1. Introduction
Named entity recognition (NER) is a fundamental technology in natural language processing (NLP), with the main functions of identifying and extracting various named entities from massive unstructured text (one of the subtasks of knowledge extraction) [
1], such as person names, location names, organization names, and domain specific words. Nowadays, named entity recognition technology has been widely applied in other tasks such as machine translation [
2], information extraction [
3], intelligent Q and A [
4], and knowledge graph construction [
5]. In the natural language processing tasks, the accuracy of named entity recognition has a significant influence on the processing efficiency of subsequent tasks.
During the early medical NER tasks, the majority employed the rule-based and dictionary-based methods [
6]. The experts in the medical field constructed the rule templates and the pattern matching and string matching were applied to identify the relevant entities, such as disease names, drug names, inspection indicators, and related symptoms. Many rules required development by professional medical experts, which are high in labor cost [
7] and low in portability. At present, with the development of deep learning and increasing computing power, more and more scholars have made use of the neural network models to process medical NER tasks [
8], treating them as sequence labeling problems. Nevertheless, in the medical NER, the most important problems currently are the lack of labeled data and the existence of polysemy and entity co-reference in a lot of specialized vocabularies, resulting in low accuracy in medical vocabulary recognition and low performance in network model recognition, which hinders the development of Chinese medical entity recognition [
9].
In the medical field, taking diabetes as an example, China has become the country with the largest number of diabetes patients in the world, where the probability of adults suffering from diabetes is 11.7%, and this probability is on the rise [
10,
11]. For patients with diabetes, most of their conditions are recorded in the form of electronic documents [
12]; nonetheless, there is a lack of efficient recognition methods to accurately identify the diabetes named entities. Accordingly, how to improve the accuracy of named entity recognition in the field of diabetes medicine has become a key issue in the field of diabetes medicine.
At present, there is less research on named entities in the field of diabetes medicine, and the labeled dataset is rare; thus, this paper crawls the information of domestic public medicine websites and related encyclopedia websites, such as “XYWY.COM”, “QIUYI.CN”, and Baidu Encyclopedia to obtain a large quantity of diabetes-related data and constructs a Chinese diabetes corpus. Further, in view of the problem of complex entity naming and entity nesting phenomena in the medical field, which leads to difficulty in entity recognition—and with most current models directly feeding the generated character vector into Bidirectional Long Short-Term Memory (BiLSTM) or Bidirectional Gate Recurrent Unit (BiGRU) to obtain the global features, which gives no consideration to local optimal features—this paper proposes a Chinese medical NER task model, uniting the local context-wise module and BiGRU with self-attention mechanism while taking into account the local and global features, and constructs a deep learning model RMBC (RoBERTa Multi-scale CNN BiGRU Self-attention CRF) for diabetes entity recognition to recognize diabetes named entities, providing effective extraction of six medical entities.
2. Related Work
There are four main methods for named entity recognition research: rule-based methods, statistics-based machine learning methods, deep learning methods, and NER methods using pre-trained models.
The origins of NER were mainly based on rule-based and dictionary-based methods [
13,
14], which relied on domain experts to manually construct corresponding rule templates and use matching methods to process text. This method requires a lot of human resources and cannot be easily transferred between different domains. After that, the methods based on statistical machine learning were introduced, including Hidden Markov Model (HMM) [
15], Maximum Entropy Model (MEM) [
16], Support Vector Machine (SVM) [
17], and Conditional Random Fields (CRF) [
18]. Among them, CRF considers the global distribution of data when normalizing, fully utilizing the feature information of internal and context, and solving the label bias problem [
19]. However, the above-mentioned machine learning methods require a large quantity of manually annotated data for feature extraction, and the size of the corpus seriously affects the recognition performance [
20]. In recent years, many deep learning methods have been applied to research on named entity recognition [
21]. For example, Hammerton et al. [
22] used a unidirectional long short-term memory (LSTM) for text recognition research and achieved very good results; therefore, LSTM-CRF became the basic structure of named entity recognition. Later, Guillaume Lample et al. [
23] proposed a neural network model that combines bidirectional long short-term memory (BiLSTM) with CRF based on this model. This structure can effectively obtain the sequential information of context, and it achieved an F1 score of 90.94% on the CoNLL-2003 dataset. It has been widely used in tasks such as named entity recognition. Collobert et al. first proposed combining the convolutional neural network (CNN) with CRF [
24]. This method assigns a fixed-size window to each word, and it extracts local information more effectively, but lacks consideration of long-distance word information. Chiu and Nichols [
25] proposed a BiLSTM-CNN model that uses CNN to learn character features and completes sequence labeling tasks through BiLSTM. The F1 value of this model on the CoNLL2003 dataset reaches 91.62%. Ma et al. [
26] proposed a model based on LSTM-CNNs-CRF to address sequence labeling problems. The model combines LSTM, CNN, and CRF models to establish an end-to-end model that does not require a large amount of data or specific task knowledge. Zhu et al. [
27] proposed a named entity recognition model that combines CNN with BiGRU. In addition, the model also introduced a multi-task learning framework to simultaneously handle both entity type classification and entity boundary recognition. Experiments on multiple datasets showed that it performed better in improving accuracy than traditional named entity recognition models. Strubel et al. [
28] proposed Iterated Dilated Convolutional Neural Networks (IDCNN), which improved traditional CNN networks by introducing regularization to address overfitting caused by an increase in the number of layers in traditional CNN networks. IDCNN also significantly improves speed and performs well in named entity recognition tasks. Zhang et al. [
29] proposed a new type of Lattice LSTM that incorporates potential word information into the traditional word-based LSTM-CRF model, which avoids error propagation caused by word segmentation errors. This method achieved good results on multiple public datasets.
