (0.9410); F1 (0.9404) |
| 2-class (COVID-19, non-COVID-19) | ACC (0.9725); F1 (0.9665) | ||||||
| 3-class (COVID-19, normal, VP) | Five-fold CV | ACC (0.8605) | |||||
| 2-class (COVID-19, non-COVID-19) | ACC (0.861) | ||||||
| 2023 | Gaur et al. [33] | X-ray | - | EfficientNetB0 | 3-class (COVID-19, normal, VP) | Test set | ACC (0.9293); F1 (0.88) |
| 2023 | Kathamuthu et al. [34] | CT | - | VGG16 | 2-class (COVID-19, normal) | Test set | ACC (0.98) |
| 2024 | Hussein et al. [5] | X-ray | - | Custom-CNN | 3-class (COVID-19, non-COVID-19, VP) 2-class (COVID-19, non-COVID-19) | Test set | ACC (0.981); F1(0.973) |
| ACC (0.998); F1 (0.998) | |||||||
| 2024 | Abdulahi et al. [6] | X-ray; CT | - | PulmoNet | 4-class (COVID-19, healthy, BP, VP) | CV on a random test sets | ACC (0.940) |
| 3-class (COVID-19, healthy, BP) | ACC (0.954) | ||||||
| 2-class (COVID-19, healthy) | ACC (0.994) | ||||||
| 2-class (pneumonia, healthy) | ACC (0.983) | ||||||
| 2024 | Talukder et al. [7] | X-ray | - | EfficientNetB4 | 4-class (COVID-19, normal, LO, VP) | Test set | ACC (0.9917); F1 (0.9914) |
| 2-class (COVID-19, normal) | ACC (1.00) | ||||||
| 2024 | Abdullah et al. [8] | X-ray | - | Hybrid DL-NN | 2-class (COVID-19, normal) | Test set | ACC (0.920; MCC (0.814) |
| 2024 | Proposed | TEM | AT2 | TFeDenseNet201 | 2-class (infected, control) | Five-fold CV | ACC (0.993); F1 (0.992); MCC (0.986) |
2. Materials and Methods
2.1. Data Preprocessing
Youk et al. [35] performed transmission electron microscopy (TEM) imaging to obtain high-resolution images pertaining to healthy and infected human lung alveolar type 2 (hAT2) cells with SARS-CoV-2. The generated images unveiled a detailed structure in the cell and other characteristics of SARS-CoV-2, including viral particles dispersed inside the cell cytoplasm. In Figure 1, we present an illustration for hAT2 images employed in this study obtained from the electron microscopy public image archive (EMPIAR) at https://www.ebi.ac.uk/empiar/EMPIAR-10533/ (accessed on 13 March 2023) and composed of 577 hAT2 images related to uninfected (control) and infected hAT2 cells [35]. The class label distribution consisted of 326 images belonging to control hAT2 cells, while the other 251 images belonged to infected hAT2 cells. As TEM imaging was employed to generate images, the 577 images had a 4096 × 4224 pixel resolution (i.e., 17.3015Mpx) and were stored as TIFF image files. In our study, we randomly selected and processed 286 TIFF images out of 577 with the use of the Image module in Python [36], obtaining 286 JPG images with a 256 × 256 pixel resolution for addressing the classification task at hand. It is worth noting that the class distribution was balanced, in which 143 images belonged to control hAT2 images while the remaining 143 images belonged to infected hAT2 images. We provide 286 preprocessed JPG images in the Supplementary Dataset.
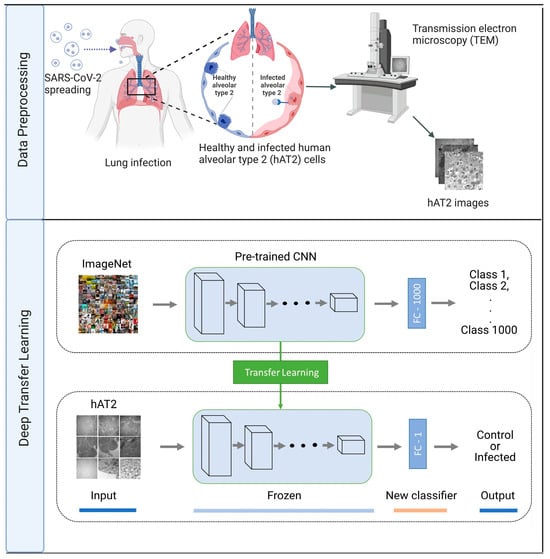
Figure 1.
Flowchart showing our deep transfer learning approach for predicting hAT2 cells infected with SARS-CoV-2. Data preprocessing: to obtain hAT2 images, healthy and infected hAT2 images using transmission electron microscopy (TEM) imaging were downloaded and processed from https://www.ebi.ac.uk/empiar/EMPIAR-10533/ (accessed on 13 March 2023). Deep transfer learning: hAT2 images were provided to a pre-trained CNN of frozen layers for feature extraction, followed by a trained new classifier to discriminate between healthy and infected hAT2 cells. Figure created with BioRender.com.
