Hypergraph-Regularized Lp Smooth Nonnegative Matrix Factorization for Data Representation
Abstract
1. Introduction
1.1. Problem Statement
1.2. Research Contribution
2. Related Work
2.1. Nonnegative Matrix Factorization
2.2. Graph Regularization Smooth Nonnegative Matrix Factorization
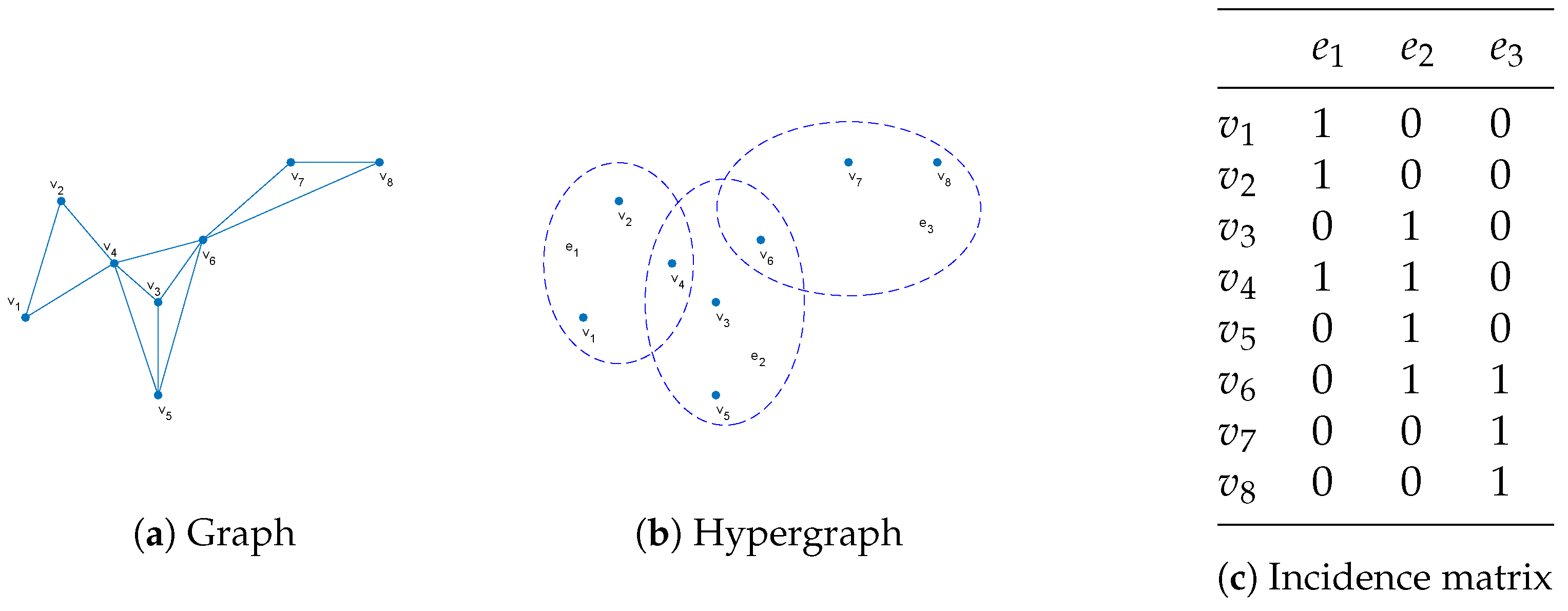
2.3. Hypergraph Learning
3. Hypergraph-Regularized Smooth Nonnegative Matrix Factorization
3.1. The Objective Function
3.2. Optimization Method
3.3. Convergence Analysis
| Algorithm 1 HGSNMF algorithm. |
| Input: Data matrix . The number of neighbors k. The algorithm |
| parameters r, p and regularization parameters , . The stopping criterion , and the maximum |
| number of iterations maxiter. Let . |
| Output: Factors and ; |
| 1: Initialize and ; |
| 2: Construct the weight matrix using (4), and calculate the |
| matrix , ; |
| 3: for maxiter do |
| 4: Update and Update according to (8), (9), respectively. |
| 5: Compute the objective function value of (5) to denote . |
| 6: if |
| Break and return . |
| 7: end if |
| 8: end for |
3.4. Computational Complexity Analysis
4. Numerical Experimentation
4.1. Data Sets
4.2. Evaluation Metrics
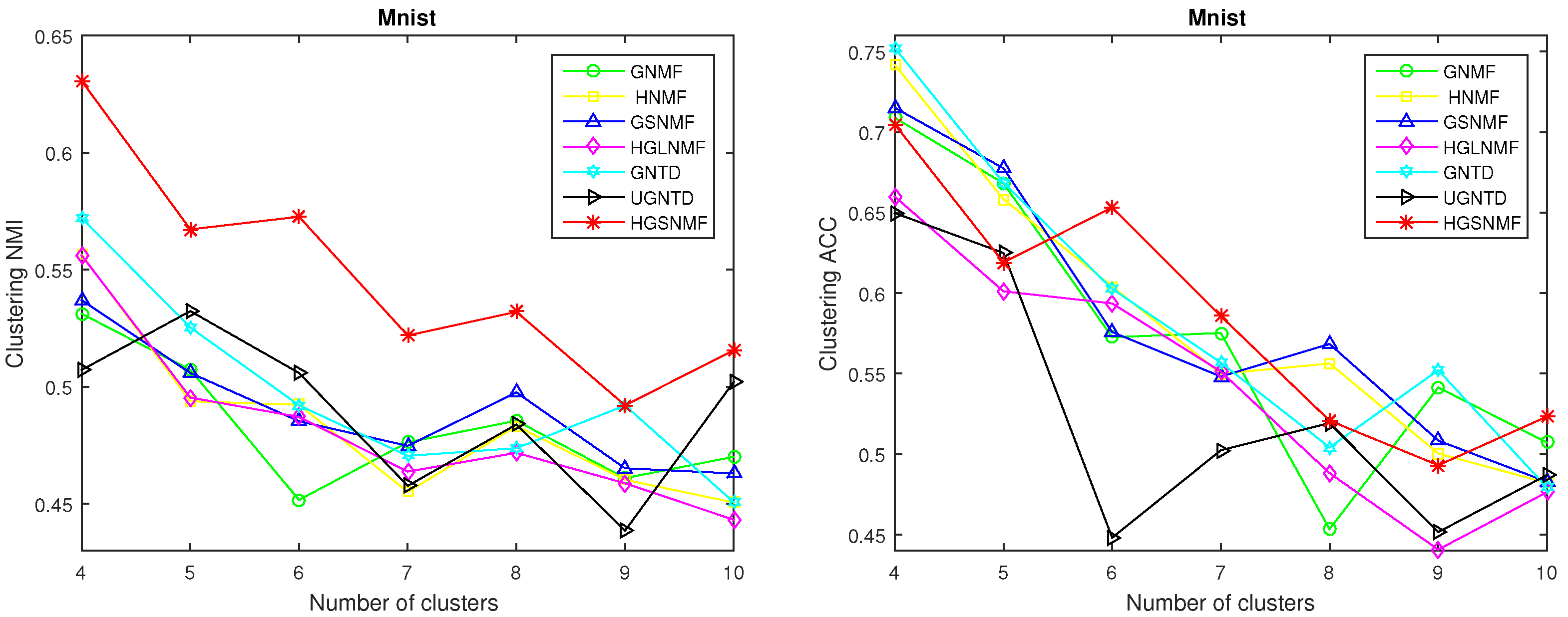
4.3. Performance Evaluations and Comparisons
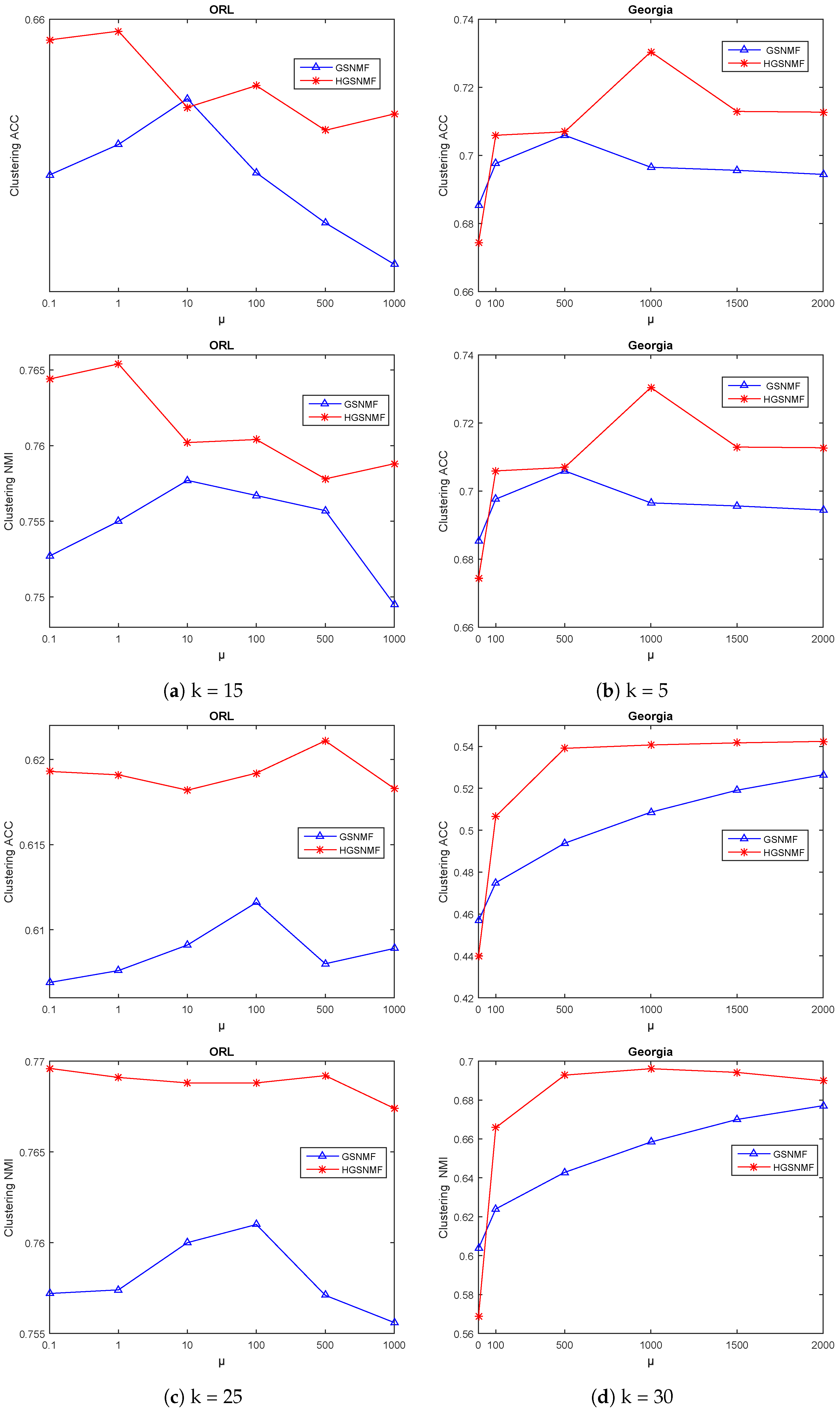
4.4. Parameters Selection
4.5. The Converage Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PCA | Principal component analysis |
| LDA | Linear discriminant analysis |
| SVD | Singular value decomposition |
| NMF | Nonnegative matrix factorization |
| HU | Hyperspectral unmixing |
| ONMF | Orthogonal nonnegative matrix tri-factorizators |
| GNMF | Graph regularized nonnegative matrix factorization |
| DNMF | Graph dual regularization nonnegative matrix factorization |
| HNMF | Hypergraph regularized nonnegative matrix factorization |
| GSNMF | Graph regularized smooth nonnegative matrix factorization |
| HGLNMF | Hypergraph regularized sparse nonnegative matrix factorization |
| MHGNMF | Nonnegative matrix factorization with mixed hypergraph regularization |
| DHPS-NMF | Dual hypergraph regularized partially shared nonnegative matrix factorization |
| HGSNMF | Hypergraph regularized smooth nonnegative matrix factorization |
| ACC | Accuracy |
| NMI | Normalized mutual information |
| MI | Mutual information |
References
- Pham, N.; Pham, L.; Nguyen, T. A new cluster tendency assessment method for fuzzy co-clustering in hyperspectral image analysis. Neurocomputing 2018, 307, 213–226. [Google Scholar] [CrossRef]
- Cai, D.; He, X.; Han, J.; Huang, T. Graph regularized nonnegative matrix factorization for data representation. IEEE Trans. Pattern Anal. Match.Intell. 2011, 33, 1548–1560. [Google Scholar]
- Li, S.; Hou, X.; Zhang, H.; Cheng, Q. Learning spatially localized, parts-based representation. In Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR 2001), Kauai, HI, USA, 8–14 December 2011; Volume 1, pp. 207–212. [Google Scholar]