However, all of the aforementioned methods have a common problem, which is the inability to handle polysemous words. These methods only focus on feature extraction between words or characters, ignoring the semantic information between contexts. Therefore, the extracted static word vectors result in lower accuracy for named entity recognition. In order to address this issue, the Google team proposed a pre-processing language model called BERT [
30] in 2018, which is used to handle word embeddings. The Bert model uses bidirectional transformers as encoders, which enhances the generalization ability of the word vector model, fully describes the relationships between characters, words, and sentences, and effectively represents the semantic information between contexts. The Bert model has become the mainstream model in the field of NLP [
31,
32].
Currently, named entity recognition technology is gradually being applied in the medical field. Compared with other fields, medical entities in Chinese named entity recognition tasks are more specialized, making their recognition more challenging. Chinese medical texts usually contain a large number of medical terms, with complex word formation and nested entities, as well as fuzzy boundaries between them. Among them, a large number of entities are mixed with numbers, symbols, Chinese and English, such as GLP-1 receptor agonists, cox-2 inhibitors, DDP-4 inhibitors, etc. In addition, in the medical field, there are some problems in addition to the difficulty of named entity recognition itself, such as the diversity of descriptions of medical entities, the lack of unified rules, and the continuous emergence of new entities with the continuous development of medical technology, which makes deep learning models have poor transferability. Due to the limited availability of publicly available annotated datasets for named entities in the medical field and the high cost of manual annotation, the training data for deep learning technology in medical entity recognition is insufficient. These issues undoubtedly increase the difficulty of named entity recognition in the medical field, leading to limited performance in entity recognition in the medical field.
To solve the current problems, some scholars have conducted research on named entity recognition in the medical field. For example, Chai et al. [
33] proposed a novel biomedical named entity recognition method by combining XLNet and CRF for noise reduction and entity recognition. This method achieved an F1 score of 89.01% in the BioCreative V CDR task, outperforming other models. In the JNLPBA task, the method achieved an F1 score of 77.39%. Guo et al. [
34] proposed a method for named entity recognition of Chinese electronic medical records using multi-task learning and transfer learning. This method used a shared deep neural network to learn multiple related tasks, including disease and drug entity recognition, and used pre-training and fine-tuning for transfer learning to improve the model’s generalization ability and robustness. The method achieved excellent performance on the CEMR-NER task of the public dataset CCKS2017, with an F1 score of 87.36%. Lee et al. [
35] proposed a multi-graph neural network method with multi-embedding enhancement for Chinese medical named entity recognition. The method achieved excellent performance on the public dataset CEMR-NER, with an F1 score of 86.95%, outperforming multiple baseline models. Liang et al. [
36] proposed a transfer learning-based method that transfers the pre-trained model in the textual entailment task to the biomedical named entity recognition task to improve the performance of biomedical NER. On the BioCreative IV dataset, the authors’ method achieved the best performance, with an F1 score of 70.63%. Chen et al. [
37] proposed a knowledge-adaptive multi-path matching network based on machine reading comprehension for the biomedical named entity recognition task. The method combines medical knowledge with text features and achieves accurate entity recognition through a multi-path matching network. On the BC2GM dataset, the method achieved the best performance, with an F1 score of 87.02%. Liu et al. [
38] designed a Med-BERT pre-training framework that combines medical corpora and specific tasks related to the field to improve the model’s performance in medical named entity recognition (NER). On the i2b2-2010 dataset, Med-BERT achieved the best F1 score of 87.02%.