2.2. Deep Transfer Learning
In Figure 1, we demonstrate how our deep transfer learning (DTL) approach was carried out. Initially, we employed five pre-trained models: DenseNet201, NasNetMobile, ResNet152V2, VGG19, and Xception. As each pre-trained model consists of a feature extraction part (i.e., interleaved convolutional and pooling layers) and a densely connected classifier for feature extraction and classification, respectively, we froze the weights (i.e., to keep the weights unaltered) of the feature extraction part while modifying the densely connected classifier, dealing with the binary class problem at hand rather than the multiclass classification of 1000 classes. Then, we extracted features from the hAT2 images by applying the feature extraction part (using the unadjusted weights of a pre-trained model), followed by training the densely connected classifiers and performing a prediction on the unseen hAT2 images. We refer to such DTL models as TFeDenseNet201, TFeNasNetMobile, TFeResNet152V2, TFeVGG19, and TFeXception (see Figure 1). In Table 2, we include information regarding the development of TFe-based models. All models have the same number of unfrozen layers because we only trained the densely connected classifier (composed of three layers) on extracted features from hAT2 images using pre-trained models. Therefore, the number of non-trainable parameters is 0. The last 3D tensor output (of shape (8, 8, 2048)) for feature extraction was the same for TFeResNet152V2 and TFeXception. Hence, both had the same number of parameters as they were flattened into a vector of 131,072 elements before feeding into the same densely connected classifier of three layers. We provide details about each TFe-based model in the Supplementary TFeModels.

Table 2.
Details about layers and parameters for TFe-based models.
For the other DTL models, we froze the weights of the bottom layers in the feature extraction part while we trained the top layers of the feature extraction layers as well as the densely connected classifier. In other words, we unaltered the weights of the first layers in the feature extraction part while adjusting the weights of subsequent layers. Moreover, we modified the densely connected classifier to tackle the binary class classification problem rather than the multiclass classification pertaining to 1000 categories. We refer to models employing such computations as TFtNasNetMobile, TFtResNet152V2, TFtVGG19, and TFtXception (see Figure 2). Table 3 demonstrates the layers and parameters used for the TFt-based models and the number of layers, including those from pre-trained models in terms of feature extraction, plus the densely connected classifier of two layers. We report the details of each model architecture in the Supplementary TFtModels.
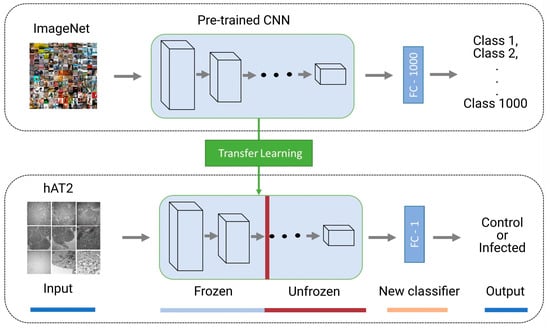
Figure 2.
Deep transfer learning composed of a pre-trained CNN with frozen and unfrozen layers for feature extraction, followed by a trained new classifier to discriminate between healthy and infected hAT2 cells.

Table 3.
Details about the layers and parameters for TFt-based models.
In the training phase, we employed the Adam optimizer with binary cross-entropy loss when updating the model parameters. It can be seen that transfer learning is ascribed to the unadjusted weights from pre-trained models that were previously trained on more than one million images from the ImageNet dataset. In terms of testing the performance to unseen hAT2 images, predictions are mapped to the infected if their values are greater than 0.5. Otherwise, predictions are mapped to the control.
3. Results
3.1. Classification Methodology
In this study, we adapted five pre-trained models: DenseNet201, NasNetMobile, ResNet152V2, VGG19, and Xception. Each pre-trained model was trained on over a million images from the ImageNet dataset for the multiclass image classification task of 1000 class labels. Then, we developed TFe-based models as follows: We utilized the feature extraction part of pre-trained models by freezing all of the layers to extract features from hAT2 images, which we then flattened and provided to a densely connected classifier (of three layers), trained from scratch for the task of classifying hAT2 cells into control (i.e., uninfected) or infected with SARS-CoV-2. For the TFt-based models, we froze the first layers in the feature extraction part of pre-trained models while we unfroze consecutive layers, in which the last output was flattened and then we provided it to a densely connected classifier of two layers, trained from scratch to address the binary classification task. For all models, we employed the Adam optimizer with the binary cross-entropy loss function. We assigned the following optimization parameters during the training phase: 0.00001 for the learning rate, 20 for the batch size, and 10 for the number of epochs. We did not use a learning rate scheduler. We evaluated the performance of each model using accuracy (ACC), F1, and Matthews correlation coefficient (MCC), calculated as follows [37,38]:
where TP is a true positive, referring to the number of infected hAT2 images that were correctly predicted as infected. FN is a false negative, referring to the number of infected hAT2 images that were incorrectly predicted as uninfected (control). TN is a true negative, referring to the number of control hAT2 images that were correctly predicted as controls. FP is a false positive, referring to the number of control hAT2 images that were incorrectly predicted as infected.