- He, X.; Yan, S.; Hu, Y.; Niyogi, P.; Zhang, H. Face recognition using laplacian faces. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 328–340. [Google Scholar]
- Liu, H.; Wu, Z.; Li, X.; Cai, D.; Huang, T. Constrained nonnegative matrix factorization for image representation. IEEE Trans. Pattern Anal. Mach.Intell. 2011, 34, 1299–1311. [Google Scholar] [CrossRef] [PubMed]
- Cutler, A.; Cutler, D.; Stevens, J. Random forests. In Ensemble Machine Learning: Methods and Applications; Springer: Berlin, Germany, 2012; pp. 157–175. [Google Scholar]
- Riedmiller, M.; Lernen, A. Multi layer perceptron. In Machine Learning Lab Special Lecture; University of Freiburg: Freiburg, Germany, 2014; pp. 7–24. [Google Scholar]
- Wu, L.; Cui, P.; Pei, J. Graph Neural Networks; Springer: Singapore, 2022. [Google Scholar]
- Feng, Y.; You, H.; Zhang, Z.; Ji, R.; Gao, Y. Hypergraph neural networks. In Proceedings of the AAAI Conference on Artificial Intelligence, Hilton, HI, USA, 27 January–1 February 2019; Volume 33, pp. 3558–3565. [Google Scholar]
- Kirby, M.; Sirovich, L. Application of the karhunen loeve procedure for the characterization of human faces. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 103–108. [Google Scholar] [CrossRef]
- Strang, G. Introduction to Linear Algebra; Wellesley-Cambridge: Wellesley, MA, USA, 2009. [Google Scholar]
- Martinez, A.; Kak, A. Pca versus lda. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 228–233. [Google Scholar] [CrossRef]
- Lee, D.; Seung, H. Learning of the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Lee, D.; Seung, H. Algorithms for nonnegative matrix factorization. In Proceedings of the International Conference on Neural Information Processing Systems, Denver, CO, USA, 28–30 November 2000; Volume 13, pp. 556–562. [Google Scholar]
- Tucker, L. Some mathematical notes on three-mode factor analysis. Psychometrika 1966, 31, 279–311. [Google Scholar] [CrossRef]
- Kim, Y.; Choi, S. Nonnegative Tucker decomposition. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007; pp. 1–8. [Google Scholar]
- Kolda, T.; Bader, B. Tensor decompositions and applications. SIAM Rev. 2009, 51, 455–500. [Google Scholar] [CrossRef]
- Che, M.; Wei, Y.; Yan, H. An efficient randomized algorithm for computing the approximate tucker decomposition. J. Sci. Comput. 2021, 88, 1–29. [Google Scholar] [CrossRef]
- Pan, J.; Ng, M.; Liu, Y.; Zhang, X.; Yan, H. Orthogonal nonnegative Tucker decomposition. SIAM J. Sci. Comput. 2021, 43, B55–B81. [Google Scholar] [CrossRef]
- Ding, C.; He, X.; Simon, H. On the equivalence of nonnegative matrix factorization and spectral clustering. In Proceedings of the 2005 SIAM International Conference on Data Mining (SDM05), Newport Beach, CA, USA, 21–23 April 2005; pp. 606–610. [Google Scholar]
- Ding, C.; Li, T.; Peng, W.; Park, H. Orthogonal nonnegative matrix tri-factorizations for clustering. In Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Philadelphia, PA, USA, 20–23 August 2006; pp. 126–135. [Google Scholar]