3. Method
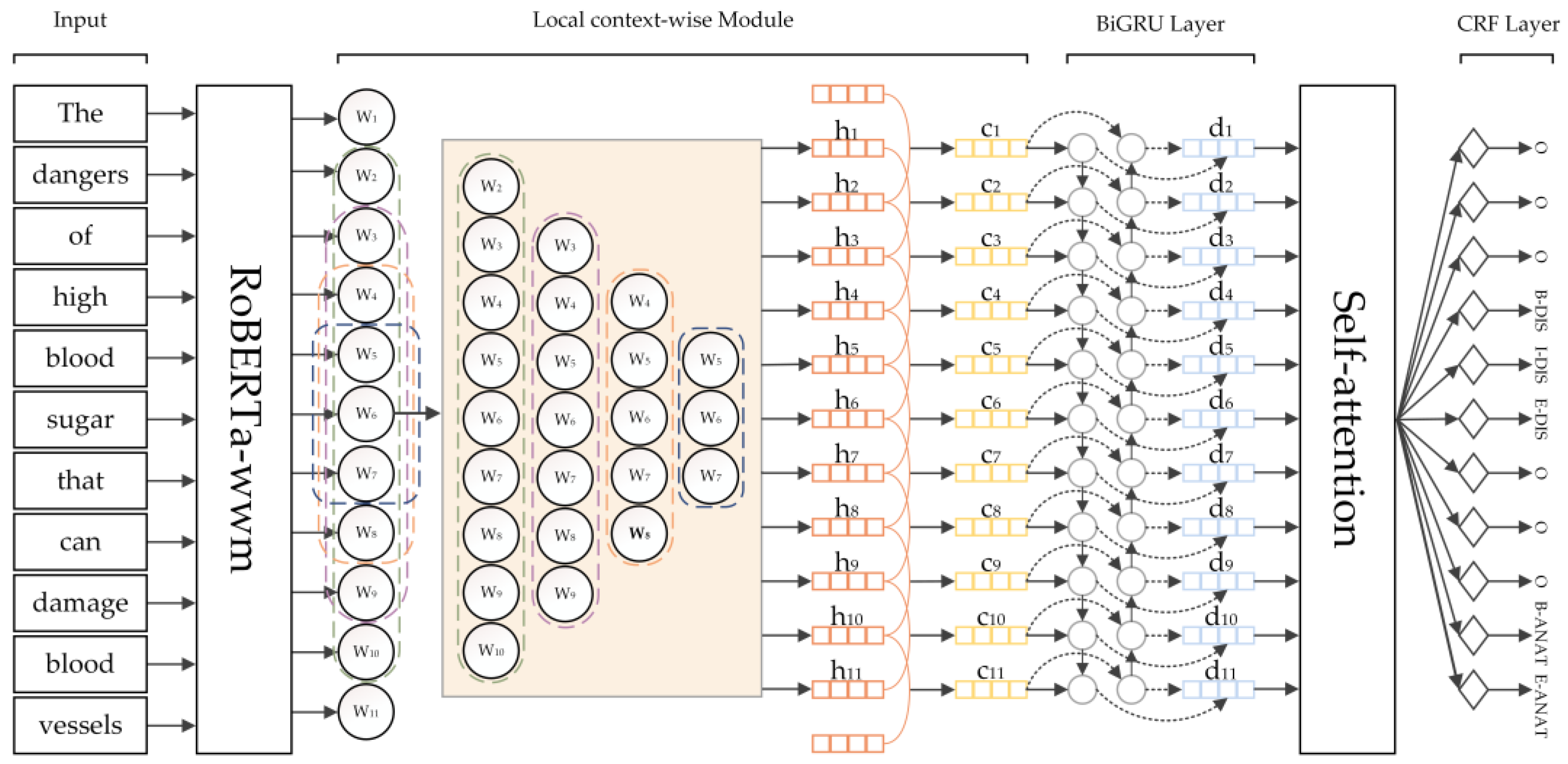
In this section, the structure of each part of the RMBC model proposed herein is introduced, including the embedding of the pre-trained model, local context-wise module, BiGRU combined self-attention layer, and CRF layer. The overall framework of the RMBC model is shown in
Figure 1; this model first employs the RoBERTa-wwm pre-trained model to extract word vectors from text data, effectively extracting the local information from text data with the help of a local context-wise module, and then introduces the self-attention mechanism BiGRU to capture the global feature information of text; finally, it inputs the CRF layer for decoding to output the tag sequence with the highest probability in order to obtain the tag category of each character.
3.1. RoBERTa-wwm Pre-Trained Language Model
Comparing ELMo [
39] and OpenAI-GPT [
40] pre-trained models, BERT is an unsupervised deep bidirectional language representation model using stack Transformer as the main architecture, which is designed through two pre-trained tasks, namely, the Masked Language Model (MLM) and Next Sentence Prediction (NSP). BERT is widely used in named entity recognition tasks for the semantic representation of pretraining.
The Joint Laboratory of HIT and iFLYTEK Research has launched the Chinese RoBERTa-wwm pre-trained language model [
41] that has been improved on the basis of RoBERTa and Chinese whole word mask technology [
42], mainly including two improvements. RoBERTa-wwm uses the dynamic mask instead of the static mask of BERT. The dynamic mask will randomly select different words for [mask] each time, which increases the randomness of model input and allows the model to learn more diverse language representations. RoBERTa-wwm adopts the whole word mask instead of the individual character mask of BERT. The whole word mask will mask the entire word rather than individual characters, which helps improve the model’s understanding of the vocabulary. As shown in
Figure 2, BERT is employed to randomly mask a single character in a sentence, while the whole word mask of RoBERTa-wwm is used to mask all characters belonging to a word. For the sentence “Patients with diabetes who experience high blood sugar levels”, the individual character mask scheme of BERT is to mask some individual characters in the word “high blood sugar levels”, such as the word “high”, and then learn the semantic representation of the character level by predicting the character to be masked. RoBERTa-wwm, on the other hand, first divides a sentence into words and then randomly masks a portion of words for prediction, such as “high blood sugar levels” and “clinical practice”; with this training method, RoBERTa-wwm can learn the semantic representation at the word level, thereby achieving the overall effect of improving the performance of the model.