For reporting the performance results on the whole dataset of 286 hAT2 images, we utilized five-fold cross-validation, in which we randomly assigned images into five folds. Then, in the first run, we assigned images of the first fold for testing while assigning the remaining examples in the other folds for training, followed by performing a prediction for the examples in the testing fold and recording the results. Such a process was repeated in the remaining four runs, in which we recorded the performance on testing folds. Then, we used the average performance calculated during the five runs as our results on five-fold cross-validation.
3.2. Implementation Details
To run the experiments, we used the Anaconda distribution, creating an environment with Python (Version 3.10.14) [15], followed by installing the following libraries: cudatoolkit (Version 11.2) [39], cudnn (Version 8.1.0) [40], and tensorflow (Version 2.8) [41]. Then, we installed Jupiter notebook [42,43] to write and execute Python code for deep learning models with Keras on our local NVIDIA GeForce RTX 2080Ti GPU with 4352 CUD cores, 11 GB of GDDR6 memory, 1545 MHz of boost clock speed, and 14 Gbps of memory clock speed. Other libraries were used to run the experiments, including pandas and NumPy for data processing, while matplotlib and Sklearn were utilized for confusion matrix visualization and performance evaluation, respectively [44,45]. Also, we utilized ggplot2 in R to visualize the results (e.g., Figure 3, Figure 4, Figure 5 and Figure 7) [46].
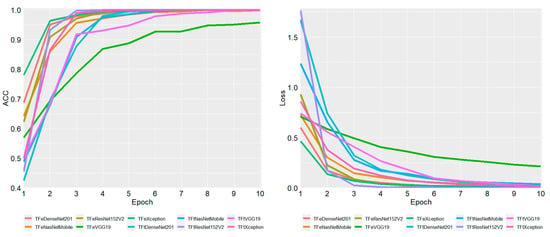
Figure 3.
Average ACC and loss of five training folds for each epoch. ACC is accuracy.
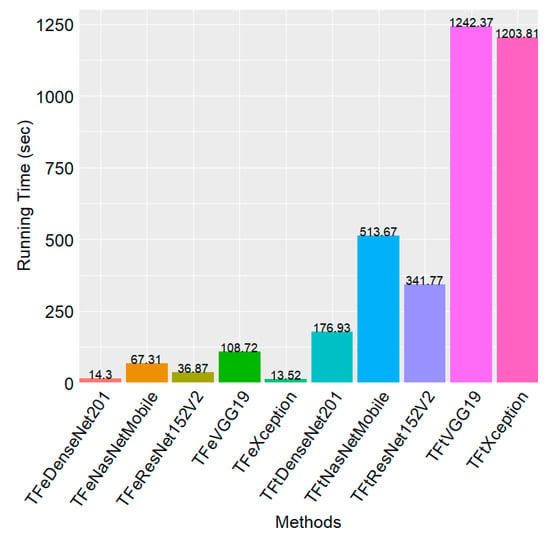
Figure 4.
Running time for each method during the training phase in the five-fold cross-validation.
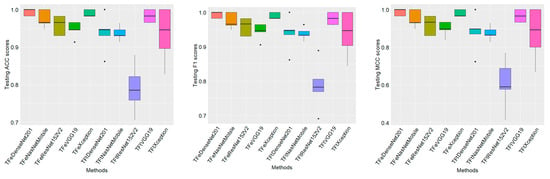
Figure 5.
Boxplots show testing (generalization) performance results of ten models during five-fold cross-validation. ACC is accuracy. MCC is the Matthews correlation coefficient.
3.3. Classification Results
3.3.1. Training Results
In Figure 3, we report the average training accuracy and loss during the five-fold cross-validation of ten models trained for 10 epochs. Accuracy is calculated in terms of correctly classified training examples, while loss is calculated in terms of binary cross-entropy. These two measures, accuracy and loss, demonstrate the ability of models to learn from data and be capable of classifying hAT2 images during testing. At the first epoch, it can be seen that TFeXception generated the highest average accuracy of 0.78 (and the lowest average loss of 0.46), followed by TFeDenseNet201, generating an average accuracy (and loss) of 0.68 (and 0.60). TFtDenseNet201 generated the lowest average accuracy (and loss) of 0.42 (and 1.67). At the second epoch, TFeXception adapted more to the training data by generating the highest average accuracy of 0.96 (and the lowest average loss of 0.13), followed by TFeDenseNet201, which achieved the second highest average accuracy of 0.95 (with an average loss of 0.17). As the number of epochs increases, the accuracy and loss increase and decrease, respectively, in which all models, except TFeVGG19, achieved an average accuracy of above 0.998 and an average loss close to 0. For each epoch, we included average accuracy and loss in the Supplementary Epoch.