- Pan, J.; Ng, M. Orthogonal nonnegative matrix factorization by sparsity and nuclear norm optimization. SIAM. J. Matrix Anal. Appl. 2018, 39, 856–875. [Google Scholar] [CrossRef]
- Guillamet, D.; Vitria, J.; Schiele, B. Introducing a weighted nonnegative matrix factorization for image classification. Pattern Recognit. Lett. 2003, 24, 2447–2454. [Google Scholar] [CrossRef]
- Tan, X.; Triggs, B. Enhanced local texture feature sets for face recognition under difficult lighting conditions. IEEE Trans. Image Process. 2010, 19, 1635–1650. [Google Scholar]
- Pauca, V.; Shahnaz, F.; Berry, M.; Plemmons, R. Text mining using nonnegative matrix factorizations. SIAM. Int. Conf. Data Min. 2004, 4, 452–456. [Google Scholar]
- Li, T.; Ding, C. The relationships among various nonnegative matrix factorization methods for clustering. IEEE. Comput. Soci. 2006, 4, 362–371. [Google Scholar]
- Liu, Y.; Jing, L.; Ng, M. Robust and non-negative collective matrix factorization for text-to-image transfer learning. IEEE Trans. Image Process. 2015, 24, 4701–4714. [Google Scholar]
- Gillis, N. Sparse and unique nonnegative matrix factorization through data preprocessing. J. Mach. Learn. Res. 2012, 1, 3349–3386. [Google Scholar]
- Gillis, N. Nonnegative Matrix Factorization; SIAM: Philadelphia, PA, USA, 2020. [Google Scholar]
- Wang, W.; Qian, Y.; Tan, Y. Hypergraph-regularized spares NMF for hyperspectral unmixing. IEEE J. Sel. Topi. Appl. Earth. Obs. Remot Sens. 2016, 9, 681–694. [Google Scholar] [CrossRef]
- Ma, Y.; Li, C.; Mei, X.; Liu, C.; Ma, J. Robust sparse hyperspectral unmixing withL2,1 norm. IEEE Trans. Geosci. Remot Sens. 2017, 55, 1227–1239. [Google Scholar] [CrossRef]
- Li, Z.; Liu, J.; Lu, H. Structure preserving non-negative matrix factorization for dimensionality reduction. Comput. Vis. Image Underst. 2013, 117, 1175–1189. [Google Scholar] [CrossRef]
- Luo, X.; Zhou, M.; Leung, H.; Xia, Y.; Zhu, Q.; You, Z.; Li, S. An incremental-and-static-combined scheme for matrix-factorization- based collaborative filtering. IEEE Trans. Autom. Sci. Eng. 2014, 13, 333–343. [Google Scholar] [CrossRef]
- Zhou, G.; Yang, Z.; Xie, S.; Yang, J. Online blind source separation using incremental nonnegative matrix factorization with volume constraint. IEEE Trans. Neur. Netw. 2011, 22, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Gillis, N. Generalized separable nonnegative matrix factorization. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 1546–1561. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Jiao, L.; Wang, F. Graph dual regularization nonnegative matrix factorization for co-clustering. Pattern Recognit. 2012, 45, 2237–2250. [Google Scholar] [CrossRef]
- Zeng, K.; Jun, Y.; Wang, C.; You, J.; Jin, T. Image clustering by hypergraph regularized nonnegatve matrix factorization. Neurocomputing 2014, 138, 209–217. [Google Scholar] [CrossRef]