3.2. Local Context-Wise Module
The convolutional neural network presents excellent performance in extracting local features; although it is primarily used for image feature extraction, more and more scholars have begun to employ CNN to solve the natural language processing problems, such as named entity recognition, in recent years.
This paper constructs a local context-wise module to extract multi-scale local features of diabetes texts. By using a multi-window attention mechanism to input character vectors, the important semantic components of local features can be effectively captured under different window sizes. The convolution layer improves the local feature awareness capability of CNN by setting multiple convolution kernels of different sizes to efficiently calculate the multiple convolution kernels in parallel, thereby fully extracting the local feature information at different scales. Next, a residual structure is leveraged to fuse the semantic information at different scales to avoid the problem of network degradation due to too deep network layers, thus improving the performance of entity recognition.
The local context-wise module encodes the character sequence output from the RoBERTa-wwm model, while implicitly grouping the related characters to capture the relevance in the local context. is utilized as the input representation for each character, and the embedded representation for the character is .
The convolution window size of CNN is set to , and each character embedding includes a position embedding that is the same as the window size . The embedded index range for this location is from 0 to , where the initial value is 1 if the current index corresponds to the location of the corresponding character in the window, otherwise it is 0; in this way, CNN can encode the location information of each character in the context into its embedding vector, thereby capturing the sequence dependency of the characters in the sequence. The embedding dimension is . In order to capture the semantic relationship between the central character and the surrounding characters, a method of combining CNN with the multi-window attention mechanism under different convolutional window sizes is applied. This method can effectively focus on the local context of each character and strengthen the semantic connection between the central character and its surrounding characters.
In the multi-window attention layer, the central character
with a window size of
is taken as the center, whose input with other surrounding characters is represented by
; such inputs ultimately generate
hidden vectors
with a length of
, with the calculation method as follows:
where
and
represents the attention weight, and the calculation formula of
is as follows:
and the score function is defined as follows:
where
,
and
is the embedding dimension.
The resulting vector sequence
undergoes convolution operations with different convolution kernel sizes, and the extracted local semantic features are represented by the following equation:
Next, to better fuse the multi-scale local context information to obtain the more effective feature information and ensure that the network depth is increased without network degradation, this paper designs a multi-scale residual convolution network structure, as shown in
Figure 3. Except for the first CNN layer, the input of each CNN layer is a fused feature vector obtained from the input and output of the previous CNN layer after residual concatenation, and finally, the multi-scale feature vectors from the output of each of its CNN layers are concatenated to obtain the output of the local context-wise module.
represents concatenation operation.
3.3. BiGRU Layer
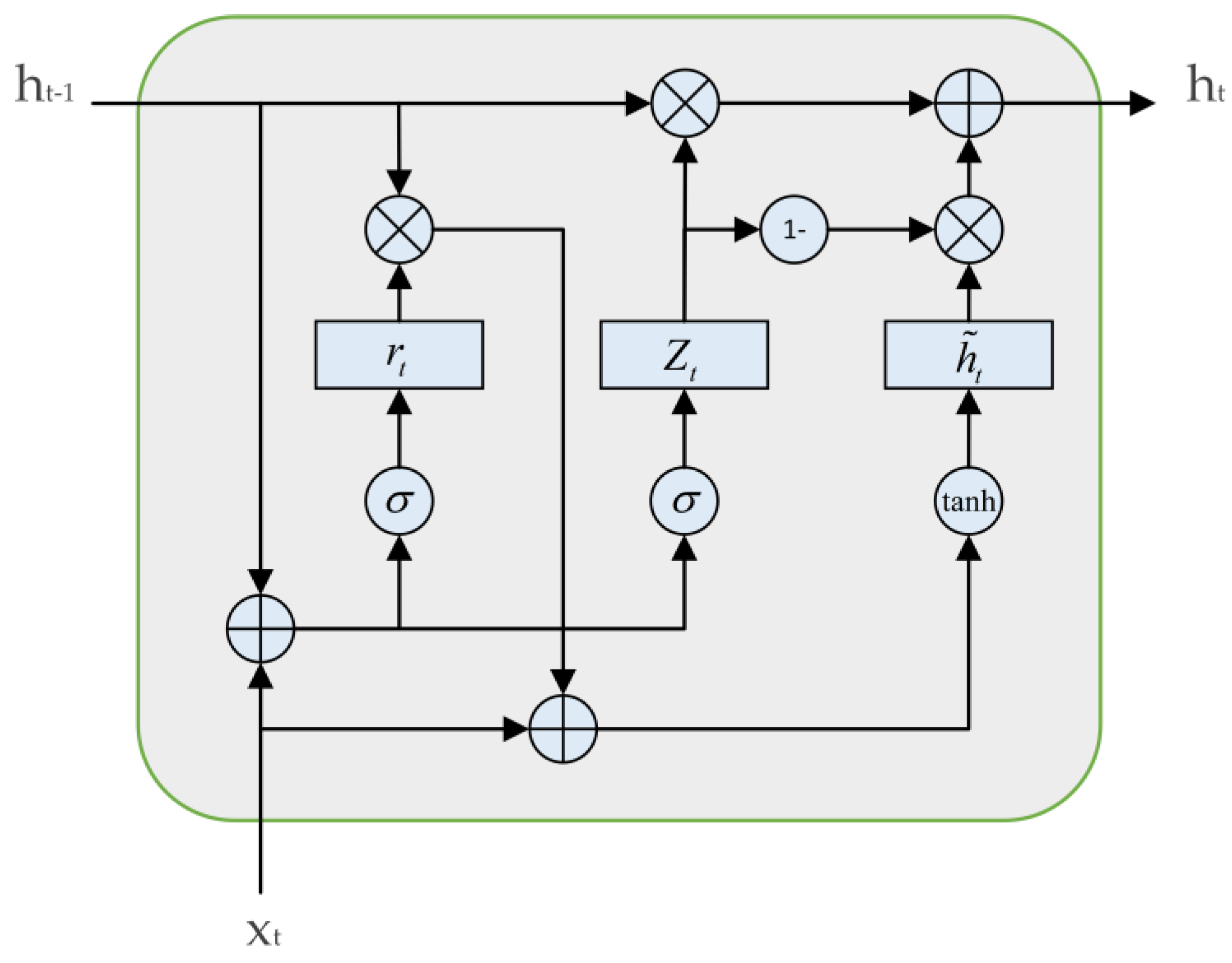