Figure 4 illustrates the total running time for each model induction process during the five-fold cross-validation. TFeXception is the fastest. Particularly, TFeXception is 89.03× faster than its counterpart, TFtXception. The second fastest method is TFeDenseNet201, which is 12.37× faster than TFtDenseNet201. The third fastest method is TFeResNet152V2, which is 9.26× faster than TFtResNet152V2. The slowest mode among the TFe-based methods is TFeVGG19, which is 11.42× faster than TFtVGG19. The running time difference between the fastest two methods, TFeXception and TFeDenseNet201, is marginal. Particularly, they differ by less than 1 s. These results demonstrate the computational efficiency of TFe-based models, attributed to fewer layers during the training phase.
3.3.2. Testing Results
In Figure 5, boxplots demonstrate the generalization (testing) results of ten models when employing five-fold cross-validation. The median results are displayed by the horizontal bold lines crossing each box. It can be noticed that TFeDenseNet201 is the best performing model, achieving the highest median of 1.00 according to the three performance measures (i.e., ACC, F1, and MCC), where the standard deviation σ is 0.009 (0.009, and 0.017) for ACC (F1 and MCC). The second-best-performing model is TFeXception, achieving a median ACC of 0.982 (σ = 0.009), a median F1 of 0.982 (σ = 0.009), and a median MCC of 0.966 (σ = 0.017). The third-best-performing model is TFtVGG19, achieving a median ACC of 0.982 (σ = 0.017), a median F1 of 0.982 (σ = 0.018), and a median MCC of 0.966 (σ = 0.030). The worst-performing model is TFtResNet152V2, yielding a median ACC of 0.785 (σ = 0.064), a median F1 of 0.782 (σ = 0.071), and a median MCC of 0.590 (σ = 0.119). Although TFtVGG19 and TFeXception achieved the same results, the standard deviation for TFtVGG19 demonstrates greater variability in performance results, attributed to degradation in performance in two testing folds when compared to TFeXception. We report the median results in the Supplementary BoxplotMed.
Table 4 records the five-fold cross-validation results, which are calculated on the basis of averaging the performance results on five testing folds. Also, we recorded the standard deviation. It can be noticed that TFeDenseNet201 is the best-performing model, achieving the highest average ACC of 0.993 (σ = 0.008), the highest average F1 of 0.992 (σ = 0.009), and the highest average MCC of 0.986 (σ = 0.018). TFeXception is the second-best-performing model, yielding an average ACC of 0.989 (σ = 0.008), an average F1 of 0.989 (σ = 0.009), and an average MCC of 0.979 (σ = 0.018). TFtVGG19 is the third-best-performing model, achieving an average ACC of 0.982 (σ = 0.015), an average F1 of 0.981 (σ = 0.018), and an average MCC of 0.966 (σ = 0.033). The worst-performing model is TFtResNet152V2, generating an average ACC of 0.790 (σ = 0.058), an average F1 of 0.787 (σ = 0.071), and an average MCC of 0.608 (σ = 0.133). These results demonstrate the superiority of TFe-based models over TFt-based models.

Table 4.
Average performance results during the five-fold cross-validation on test folds for ten models. ACC is accuracy. MCC is the Matthews correlation coefficient. Bold refers to a model achieving the highest performance results.
Figure 6 illustrates the combined confusion matrices of test predictions when running five-fold cross-validation. The best-performing model, TFeDeNseNet201, accurately predicted a total of 284 out of 286 hAT2 images while incorrectly predicting 2 images in which the ground truth was infected (positive) and predicted as control (negative), which counted as 2 FN. The second-best-performing model, TFeXception, accurately predicted 283 hAT2 images, while 3 images were incorrectly predicted as negative when their actual label was positive, which counted as 3 FN. TFtVGG19, the third-best-performing model, accurately predicted 281 hAT2 images and incorrectly predicted 5 as negative hAT2 images when their actual label was positive, which counted as 5 FN. The worst-performing model, TFtResNet152V2, accurately predicted 226 hAT2 images. A total of 31 hAT2 images were predicted to be positive when their label was negative, which counted as 31 FP. Moreover, 29 hAT2 images were predicted to be negative when their actual label was positive, which counted as 29 FN.
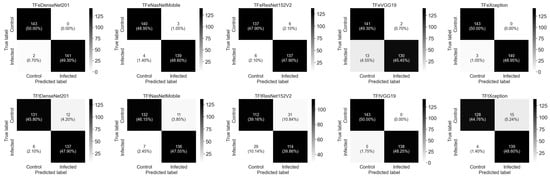
Figure 6.
The combined confusion matrices of five testing folds during the running of five-fold cross-validation for ten models.
Figure 7 depicts computational insights for the generalization ability of ten studied models, applied to testing. It can be seen from the boxplots and strip charts that the prediction differences for TFeDenseNet201 between the control hAT2 against infected hAT2 were statistically significant (P < , obtained from a t-test), suggesting that the TFeDenseNet201 model is a general predictor for control or infected hAT2 prediction. The prediction differences for the second- and third-best-performing models (i.e., TFeXception and TFtVGG19) were not significant (P = 0.4838 and P = 0.1442, respectively, obtained from a t-test). These results suggest that these two models are specific rather than general predictors for hAT2. The other two models, TFeVGG19 and TFtNasNetMobile, exhibited significant prediction differences between control and infected hAT2 (P = and P = , respectively, obtained from a t-test), although their prediction performance does not outperform TFeDenseNet201. The prediction differences for all other models between control and infected hAT2 were not significant, suggesting that these models are specific predictors, and we included their p-values in the Supplementary BoxStripPval.