- Leng, C.; Zhang, H.; Cai, G.; Cheng, I.; Basu, A. Graph regularized Lp smooth nonnegative matrix factorization for data representation. IEEE/CAA J. Autom. 2019, 6, 584–595. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhou, G.; Zhang, Y.; Xie, S. Graph regularized nonnegative tucker decomposition for tensor data representation. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 8613–8617. [Google Scholar]
- Qiu, Y.; Zhou, G.; Wang, Y.; Zhang, Y.; Xie, S. A generalized graph regularized non-negative Tucker decomposition framework for tensor data representation. IEEE Trans. Cybern. 2022, 52, 594–607. [Google Scholar] [CrossRef]
- Wood, G.; Jennings, L. On the use of spline functions for data smoothing. J. Biomech. 1979, 12, 477–479. [Google Scholar] [CrossRef]
- Lyons, J. Differentiation of solutions of nonlocal boundary value problems with respect to boundary data. Electron. J. Qual. Theory Differ. Equ. 2001, 51, 1–11. [Google Scholar] [CrossRef]
- Xu, L. Data smoothing regularization, multi-sets-learning, and problem solving strategies. Neural Netw. 2003, 16, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Huang, J.; Scholkopf, B. Learning with Hypergraphs: Clustering, Classification, and Embdding; MIT Press: Cambridge, MA, USA, 2006; Volume 19, pp. 1601–1608. [Google Scholar]
- Gao, Y.; Zhang, Z.; Lin, H.; Zhao, X.; Du, S.; Zou, C. Hypergraph learning: Methods and practices. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 44, 2548–2566. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Liu, Q.; Lv, F.; Gong, Y.; Metaxax, D. Unsupervised image categorization by hypergraph partition. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 17–24. [Google Scholar]
- Yu, J.; Tao, D.; Wang, M. Adaptive hypergraph learning and its application in image classification. IEEE Trans. Image Process. 2012, 21, 3262–3272. [Google Scholar] [PubMed]
- Hong, C.; Yu, J.; Li, J.; Chen, X. Multi-view hypergraph learning by patch alignment framework. Neurocomputing 2013, 118, 79–86. [Google Scholar] [CrossRef]
- Wang, C.; Yu, J.; Tao, D. High-level attributes modeling for indoor scenes classifiation. Neurocomputing 2013, 121, 337–343. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y. A survey on hyperlink prediction. arXiv 2022, arXiv:2207.02911. [Google Scholar]
- Yin, W.; Qu, Y.; Ma, Z.; Liu, Q. Hyperntf: A hypergraph regularized nonnegative tensor factorization for dimensionality reduction. Neurocomputing 2022, 512, 190–202. [Google Scholar] [CrossRef]
- Wu, W.; Kwong, S.; Zhou, Y. Nonnegative matrix factorization with mixed hypergraph regularization for community detection. Inf. Sci. 2018, 435, 263–281. [Google Scholar] [CrossRef]
- Zhang, D. Semi-supervised multi-view clustering with dual hypergraph regularized partially shared nonnegative matrix factorization. Sci. China Technol. Sci. 2022, 65, 1349–1365. [Google Scholar] [CrossRef]