The gate recurrent unit (GRU) and the long short-term memory network (LSTM) are both improvements of the recurrent neural network (RNN). They can effectively solve the gradient vanishing or exploding problems faced by traditional RNNs. In the design of LSTM, three gate units are used: input gate, forget gate, and output gate. In contrast, the structure of GRU is simpler, requires less computation, and has faster training speed. GRU includes two gate units: update gate and reset gate. The update gate replaces the input gate and forget gate in LSTM. The update gate
controls its gate state to determine how much information
from the previous time step will be passed to the current time step
, and selectively accepts information from candidate states
based on its gate state. Reset gate
is responsible for managing how to merge the candidate state
with the information from the previous time step
. The formulae for calculating the various states of a GRU unit are as follows:
and the structure of a GRU is shown in
Figure 4.
Unidirectional GRU can only receive text sequences in the forward direction and obtain upstream information, while ignoring downstream information. However, in text, the front and back information are interrelated, so unidirectional processing easily loses important information. To solve this problem, this paper adopts the Bidirectional Gated Recurrent Unit (BiGRU) structure. BiGRU consists of a forward hidden layer and a backward hidden layer, which can simultaneously obtain two different vector representations of the current input information and combine them into the input information at the current time step, thereby better performing deep feature extraction on text and understanding the dependency relationship between the contexts.
3.4. Self-Attention Layer
During the feature extraction process, BiGRU uses gating mechanisms to selectively retain or discard context information at the current time step, without specifically distinguishing the influence of different characters on the current position. For example, in the sentence “Even mild hypoglycemia can potentially cause physical injuries such as falls and fractures in patients, leading to hospitalization and increasing their psychological and economic burden”, the contribution of the character “hypoglycemia” to identifying diseases’ “falls” and “fractures” is obviously greater than the character “mild”. Therefore, we believe that the BiGRU model has a problem of attention dispersion in recognition, meaning that it does not assign different weights to different characters according to their importance, resulting in a lack of proper distinction between important and regular information during encoding.
To address this issue, this paper employs a self-attention mechanism to filter key information from the input text. Similar to the self-attention mechanism in Transformer, it only focuses on the relationships between characters within the input sequence in order to identify the connections between different characters and select the most representative and critical words and phrases. Its calculation formula is:
where
represents the query matrix,
represents the key matrix,
represents the value matrix, and
represents the dimension of
and
. The attention mechanism calculates the similarity scores between the query matrix
and all key matrices
. To prevent these scores from being too large in high dimensions, they are scaled by dividing them by a scaling factor
. This approach can avoid numerical issues that may occur when calculating the softmax function. Next, the softmax function is used to transform these scores into normalized weights, which are then applied to the value matrix
to obtain a weighted vector representation. Finally, these weighted vector representations are used to express the importance of different parts of the input sequence. The calculation process of the self-attention mechanism is shown in
Figure 5.