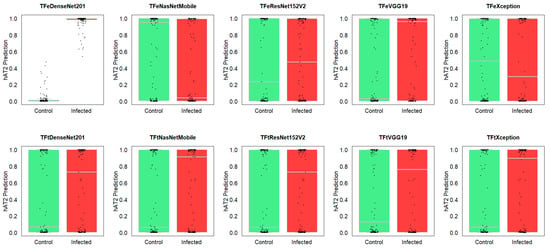
Figure 7.
Boxplots and strip charts of predicted hAT2 for control and infected cells on the whole testing folds related to ten models.
4. Discussion
To discriminate between control and infected hAT2 cells, our approach consisted of two parts: data preprocessing followed by DTL. In the data preprocessing, we randomly pulled a sample of 286 images pertaining to infected and control hAT2 cells, followed by using the Image library in Python to convert 286 TIFF images of a 4096 × 4224 pixel resolution to 286 jpg images of a 256 × 256 pixel resolution, which are included in the Supplementary Dataset. Then, we conducted two DTL computations using five pre-trained models, namely DenseNet201, NasNetMobile, ResNet152V2, VGG19, and Xception. The first DTL transferred the knowledge by applying the feature extraction part of pre-trained models to hAT2 images in the training set, extracting features that are flattened and providing them as inputs to a densely connected classifier of three layers, trained from scratch to deal with two class labels (i.e., control and infected). We refer to models using this type of transfer learning computation as TFeDenseNet201, TFeNasNetMobile, TFeResNet152V2, TFeVGG19, and TFeXception. The second DTL computation involves the use of the first layers in pre-trained models for feature extraction while training from scratch the next layers, including the densely connected classifier, dealing with discriminating between control and infected hAT2 images. Such computation leads to another five DTL models, including TFtDenseNet201, TFtNasNetMobile, TFtResNet152V2, TFtVGG19, and TFtXception.
When we employed five-fold cross-validation, we divided the dataset into training and testing, in which the former had examples from four folds while the latter had examples from the remaining one fold. Then, we performed training, monitoring the accuracy and loss on the training fold. After finishing ten epochs, we recorded the loss and accuracy results, and we induced a total of ten DTL models. Then, we applied each DTL to examples in the testing fold and recorded the performance results. We repeated such a process for four more runs, recording the results, followed by taking the average loss and accuracy (see Figure 3) and reporting the average testing results and standard deviation (see Table 4). The experimental results demonstrate the feasibility of DTL in tackling the studied classification task, in which TFeDenseNet201 generated the highest average ACC of 0.993, the highest average F1 of 0.992, and the highest average MCC of 0.986.
It is worth noting that the frozen layers in DTL contributed to reducing the number of trainable layers and, thereby, the mitigation of overfitting. We introduced the dropout layer in the densely connected classifier of TFe-based models to reduce overfitting. In TFe-based models, we just trained the densely connected classifier composed of three layers, while we trained the top layers in the feature extraction part in addition to the densely connected classifier for TFt-based models. Therefore, we had three unfrozen layers in TFe-based models (see Table 2) while having a different number of unfrozen layers for TFt-based models (see Table 3).
For the extracted features via studied models when considering TFeVGG19, the output shape of the feature extraction part is (none, 8, 8, 512), which is flattened to an output shape of (none, 32768) and provided to a trained densely connected classifier from scratch in which the first dense layer has an output shape of (none, 256) and the number of parameters is 8388864. The second dense layer produces an output shape of (none, 1), and the number of parameters is 257. In TFtVGG19, we froze the first layers while unfreezing the top layers that were jointly trained with the densely connected classifier. It is evident that TFe-based models are faster because the extracted features are provided to train a densely connected classifier, although both TFeVGG19 and TFtVGG19 have the same output shape fed as an input to the densely connected classifier. Moreover, the number of parameters in densely connected classifiers for TFe-based models is equal to the number of parameters in densely connected classifiers for counterpart models in TFt-based models. We provide details about all of the models in the Supplementary TFeModels and TFeModels.
In Table 1, we present twenty existing studies to show that our study is unique in terms of the scanning method (see the scanning column) to obtain images as well as the studied cell type (see the cell type column). It is evident that no DL study, to the best of our knowledge, coincides with our work, although all of these studies share the same classification task of discriminating between healthy and infected patients with SARS-CoV-2. Therefore, we are the first, to the best of our knowledge, to investigate COVID-19 classification using DL at the lung cellular level concerning AT2 cells in response to SARS-CoV-2 infection.
In terms of the computational training running time, the two DTL computations were efficient. The DTL computation inducing TFe-based models was more efficient, attributed to the training of just the densely connected classifier. Therefore, it is no surprise that the training time to induce DTL models was longer for computations involving the induction of TFt-based models than for computations involved in inducing TFe-based models (see Figure 4). The slowest TFe-based model was TFeVGG19, which was 11.42× faster than TFtVGG19, the TFt-based model. These results demonstrate that DTL computation-inducing TFe-based models can efficiently address the task of discriminating between infected and control hAT2 cells.