- Huang, H.; Zhou, G.; Liang, N.; Zhao, Q.; Xie, S. Diverse deep matrix factorization with hypergraph regularization for multiview data representation. IEEE/CAA J. Autom. Sin. 2022, 34, 1–44. [Google Scholar] [CrossRef]
- Cai, D.; He, X.; Han, J. Documen clustering using locality preserving indexing. IEEE Trans. Knowl. Data Eng. 2005, 17, 1624–1637. [Google Scholar] [CrossRef]
- Lovasz, L.; Plummer, M. Matching Theory; American Mathematical Society: Providence, RI, USA, 2009; Volume 367. [Google Scholar]






| fladd | flmlt | fldiv | Overall | |
|---|---|---|---|---|
| NMF | 2+2 | |||
| GNMF | 2+2 | 2+2 | ||
| HNMF | 2+2 | 2+2 | ||
| GSNMF | 2+2 | 2+2 | ||
| HGLNMF | 2+2 | 2+2 | ||
| HGSNMF | 2+2 | 2+2 |
| Data Sets | Samples | Features | Classes |
|---|---|---|---|
| COIL20 | 1440 | 1024 | 20 |
| YALE | 165 | 1024 | 15 |
| ORL | 400 | 1024 | 40 |
| Georgia | 750 | 1024 | 50 |
| Mnist | 500 | 784 | 10 |
| k | K-Means | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|---|
| 4 | 65.13 ± 16.60 | 69.63 ± 15.76 | 72.86 ± 14.86 | 79.53 ± 13.65 | 73.79 ± 13.51 | 79.52 ± 13.64 | |
| 6 | 67.70 ± 9.79 | 69.65 ± 11.27 | 72.79 ± 10.70 | 80.72 ± 10.58 | 68.63 ± 10.00 | 80.76 ± 10.54 | |
| 8 | 70.56 ± 5.89 | 69.44 ± 8.78 | 71.53 ± 8.60 | 80.94 ± 7.35 | 73.55 ± 7.28 | 81.08 ± 7.38 | |
| 10 | 76.02 ± 7.12 | 70.13 ± 6.95 | 76.01 ± 5.48 | 82.99 ± 5.84 | 76.36 ± 5.92 | 83.00 ± 5.75 | |
| 12 | 73.21 ± 4.82 | 70.91 ± 5.50 | 77.12 ± 5.68 | 82.16 ± 5.03 | 75.70 ± 5.33 | 82.31 ± 5.08 | |
| 14 | 74.10 ± 4.31 | 70.41 ± 4.66 | 77.06 ± 4.75 | 82.20 ± 4.24 | 76.88 ± 4.63 | 82.21 ± 4.19 | |
| 16 | 74.85 ± 3.65 | 72.70 ± 4.04 | 79.38 ± 4.52 | 84.05 ± 3.95 | 79.04 ± 4.27 | 83.99 ± 3.97 | |
| 18 | 73.28 ± 3.08 | 71.49 ± 3.00 | 78.03 ± 3.65 | 84.61 ± 3.15 | 79.36 ± 3.47 | 84.70 ± 3.16 | |
| 20 | 73.83 ± 2.52 | 71.95 ± 2.76 | 79.20 ± 3.05 | 84.08 ± 2.79 | 78.84 ± 2.76 | 84.05 ± 2.71 | |
| Avg. | 72.0 | 71.70 | 76.00 | 82.36 | 75.80 | 82.40 |
| k | K-Means | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|---|
| 4 | 71.45 ± 14.82 | 72.35 ± 16.50 | 73.41 ± 14.85 | 75.78 ± 17.88 | 75.33 ± 15.74 | 75.77 ± 17.88 | |
| 6 | 67.05 ± 10.72 | 68.87 ± 11.80 | 69.65 ± 12.61 | 74.42 ± 13.78 | 68.04 ± 10.78 | 74.44 ± 13.78 | |
| 8 | 64.56 ± 64.56 | 65.39 ± 9.95 | 64.35 ± 10.93 | 71.06 ± 11.77 | 67.36 ± 9.73 | 70.71 ± 11.75 | |
| 10 | 67.38 ± 9.76 | 63.27 ± 7.96 | 66.76 ± 7.47 | 70.73 ± 10.04 | 67.07 ± 8.44 | 70.61 ± 5.75 | |
| 12 | 63.73 ± 7.09 | 63.19 ± 7.02 | 66.81 ± 8.30 | 68.63 ± 8.43 | 65.81 ± 8.43 | 69.00 ± 5.08 | |
| 14 | 62.48 ± 6.42 | 60.01 ± 6.16 | 65.18 ± 7.57 | 68.04 ± 8.33 | 64.21 ± 7.77 | 68.03 ± 8.18 | |
| 16 | 61.78 ± 5.69 | 62.18 ± 6.43 | 65.47 ± 7.25 | 69.09 ± 7.62 | 66.03 ± 7.50 | 68.85 ± 7.66 | |
| 18 | 59.15 ± 6.18 | 59.68 ± 5.44 | 63.39 ± 6.86 | 69.29 ± 6.51 | 65.84 ± 6.70 | 69.65 ± 6.59 | |
| 20 | 58.18 ± 5.43 | 59.11 ± 4.60 | 63.86 ± 6.24 | 63.95 ± 6.00 | 68.56 ± 6.34 | 68.19 ± 6.83 | |
| Avg. | 63.97 | 63.78 | 66.54 | 70.64 | 67.07 | 70.63 |
| k | K-Means | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|---|