We take
as an example to explain the calculation process of self-attention mechanism. Here,
,
, and
are weight matrices initialized for the self-attention mechanism, which, respectively, are dot multiplied with the input feature vector
to obtain the
,
, and
matrices. The attention score
between feature
and feature
is calculated using Formula (11):
Finally, the correlation
between
and
is obtained using Formula (12):
3.5. CRF Layer
CRF is a discriminative model based on conditional probability, which can model the conditional probability distribution of a set of random variables given the input variables. Because CRF can fully consider the relationship between contextual labels, it is widely used in sequence labeling tasks. In particular, in named entity recognition, there are strong constraint relationships between labels, and CRF can obtain the globally optimal label sequence. The probability score of output label sequence
given input sequence
is shown in Formula (13):
where
is the transition matrix,
represents the probability of transition from label
to label
, and
represents the probability of labeling the
character of input sequence
with label
. The conditional probability distribution of sequence y is shown in Formula (14):
In the training process of CRF, the maximum likelihood method is used to maximize the probability of the correct label sequence
, as shown in Formula (15):
The highest scoring label sequence obtained using the Viterbi algorithm is the globally optimal result output by the CRF, as shown in Formula (16):
4. Experiments
This section introduces the related experimental processes, including the construction of the dataset, annotation strategy, experimental environment, evaluation metrics, and finally, the analysis and comparison of the results and models.
4.1. Dataset Construction and Strategy
In the existing Chinese named entity recognition datasets, the specific medical field dataset for diabetes is relatively scarce. Therefore, we are committed to creating a named entity recognition dataset specifically for the diabetes field. The data for this experiment mainly come from domestic public medical and health websites and related encyclopedic websites. Websites such as “XYWY.COM” and “QIUYI.CN” were used to collect 9000 pieces of data information; we preprocessed the raw data to facilitate text recognition in subsequent work. In our experiments, text preprocessing mainly includes two stages: data cleaning and sentence segmentation. Data cleaning involves tasks such as removing meaningless characters, modifying text formats, handling missing data, and splitting long text sentences to obtain text that can be labeled with entities. The goal of data cleaning is to convert continuous raw text into a labeled sequence that only contains information such as words, punctuation marks, numbers, and spaces, while sentence segmentation identifies sentence boundaries using punctuation marks such as periods and divides the cleaned text into standardized sentences.