In the pre-trained models, the final layer in the densely connected classifier had a softmax activation for the last layer pertaining to 1000 classes. In our study, we replaced the densely connected classifier in which the final layer had a sigmoid activation producing probabilities. When a probability was greater than 0.5, we classified an image as an infected hAT2 cell. Otherwise, we classified the image as a control (healthy) hAT2 cell. Obtaining DTL models in terms of the second computation is time-consuming, and utilizing ten-fold cross-validation requires a training process to be performed ten times, attributed to the ten runs in ten-fold cross-validation. Therefore, we utilized five-fold cross-validation as a less time-consuming option in which we have five runs, where in each run, we performed one training process to achieve a model and perform predictions for the testing set.
5. Conclusions and Future Work
To address the novel target task of classifying control and infected human alveolar type II (hAT2) cells with SARS-CoV-2, we assessed and presented ten deep transfer learning (DTL) models, derived as follows: First, we downloaded and processed a total of 286 images from the electron microscopy public image archive, pertaining to control and infected hAT2 cells with SARS-CoV-2. Second, we utilized five pre-trained models (DenseNet201, NasNetMobile, ResNet152V2, VGG19, and Xception), previously trained on more than a million images from the ImageNet database. Then, we applied the feature extraction part in pre-trained models to extract features from hAT2 images in the training set, followed by performing a flattening step before providing the feature vectors to a modified densely connected classifier with the Adam optimizer, trained from scratch to discriminate between control and infected samples. Another DTL computation involved freezing the first layers in the feature extraction part of pre-trained models while unfreezing and training the next layers, including a modified densely connected classifier coupled with the Adam optimizer. The experimental results on the entire dataset of 286 hAT2 images employing five-fold cross-validation demonstrate (1) the efficiency of the TFeDenseNet201 model, which was 12.37× faster than its counterpart, TFtDenseNet201, during the training time; (2) the superiority of TFeDenseNet201, achieving the highest average ACC of 0.993 (σ = 0.008), the highest average F1 of 0.992 (σ = 0.009), and the highest average MCC of 0.986 (σ = 0.018), outperforming its counterpart,TFtDenseNet201, which achieved an average ACC of 0.937 (σ = 0.044), an average F1 of 0.938 (σ = 0.049), and an average MCC of 0.877(σ = 0.099); (3) the significant results achieved via TFeDenseNet201 (P < , obtained from a t-test), while TFtDenseNet201 did not establish significance (P = 0.093, obtained from a t-test); and (4) the feasibility and reliability of the presented TFeDenseNet201 (among other DTL models) as an assisting AI tool for classifying hAT2 cells based on TEM images.
Future work can include the following: (1) inducing medical-based imaging models derived from DTL and thereby efficiently addressing different target tasks pertaining to neurological disorders [47]; (2) developing ensemble models using DTL methods and evaluating their generalization performance; and (3) integrating genomic, clinical information, and features using DTL models to improve the prediction performance in problems from biology and medicine.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/math12101573/s1. The README file provides a quick guide about each file included in this study.
Author Contributions
T.T. conceived and designed the study. T.T. performed the analysis. T.T. and Y.-h.T. evaluated the results and discussions. T.T., S.A.H. and Y.-h.T. wrote the manuscript. T.T. supervised the study. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The hAT2 dataset in this study is available at https://www.ebi.ac.uk/empiar/EMPIAR-10533/; accessed on 13 March 2023.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| DL | Deep Learning |
| DTL | Deep Transfer Learning |
| DenseNet | Dense Convolutional Network |
| NasNet | Neural Architecture Search Network |
| ResNet | Residual Neural Network |
| VGG | Visual Geometry Group |
| Xception | Extreme Inception |
| CNN | Convolutional Neural Network |
| hAT2 | Human Alveolar Type II |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| COVID-19 | Coronavirus Disease 19 |
| Adam | Adaptive Moment Estimation |
| EMPIAR | Electron Microscopy Public Image Archive |
| CT | Computerized Tomography |