| 3 | 40.18 ± 23.03 | 28.96 ± 11.71 | 28.81 ± 12.06 | 36.08 ± 12.82 | 37.80 ± 16.40 | 36.12 ± 12.89 | |
| 5 | 35.72 ± 12.87 | 38.23 ± 10.25 | 38.48 ± 10.02 | 39.37 ± 8.83 | 40.01 ± 10.61 | 39.35 ± 8.85 | |
| 7 | 38.38 ± 6.51 | 38.17 ± 7.57 | 39.07 ± 6.84 | 39.33 ± 6.46 | 39.38 ± 5.23 | 42.32 ± 7.39 | |
| 9 | 41.80 ± 3.87 | 40.56 ± 5.00 | 38.93 ± 5.05 | 38.85 ± 4.88 | 39.18 ± 5.23 | 40.21 ± 4.35 | |
| 11 | 39.80 ± 4.40 | 41.82 ± 4.45 | 42.05 ± 4.25 | 43.88 ± 4.30 | 44.08 ± 4.62 | 44.04 ± 4.61 | |
| 13 | 44.13 ± 4.63 | 44.17 ± 3.03 | 44.59 ± 3.43 | 44.24 ± 3.12 | 44.53 ± 2.80 | 44.29 ± 3.07 | |
| 14 | 44.34 ± 3.84 | 43.27 ± 2.92 | 43.31 ± 3.10 | 44.21 ± 3.39 | 44.82 ± 3.02 | 44.21 ± 3.32 | |
| 15 | 44.48 ± 2.92 | 43.91 ± 2.90 | 44.36 ± 2.72 | 45.37 ± 2.38 | 45.32 ± 2.62 | 45.47 ± 2.35 | |
| Avg. | 41.76 | 40.07 | 40.04 | 41.40 | 41.84 | 41.51 |
| k | K-Means | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|---|
| 3 | 62.24 ± 14.91 | 59.30 ± 8.16 | 59.12 ± 8.11 | 61.49 ± 8.20 | 62.64 ± 11.14 | 61.91 ± 8.61 | |
| 5 | 50.24 ± 10.66 | 54.15 ± 8.92 | 53.78 ± 8.27 | 54.95 ± 7.83 | 54.82 ± 9.04 | 54.84 ± 7.82 | |
| 7 | 49.43 ± 6.26 | 47.40 ± 6.33 | 46.92 ± 6.90 | 47.21 ± 6.81 | 46.65 ± 6.14 | 47.44 ± 6.62 | |
| 9 | 44.40 ± 7.02 | 43.12 ± 4.87 | 42.48 ± 5.45 | 42.15 ± 4.74 | 42.81 ± 5.49 | 43.68 ± 4.98 | |
| 11 | 39.12 ± 4.80 | 41.37 ± 5.06 | 41.74 ± 4.65 | 43.43 ± 5.00 | 43.07 ± 5.29 | 43.50 ± 5.12 | |
| 13 | 40.41 ± 4.81 | 41.18 ± 3.73 | 41.11 ± 4.10 | 40.76 ± 3.83 | 41.05 ± 3.61 | 40.78 ± 3.91 | |
| 14 | 39.46 ± 4.26 | 39.05 ± 4.21 | 38.27 ± 3.72 | 39.92 ± 4.21 | 40.03 ± 3.75 | 39.94 ± 4.22 | |
| 15 | 38.52 ± 3.30 | 38.72 ± 3.51 | 38.67 ± 3.21 | 40.25 ± 3.39 | 39.52 ± 3.14 | 40.44 ± 3.24 | |
| Avg. | 45.41 | 45.76 | 45.34 | 46.31 | 46.24 | 46.46 |
| k | K-Means | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|---|
| 5 | 66.24 ± 12.00 | 67.18 ± 12.07 | 68.81 ± 11.16 | 68.58 ± 12.77 | 66.51 ± 13.18 | 68.97 ± 13.01 | |
| 10 | 70.22 ± 6.63 | 73.59 ± 5.90 | 72.11 ± 6.70 | 71.95 ± 5.92 | 72.39 ± 6.59 | 73.29 ± 6.55 | |
| 15 | 68.46 ± 4.21 | 75.23 ± 5.01 | 76.14 ± 5.18 | 75.26 ± 5.62 | 75.67 ± 5.27 | 75.26 ± 5.62 | |
| 20 | 69.87 ± 4.75 | 74.21 ± 4.34 | 74.44 ± 4.78 | 75.24 ± 4.25 | 75.49 ± 3.76 | 75.46 ± 4.19 | |
| 25 | 71.13 ± 3.48 | 75.51 ± 2.69 | 75.88 ± 3.13 | 76.03 ± 3.29 | 76.10 ± 3.17 | 76.06 ± 3.12 | |
| 30 | 71.03 ± 2.81 | 75.34 ± 3.12 | 75.55 ± 2.81 | 74.60 ± 2.67 | 74.69 ± 2.65 | 75.88 ± 2.80 | |
| 35 | 71.07 ± 1.82 | 75.07 ± 2.23 | 74.96 ± 2.06 | 74.46 ± 1.87 | 75.85 ± 2.18 | 74.52 ± 1.91 | |
| 40 | 71.45 ± 2.06 | 75.05 ± 1.90 | 75.26 ± 1.82 | 74.54 ± 1.87 | 75.40 ± 1.91 | 74.54 ± 1.91 | |
| Avg. | 69.93 | 73.90 | 74.35 | 73.85 | 74.11 | 73.99 |
| k | K-Means | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|---|
| 5 | 67.32 ± 14.91 | 68.12 ± 12.11 | 68.76 ± 12.31 | 68.70 ± 12.27 | 67.36 ± 12.72 | 68.97 ± 13.01 | |
| 10 | 62.72 ± 10.66 | 65.85 ± 9.86 | 64.05 ± 7.87 | 64.59 ± 7.22 | 64.46 ± 7.82 | 65.42 ± 7.50 | |