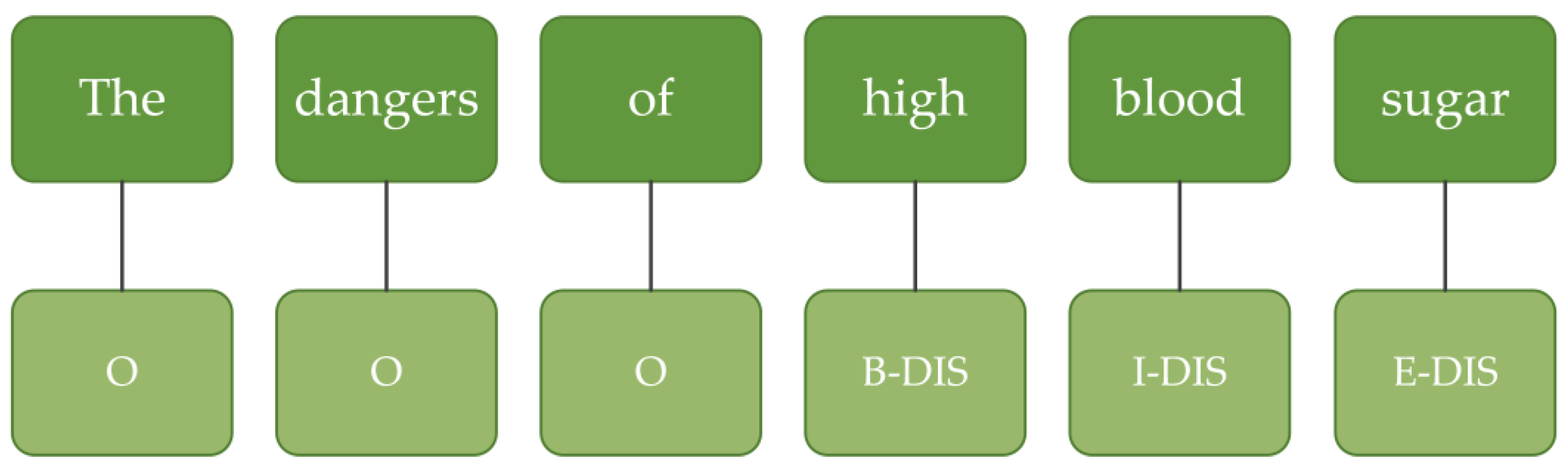
The data were annotated using the BIOES annotation mode, where B stands for the beginning of the entity, I stands for inside the entity, E stands for the end of the entity, S stands for a single character that is itself an entity, and O stands for outside the entity, indicating that it does not belong to any type. An example of the BIOES format is shown in
Figure 6. Finally, the dataset was divided into training, validation, and test sets in a ratio of 8:1:1. The number distribution of all types of entities in the training, validation, and test sets is shown in
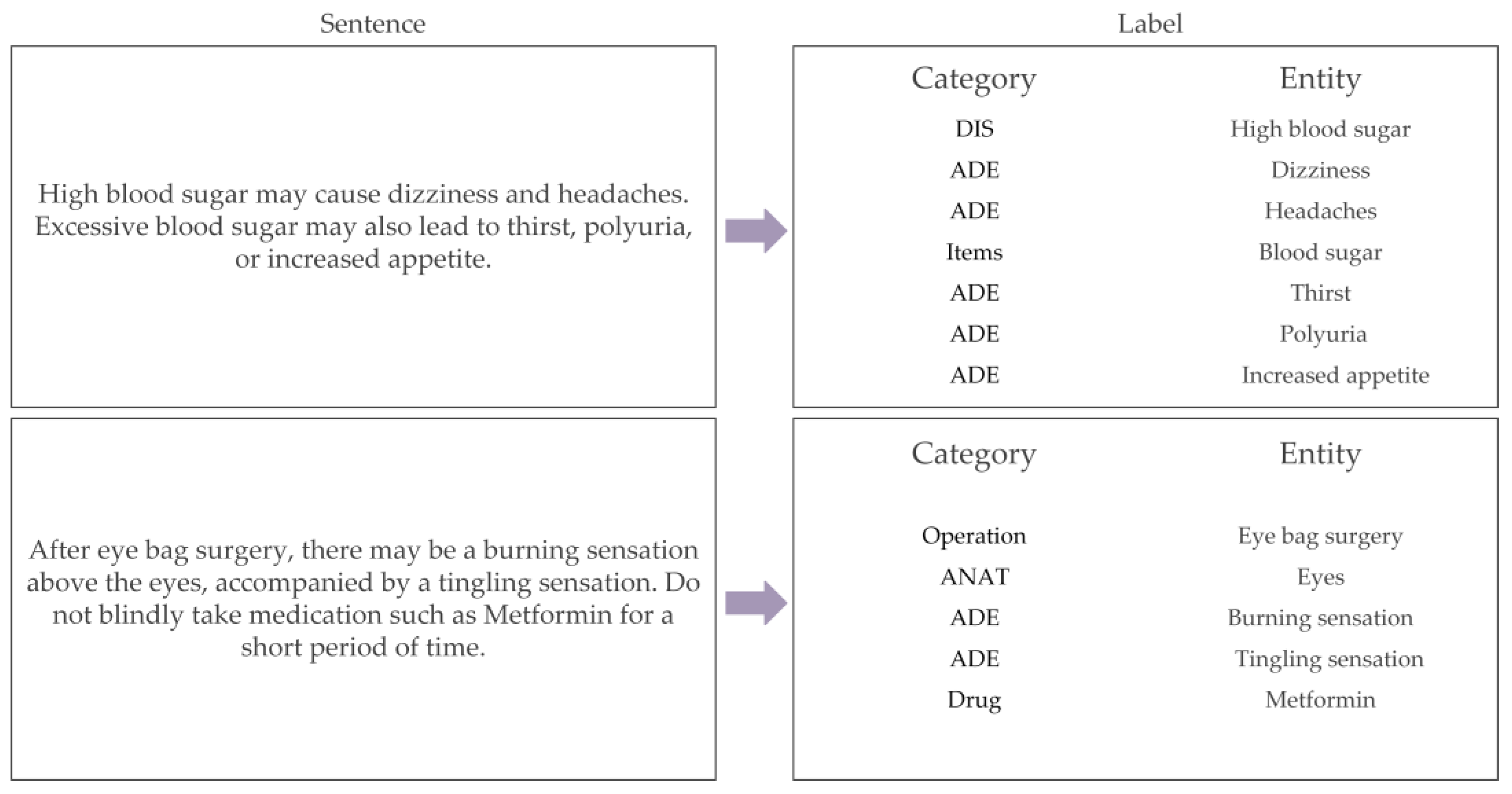
Table 1. For the classification of medical entities, they were divided into examination indicators, drug names, adverse reactions, anatomy, operation, and diseases. Some detailed examples of the DNER dataset are shown in
Figure 7.
In order to verify the generalization ability of the model, this article also conducted experiments using two public datasets, the diabetes dataset provided by the Ruijin Hospital and introduced by Alibaba Cloud Tianchi Laboratory, and CLUENER2020 [
43].
The Ruijin Hospital diabetes dataset is a resource containing clinical data, sourced from a well-known Chinese diabetes research journal, and spanning over seven years. It includes the most extensive research and hot topics in the field of diabetes. This dataset includes 15 medical entities, such as examination methods, etiology, clinical manifestations, drug administration methods, and locations, among others. The release of this dataset can provide important reference information for researchers and medical workers in the field of diabetes and has a positive significance for promoting the prevention, treatment, and management of diabetes.
The CLUENER2020 dataset is a fine-grained Chinese named entity recognition (NER) dataset that includes ten entity types: address, book, company, game, government, movie, name, organization, position, and scene. This dataset covers scenarios in which the same entity may belong to different categories. Compared to other Chinese NER datasets such as People’s Daily and Weibo, the CLUENER2020 dataset is more challenging and better reflects real-world scenarios.
4.2. Evaluation Metrics
This experiment uses three evaluation metrics commonly used in named entity recognition—precision (
), recall (
), and F1 score (
)—to evaluate the performance of the model. The specific formulae are as follows:
where
represents true positives, which is the number of positive samples predicted as positive;
represents false positives, which is the number of negative samples predicted as positive; and
represents false negatives, which is the number of positive samples predicted as negative.