| TEM | Transmission Electron Microscopy |
| ACC | Accuracy |
| MCC | Matthews Correlation Coefficient |
References
- Van Slambrouck, J.; Khan, M.; Verbeken, E.; Choi, S.; Geudens, V.; Vanluyten, C.; Feys, S.; Vanhulle, E.; Wollants, E.; Vermeire, K. Visualising SARS-CoV-2 infection of the lung in deceased COVID-19 patients. EBioMedicine 2023, 92, 104608. [Google Scholar] [CrossRef] [PubMed]
- Gerard, L.; Lecocq, M.; Bouzin, C.; Hoton, D.; Schmit, G.; Pereira, J.P.; Montiel, V.; Plante-Bordeneuve, T.; Laterre, P.-F.; Pilette, C. Increased angiotensin-converting enzyme 2 and loss of alveolar type II cells in COVID-19–related acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2021, 204, 1024–1034. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, Y.; Turki, T. A new advanced in silico drug discovery method for novel coronavirus (SARS-CoV-2) with tensor decomposition-based unsupervised feature extraction. PLoS ONE 2020, 15, e0238907. [Google Scholar] [CrossRef] [PubMed]
- Kathiriya, J.J.; Wang, C.; Zhou, M.; Brumwell, A.; Cassandras, M.; Le Saux, C.J.; Cohen, M.; Alysandratos, K.-D.; Wang, B.; Wolters, P. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5+ basal cells. Nat. Cell Biol. 2022, 24, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Hussein, A.M.; Sharifai, A.G.; Alia, O.M.d.; Abualigah, L.; Almotairi, K.H.; Abujayyab, S.K.; Gandomi, A.H. Auto-detection of the coronavirus disease by using deep convolutional neural networks and X-ray photographs. Sci. Rep. 2024, 14, 534. [Google Scholar] [CrossRef]
- Abdulahi, A.T.; Ogundokun, R.O.; Adenike, A.R.; Shah, M.A.; Ahmed, Y.K. PulmoNet: A novel deep learning based pulmonary diseases detection model. BMC Med. Imaging 2024, 24, 51. [Google Scholar] [CrossRef] [PubMed]
- Talukder, M.A.; Layek, M.A.; Kazi, M.; Uddin, M.A.; Aryal, S. Empowering COVID-19 detection: Optimizing performance through fine-tuned efficientnet deep learning architecture. Comput. Biol. Med. 2024, 168, 107789. [Google Scholar] [CrossRef]
- Abdullah, M.; Kedir, B.; Takore, T.T. A Hybrid Deep Learning CNN model for COVID-19 detection from chest X-rays. Heliyon 2024, 10, e26938. [Google Scholar] [CrossRef]
- Haennah, J.J.; Christopher, C.S.; King, G.G. Prediction of the COVID disease using lung CT images by deep learning algorithm: DETS-optimized Resnet 101 classifier. Front. Med. 2023, 10, 1157000. [Google Scholar] [CrossRef]
- Celik, G. Detection of COVID-19 and other pneumonia cases from CT and X-ray chest images using deep learning based on feature reuse residual block and depthwise dilated convolutions neural network. Appl. Soft Comput. 2023, 133, 109906. [Google Scholar] [CrossRef]
- Park, D.; Jang, R.; Chung, M.J.; An, H.J.; Bak, S.; Choi, E.; Hwang, D. Development and validation of a hybrid deep learning–machine learning approach for severity assessment of COVID-19 and other pneumonias. Sci. Rep. 2023, 13, 13420. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Umemura, Y.; Shi, S.; Inoue, S.; Honda, S.; Matsuzawa, Y.; Hirano, Y.; Kikuyama, A.; Yamakawa, M.; Gyobu, T.; et al. “KAIZEN” method realizing implementation of deep-learning models for COVID-19 CT diagnosis in real world hospitals. Sci. Rep. 2024, 14, 1672. [Google Scholar] [CrossRef] [PubMed]
- Salama, G.M.; Mohamed, A.; Abd-Ellah, M.K. COVID-19 classification based on a deep learning and machine learning fusion technique using chest CT images. Neural Comput. Appl. 2024, 36, 5347–5365. [Google Scholar] [CrossRef]
- Ju, H.; Cui, Y.; Su, Q.; Juan, L.; Manavalan, B. CODE-NET: A deep learning model for COVID-19 detection. Comput. Biol. Med. 2024, 171, 108229. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Deep Learning with Python; Simon and Schuster: New York, NY, USA, 2021. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 June 2017; pp. 4700–4708. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8697–8710. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; Proceedings, Part IV 14. pp. 630–645. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Oğuz, Ç.; Yağanoğlu, M. Detection of COVID-19 using deep learning techniques and classification methods. Inf. Process. Manag. 2022, 59, 103025. [Google Scholar] [CrossRef] [PubMed]
- Haghanifar, A.; Majdabadi, M.M.; Choi, Y.; Deivalakshmi, S.; Ko, S. Covid-cxnet: Detecting COVID-19 in frontal chest X-ray images using deep learning. Multimed. Tools Appl. 2022, 81, 30615–30645. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, A.; Bhaik, D.; Kumar, S.; Thakur, P.; Sharma, R.; Pachori, R.B. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed. Signal Process. Control 2022, 71, 103182. [Google Scholar] [CrossRef]