| 15 | 56.19 ± 5.80 | 63.55 ± 6.85 | 64.84 ± 6.96 | 63.99 ± 7.44 | 64.59 ± 7.32 | 63.99 ± 7.44 | |
| 20 | 55.29 ± 6.25 | 61.21 ± 5.83 | 61.56 ± 6.21 | 62.44 ± 5.87 | 62.67 ± 5.30 | 62.52 ± 5.57 | |
| 25 | 54.15 ± 4.50 | 60.58 ± 3.98 | 61.01 ± 4.84 | 61.31 ± 4.66 | 61.16 ± 4.96 | 61.32 ± 4.80 | |
| 30 | 52.52 ± 4.29 | 58.57 ± 4.67 | 58.88 ± 4.52 | 58.07 ± 4.42 | 58.38 ± 4.28 | 59.50 ± 4.20 | |
| 35 | 51.30 ± 3.20 | 57.83 ± 3.62 | 57.22 ± 3.46 | 56.95 ± 3.28 | 57.05 ± 3.21 | 58.35 ± 4.06 | |
| 40 | 50.68 ± 3.43 | 56.57 ± 3.39 | 56.73 ± 3.15 | 55.88 ± 3.32 | 57.20 ± 3.46 | 55.88 ± 3.37 | |
| Avg. | 56.27 | 61.57 | 61.86 | 61.42 | 62.03 | 61.57 |
| k | K-Means | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|---|
| 5 | 67.05 ± 11.33 | 59.40 ± 11.79 | 63.00 ± 12.27 | 60.93 ± 11.93 | 64.46 ± 10.83 | 60.99 ± 11.94 | |
| 10 | 60.25 ± 8.93 | 61.24 ± 8.51 | 57.48 ± 10.50 | 61.59 ± 10.12 | 57.63 ± 10.67 | 65.91 ± 9.15 | |
| 15 | 64.64 ± 5.32 | 60.57 ± 4.39 | 62.46 ± 4.85 | 61.89 ± 5.12 | 64.02 ± 5.72 | 61.99 ± 5.02 | |
| 20 | 67.12 ± 4.30 | 60.60 ± 3.71 | 62.58 ± 3.91 | 60.98 ± 3.81 | 64.44 ± 3.18 | 61.08 ± 3.82 | |
| 25 | 66.30 ± 3.31 | 59.31 ± 2.73 | 61.35 ± 2.62 | 61.33 ± 3.22 | 64.83 ± 2.98 | 61.32 ± 3.68 | |
| 30 | 66.01 ± 3.13 | 60.26 ± 2.56 | 63.20 ± 2.25 | 60.52 ± 3.04 | 64.61 ± 2.97 | 60.47 ± 2.99 | |
| 35 | 65.10 ± 2.13 | 59.93 ± 2.33 | 63.3 ± 1.80 | 59.21 ± 2.21 | 63.27 ± 2.37 | 59.20 ± 2.22 | |
| 40 | 66.06 ± 2.20 | 59.58 ± 2.34 | 62.84 ± 1.82 | 58.61 ± 2.38 | 63.57 ± 1.98 | 58.62 ± 2.48 | |
| 45 | 66.17 ± 1.35 | 59.99 ± 1.75 | 62.92 ± 1.55 | 58.22 ± 1.90 | 62.92 ± 1.66 | 58.25 ± 1.99 | |
| 50 | 66.36 ± 1.32 | 59.05 ± 1.56 | 62.11 ± 1.51 | 58.19 ± 1.33 | 63.18 ± 1.24 | 58.19 ± 1.25 | |
| Avg. | 66.26 | 59.90 | 62.50 | 59.74 | 63.69 | 59.77 |
| k | K-Means | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|---|
| 5 | 68.73 ± 11.50 | 66.68 ± 10.96 | 69.52 ± 11.15 | 68.00 ± 10.33 | 69.76 ± 10.50 | 67.68 ± 10.47 | |
| 10 | 61.71 ± 8.56 | 57.79 ± 8.48 | 59.25 ± 8.44 | 55.73 ± 9.09 | 59.07 ± 8.94 | 55.83 ± 9.23 | |
| 15 | 55.38 ± 6.27 | 53.33 ± 4.41 | 55.05 ± 5.38 | 55.07 ± 5.41 | 56.08 ± 6.15 | 55.21 ± 5.38 | |
| 20 | 55.14 ± 5.07 | 50.55 ± 4.58 | 52.30 ± 4.60 | 50.22 ± 4.45 | 54.93 ± 4.36 | 50.37 ± 4.40 | |
| 25 | 51.82 ± 4.40 | 46.8¡¤ ± 3.59 | 48.50 ± 3.42 | 48.25 ± 4.38 | 52.10 ± 4.09 | 48.311 ± 4.30 | |
| 30 | 49.67 ± 4.12 | 45.20 ± 3.49 | 48.31 ± 3.13 | 45.82 ± 3.48 | 50.17 ± 3.82 | 45.68 ± 3.33 | |
| 35 | 47.80 ± 3.28 | 43.78 ± 3.12 | 47.09 ± 2.92 | 42.57 ± 3.03 | 47.19 ± 3.31 | 42.53 ± 3.02 | |
| 40 | 47.88 ± 3.28 | 41.93 ± 2.95 | 45.35 ± 2.81 | 40.47 ± 3.26 | 46.27 ± 3.02 | 40.41 ± 3.45 | |
| 45 | 47.39 ± 2.26 | 41.07 ± 2.52 | 43.93 ± 2.44 | 38.53 ± 2.49 | 44.34 ± 2.50 | 38.58 ± 2.59 | |
| 50 | 46.18 ± 2.19 | 38.94 ± 2.22 | 42.14 ± 2.40 | 37.24 ± 1.92 | 43.78 ± 2.32 | 37.26 ± 1.90 | |
| Avg. | 53.17 | 48.61 | 51.14 | 48.19 | 52.37 | 48.19 |
| k | GNMF | HNMF | GSNMF | HGLNMF | GNTD | UGNTD | HGSNMF |