4.3. Experimental Environment
All experiments in this article were conducted on a Linux system, using the development environment of Python 3.6 and the development framework of PyTorch 1.6.0. The optimizer used was AdamW, and the training was conducted on a single RTX A4000 GPU.
4.4. Experimental Results and Analysis
In order to verify the performance of the proposed model for named entity recognition in the field of diabetes, a comparative experiment has been conducted with six mainstream models on the private diabetes dataset. The experimental results are shown in
Table 2.
The BiLSTM-CRF model is our baseline method which is widely used in named entity recognition tasks. The BERT-CRF model utilizes the pre-trained BERT model to learn features of character sequences. The obtained sequence state scores are then fed into a CRF decoder to generate entity label sequences. The BERT-BiLSTM-CRF model is an extension of the BiLSTM-CRF model that incorporates the pre-trained BERT model. The recognition performance of this model is significantly improved compared to BiLSTM-CRF and BERT-CRF, mainly because BERT can consider contextual information, but it ignores the dependencies between words; therefore, BERT combined with BiLSTM learns the sequential relationship on observations, which performs better than BiLSTM-CRF and BERT-CRF in terms of performance. Compared to BiGRU, BiLSTM has a slightly larger parameter quantity and a relatively slow operating speed, which, however, has a smaller impact on accuracy. BERT-BiLSTM-IDCNN-CRF adds an IDCNN layer on the basis of BERT-BiLSTM-CRF to obtain the local information of sentences, alleviating the deficiency of BERT-BiLSTM-CRF considering only the global information. The RMBC model proposed herein takes full account the extraction of local and global features, which relies on the local feature awareness module to extract the local information of sentences, presenting better feature extraction than the IDCNN layer, and adds a self-attention mechanism on the basis of the BiGRU layer to solve the problem of unreasonable weight allocation of characters in BiGRU.
Table 2 shows that the models proposed herein are superior to various contrast models in terms of F1 score. Compared to the baseline model BiLSTM-CRF, the F1 score grows by 14.79%; compared with other models, this model has been improved to different degrees, suggesting that this model demonstrates a good recognition effect in the field of diabetes medicine.
The recognition results on the Ruijin diabetes dataset are shown in
Table 3. In the diabetes dataset of Ruijin Hospital, “hypoglycemia” in the “For patients who are older, have had diabetes for a long time, and have high-risk factors for hypoglycemia, the HbA1c target should be controlled to <7.5% or <8.0%.” sentence is recognized as a disease, which conforms to the entity category in the sentence. Yet, the entity “hypoglycemia” belongs to multiple categories; for example, in the “Insulin secretagogues also have potential adverse reactions of hypoglycemia and weight gain, so it is not recommended to use them in combination with insulin other than basal insulin.” sentence, the entity “hypoglycemia” belongs to the adverse reaction category. For difficult-to-recognize entities such as these, the RMBC model proposed in this paper can fully consider the semantic relationship between contexts by combining multi-scale local features and overcome the limitation of long-distance dependencies. Compared with other models, it has a significant improvement in performance in dealing with difficult-to-recognize entities. The experimental results also verify from one side that the model proposed in this paper has excellent recognition performance on the public diabetes dataset.
In order to verify the generalization of RMBC, compared on the Chinese fine-grained dataset CLUENER2020, which contains Chinese texts from ten different news domains, it has high quality and accuracy. Based on the recognition results in
Table 4, the RMBC model proposed in this paper has shown significant improvement in performance compared to other mainstream models. Our model achieved an F1 score of 81.79% on the CLUENER2020 dataset, which is 2.97% higher than the public baseline model BERT-CRF, and 11.79% higher than the baseline model BiLSTM-CRF proposed in this paper. Therefore, it further validates that the proposed model in this paper can effectively capture both local and global information through the multi-scale local context-wise module and the combination of self-attention mechanism and BiGRU, which can better distinguish the categories of different entities in different scenarios and presents excellent recognition effects. Experimental results indicate that the proposed RMBC model in this paper not only presents excellent recognition effect in the field of diabetes medicine but also presents good generalization capability for datasets in different fields.
4.5. Ablation Study
In order to verify the effectiveness of the local context-wise module, this paper conducts an ablation study. Model1 represents RoBERTa-BiGRU-CRF model without adding a local context-wise module. Model2 represents the local context-wise module under multi-scale residual convolution without adding the multi-window attention. Model3 represents a local context-wise module in a single scale convolution.
The results of the ablation study are shown in
Table 5; the F1 score of Model1, without adding a local context-wise module, decrease by 3.7%, 3.83%, and 0.95% on the three datasets, respectively; moreover, the F1 scores of Model2 and Model3 on the three datasets with partial local context-wise modules added are higher than Model1; nevertheless, the F1 scores are lower than RMBC, indicating that each part of the proposed local context-wise module can improve the recognition performance, the multi-window attention can effectively capture the important semantic components of local features, and the multi-scale residual convolution can fully integrate the context information at different scales, fully reflecting the importance of multi-window attention and multi-scale residual convolution in the local context-wise module. This proves that the local context-wise module can effectively capture the semantic information of the local context during the recognition process and plays a crucial role in the recognition process.