- Chouat, I.; Echtioui, A.; Khemakhem, R.; Zouch, W.; Ghorbel, M.; Hamida, A.B. COVID-19 detection in CT and CXR images using deep learning models. Biogerontology 2022, 23, 65–84. [Google Scholar] [CrossRef]
- Asif, S.; Zhao, M.; Tang, F.; Zhu, Y. A deep learning-based framework for detecting COVID-19 patients using chest X-rays. Multimed. Syst. 2022, 28, 1495–1513. [Google Scholar] [CrossRef]
- Ullah, N.; Khan, J.A.; Almakdi, S.; Khan, M.S.; Alshehri, M.; Alboaneen, D.; Raza, A. A novel CovidDetNet deep learning model for effective COVID-19 infection detection using chest radiograph images. Appl. Sci. 2022, 12, 6269. [Google Scholar] [CrossRef]
- Zouch, W.; Sagga, D.; Echtioui, A.; Khemakhem, R.; Ghorbel, M.; Mhiri, C.; Hamida, A.B. Detection of COVID-19 from CT and Chest X-ray Images Using Deep Learning Models. Ann. Biomed. Eng. 2022, 50, 825–835. [Google Scholar] [CrossRef]
- Ayalew, A.M.; Salau, A.O.; Tamyalew, Y.; Abeje, B.T.; Woreta, N. X-Ray image-based COVID-19 detection using deep learning. Multimed. Tools Appl. 2023, 82, 44507–44525. [Google Scholar] [CrossRef]
- Constantinou, M.; Exarchos, T.; Vrahatis, A.G.; Vlamos, P. COVID-19 classification on chest X-ray images using deep learning methods. Int. J. Environ. Res. Public Health 2023, 20, 2035. [Google Scholar] [CrossRef]
- Patro, K.K.; Allam, J.P.; Hammad, M.; Tadeusiewicz, R.; Pławiak, P. SCovNet: A skip connection-based feature union deep learning technique with statistical approach analysis for the detection of COVID-19. Biocybern. Biomed. Eng. 2023, 43, 352–368. [Google Scholar] [CrossRef]
- Zhu, H.; Zhu, Z.; Wang, S.; Zhang, Y. CovC-ReDRNet: A Deep Learning Model for COVID-19 Classification. Mach. Learn. Knowl. Extr. 2023, 5, 684–712. [Google Scholar] [CrossRef]
- Chakraborty, G.S.; Batra, S.; Singh, A.; Muhammad, G.; Torres, V.Y.; Mahajan, M. A Novel Deep Learning-Based Classification Framework for COVID-19 Assisted with Weighted Average Ensemble Modeling. Diagnostics 2023, 13, 1806. [Google Scholar] [CrossRef]
- Gaur, L.; Bhatia, U.; Jhanjhi, N.; Muhammad, G.; Masud, M. Medical image-based detection of COVID-19 using Deep Convolution Neural Networks. Multimed. Syst. 2023, 29, 1729–1738. [Google Scholar] [CrossRef]
- Kathamuthu, N.D.; Subramaniam, S.; Le, Q.H.; Muthusamy, S.; Panchal, H.; Sundararajan, S.C.M.; Alrubaie, A.J.; Zahra, M.M.A. A deep transfer learning-based convolution neural network model for COVID-19 detection using computed tomography scan images for medical applications. Adv. Eng. Softw. 2023, 175, 103317. [Google Scholar] [CrossRef]
- Youk, J.; Kim, T.; Evans, K.V.; Jeong, Y.-I.; Hur, Y.; Hong, S.P.; Kim, J.H.; Yi, K.; Kim, S.Y.; Na, K.J. Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell 2020, 27, 905–919.e10. [Google Scholar] [CrossRef]
- Clark, A. Pillow (pil fork) documentation, readthedocs. 2015.
- Alghamdi, S.; Turki, T. A novel interpretable deep transfer learning combining diverse learnable parameters for improved T2D prediction based on single-cell gene regulatory networks. Sci. Rep. 2024, 14, 4491. [Google Scholar] [CrossRef]
- Turki, T.; Wei, Z. Boosting support vector machines for cancer discrimination tasks. Comput. Biol. Med. 2018, 101, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Fatica, M. CUDA toolkit and libraries. In Proceedings of the 2008 IEEE Hot Chips 20 Symposium (HCS), Stanford, CA, USA, 24–26 August 2008; pp. 1–22. [Google Scholar]
- Chetlur, S.; Woolley, C.; Vandermersch, P.; Cohen, J.; Tran, J.; Catanzaro, B.; Shelhamer, E. cuDNN: Efficient primitives for deep learning. arXiv 2014, arXiv:1410.0759. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M. TensorFlow: A system for Large-Scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Granger, B.E.; Pérez, F. Jupyter: Thinking and storytelling with code and data. Comput. Sci. Eng. 2021, 23, 7–14. [Google Scholar] [CrossRef]
- Perez, F.; Granger, B.E. Project Jupyter: Computational narratives as the engine of collaborative data science. Retrieved Sept. 2015, 11, 108. [Google Scholar]
- McKinney, W. Python for Data Analysis; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2022. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wickham, H.; Chang, W.; Wickham, M.H. Package ‘ggplot2’. Create elegant data visualisations using the grammar of graphics. Version 2016, 2, 1–189. [Google Scholar]
- Suganyadevi, S.; Pershiya, A.S.; Balasamy, K.; Seethalakshmi, V.; Bala, S.; Arora, K. Deep Learning Based Alzheimer Disease Diagnosis: A Comprehensive Review. SN Comput. Sci. 2024, 5, 391. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).