|---|---|---|---|---|---|---|---|
| 2 | 58.89 ± 23.94 | 57.08 ± 24.38 | 42.94 ± 12.91 | 57.34 ± 24.27 | 67.11 ± 18.27 | 53.17 ± 36.19 | |
| 4 | 53.12 ± 12.67 | 55.64 ± 14.51 | 53.70 ± 12.26 | 55.61 ± 14.04 | 57.17 ± 11.60 | 56.32 ± 11.42 | |
| 6 | 45.17 ± 4.90 | 49.23 ± 5.85 | 48.53 ± 6.11 | 48.72 ± 5.99 | 49.20 ± 5.01 | 56.27 ± 7.69 | |
| 7 | 47.63 ± 6.82 | 45.51 ± 4.82 | 47.48 ± 4.48 | 46.88 ± 5.46 | 47.05 ± 5.14 | 55.69 ± 7.31 | |
| 8 | 48.55 ± 3.86 | 48.32 ± 4.38 | 49.76 ± 4.87 | 47.22 ± 3.83 | 47.38 ± 3.38 | 57.30 ± 6.01 | |
| 10 | 47.01 ± 4.39 | 45.06 ± 2.21 | 46.30 ± 3.65 | 44.46 ± 2.81 | 45.07 ± 4.08 | 56.11 ± 4.73 | |
| Avg. | 50.06 | 50.14 | 48.12 | 50.04 | 52.16 | 55.81 |
| k | GNMF | HNMF | GSNMF | HGLNMF | GNTD | UGNTD | HGSNMF |
|---|---|---|---|---|---|---|---|
| 2 | 80.07 ± 28.05 | 87.96 ± 13.17 | 80.50 ± 17.55 | 88.13 ± 12.96 | 82.73 ± 17.28 | 91.57 ± 13.32 | |
| 4 | 70.89 ± 11.28 | 74.17 ± 12.67 | 71.51 ± 16.93 | 65.99 ± 24.92 | 67.74 ± 11.50 | 71.32 ± 26.19 | |
| 6 | 57.26 ± 5.48 | 60.41 ± 7.06 | 57.58 ± 7.37 | 59.36 ± 7.64 | 60.50 ± 6.74 | 44.47 ± 21.53 | |
| 7 | 57.52 ± 7.66 | 54.99 ± 5.84 | 54.82 ± 3.97 | 55.54 ± 5.69 | 55.68 ± 5.24 | 56.34 ± 7.57 | |
| 8 | 45.34 ± 2.08 | 55.63 ± 5.18 | 56.84 ± 5.27 | 49.11 ± 15.91 | 55.21 ± 4.34 | 56.78 ± 19.12 | |
| 10 | 50.72 ± 5.42 | 48.21 ± 3.68 | 48.28 ± 6.05 | 48.02 ± 4.00 | 47.93 ± 4.53 | 51.01 ± 6.48 | |
| Avg. | 60.30 | 63.56 | 61.59 | 60.86 | 60.05 | 68 |
| k | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|
| 3 | 1.15 | 2.51 | 3.14 | 4.08 | 3.15 | |
| 11 | 9.21 | 20.75 | 21.31 | 35.62 | 14.77 | |
| 13 | 17.63 | 41.12 | 42.18 | 67.89 | 28.70 | |
| 14 | 20.64 | 49.80 | 46.75 | 80.06 | 35.45 | |
| 15 | 22.52 | 55.35 | 54.18 | 96.94 | 43.62 |
| k | NMF | GNMF | HNMF | GSNMF | HGLNMF | HGSNMF |
|---|---|---|---|---|---|---|
| 10 | 25.39 | 47.79 | 35.77 | 69.7 | 45.07 | |
| 15 | 40.36 | 94.57 | 67.58 | 134.67 | 88.01 | |
| 20 | 56.96 | 123.30 | 89.73 | 183.88 | 120.07 | |
| 25 | 90.97 | 233.04 | 151.94 | 312.03 | 201.96 | |
| 30 | 114.43 | 343.62 | 196.97 | 429.94 | 214.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Lu, L.; Liu, Q.; Chen, Z. Hypergraph-Regularized Lp Smooth Nonnegative Matrix Factorization for Data Representation. Mathematics 2023, 11, 2821. https://doi.org/10.3390/math11132821
Xu Y, Lu L, Liu Q, Chen Z. Hypergraph-Regularized Lp Smooth Nonnegative Matrix Factorization for Data Representation. Mathematics. 2023; 11(13):2821. https://doi.org/10.3390/math11132821
Chicago/Turabian StyleXu, Yunxia, Linzhang Lu, Qilong Liu, and Zhen Chen. 2023. "Hypergraph-Regularized Lp Smooth Nonnegative Matrix Factorization for Data Representation" Mathematics 11, no. 13: 2821. https://doi.org/10.3390/math11132821
APA StyleXu, Y., Lu, L., Liu, Q., & Chen, Z. (2023). Hypergraph-Regularized Lp Smooth Nonnegative Matrix Factorization for Data Representation. Mathematics, 11(13), 2821. https://doi.org/10.3390/math11132821






