ELCT-YOLO: An Efficient One-Stage Model for Automatic Lung Tumor Detection Based on CT Images
Abstract
1. Introduction
- In order to solve the problem of multi-scale detection, a novel neck structure, called DENeck, is designed and proposed to effectively model the dependency between feature layers and improve the detection performance by using complementary features with similar semantic information. In addition, compared with the original FPN structure, the design of DENeck is more efficient in terms of the number of parameters used.
- A novel CRS structure is designed and proposed to improve the robustness of variable-size tumor detection by collecting rich context information. At the same time, an effective receptive field is constructed to refine the tumor features.
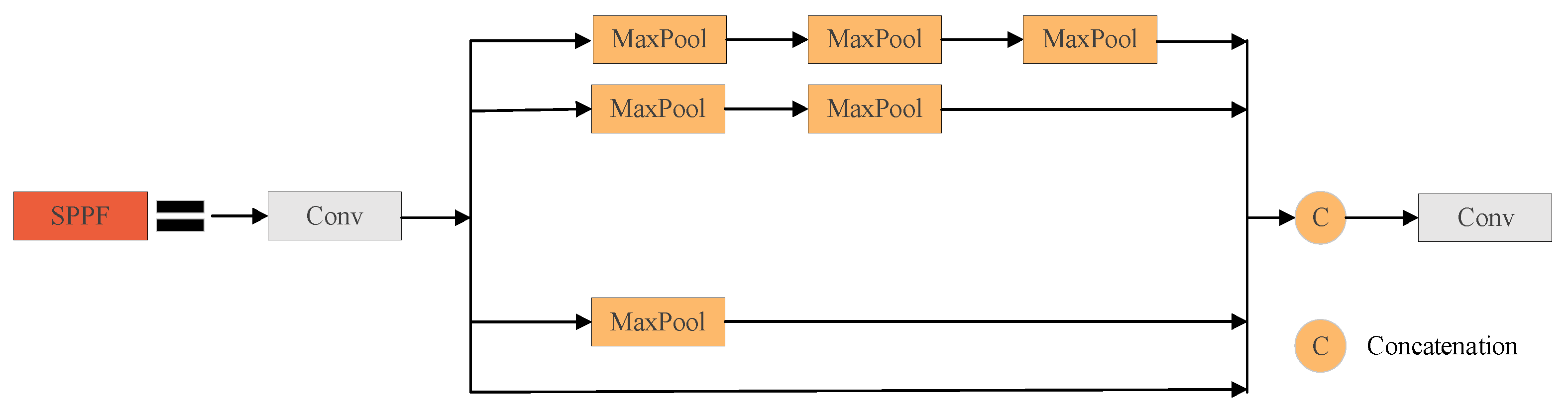
- It is proposed to integrate the spatial pyramid pooling—fast (SPPF) module of YOLOv5 [25] at the top of the original YOLOv7-tiny backbone network in order to extract important context features by utilizing a smaller number of parameters and using multiple small-size cascaded pooling kernels, in order to increase further the model’s operational speed and enrich the representation ability of feature maps.
2. Related Work
2.1. The YOLO Family
2.2. Multi-Scale Challenge and FPN
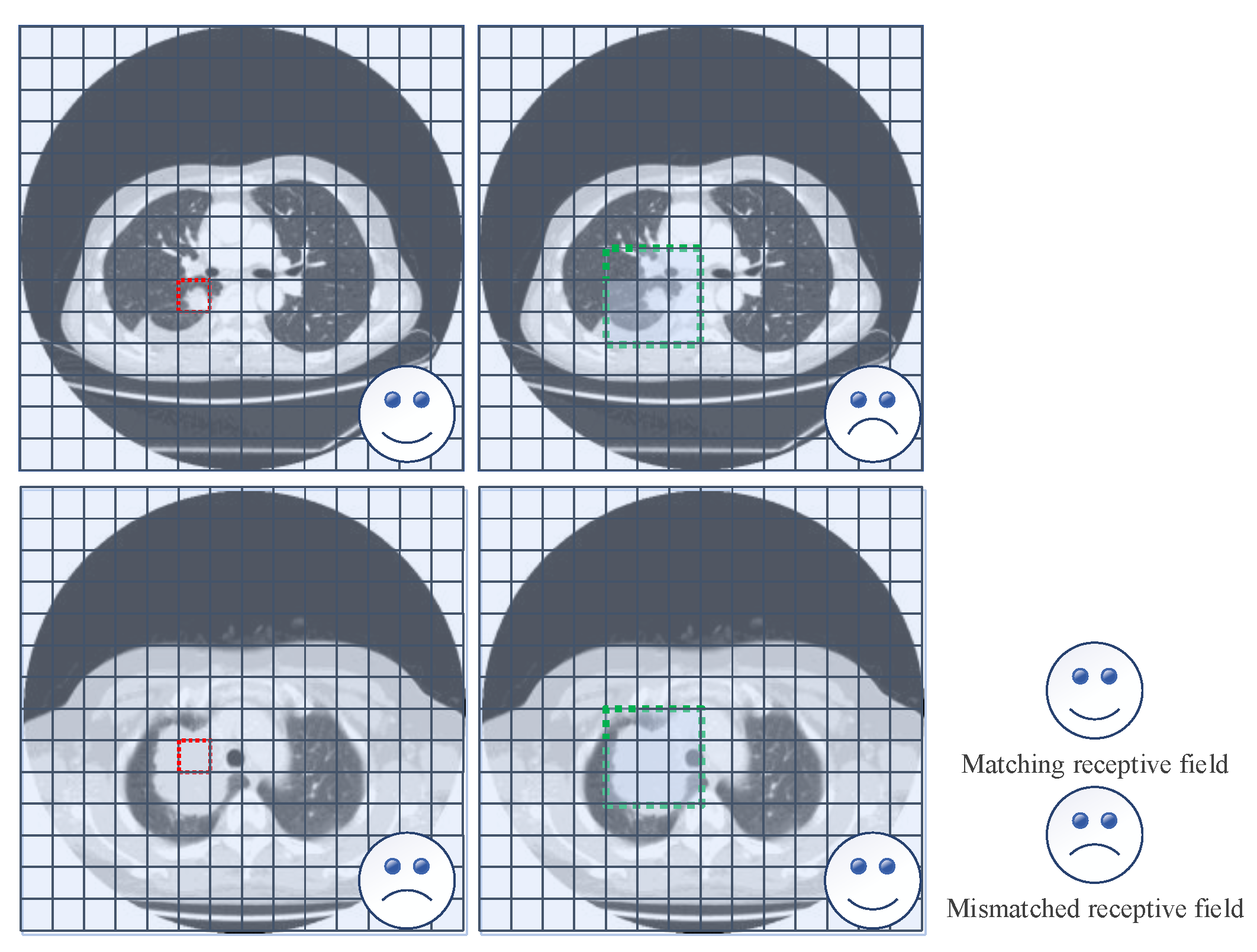
2.3. Exploring Context Information by Using Enlarged Receptive Field
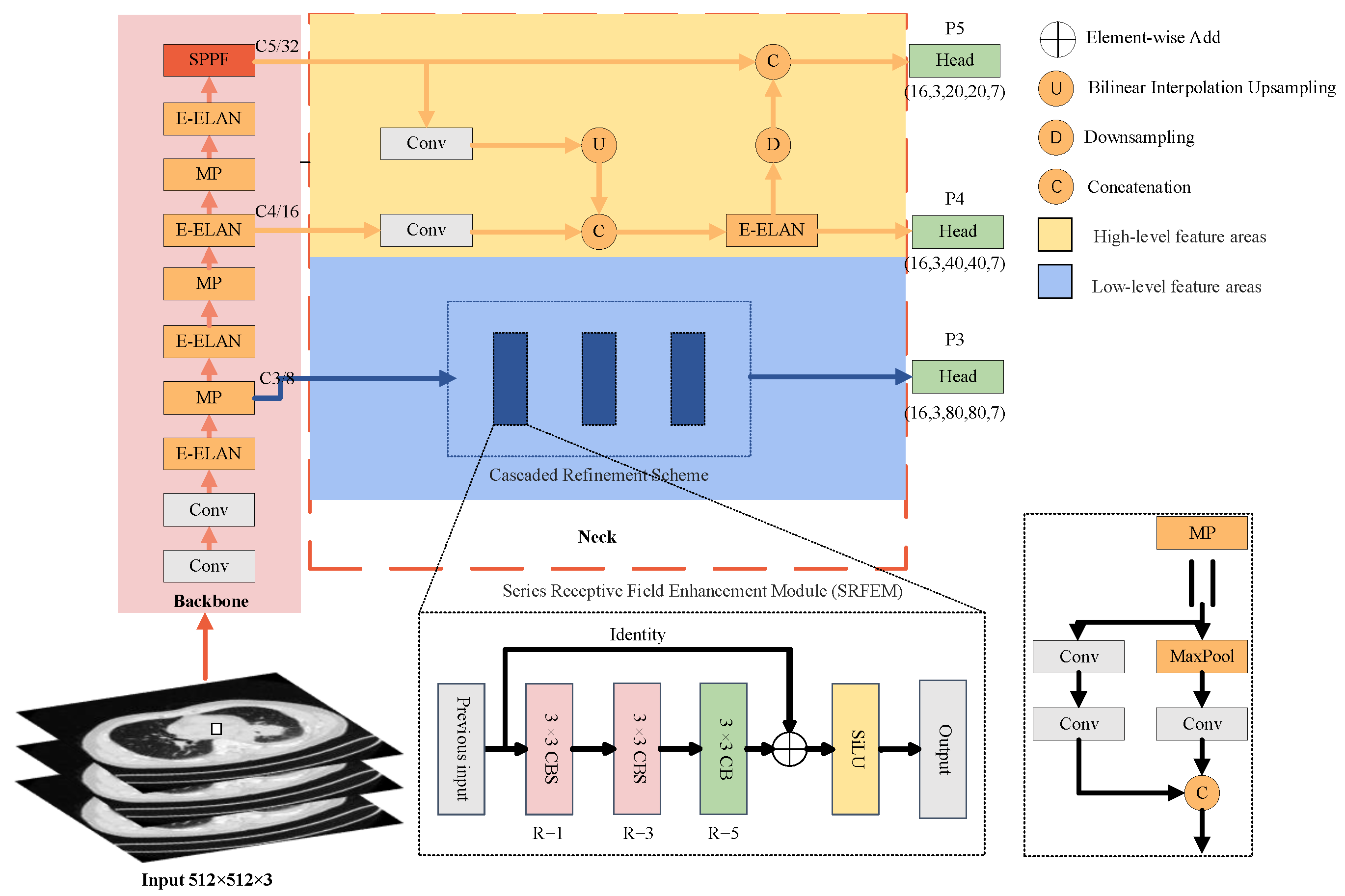
3. Proposed Model
3.1. Overview
3.2. Decoupled Neck (DENeck)
3.2.1. Motivation
3.2.2. Structure
3.3. Cascaded Refinement Scheme (CRS)
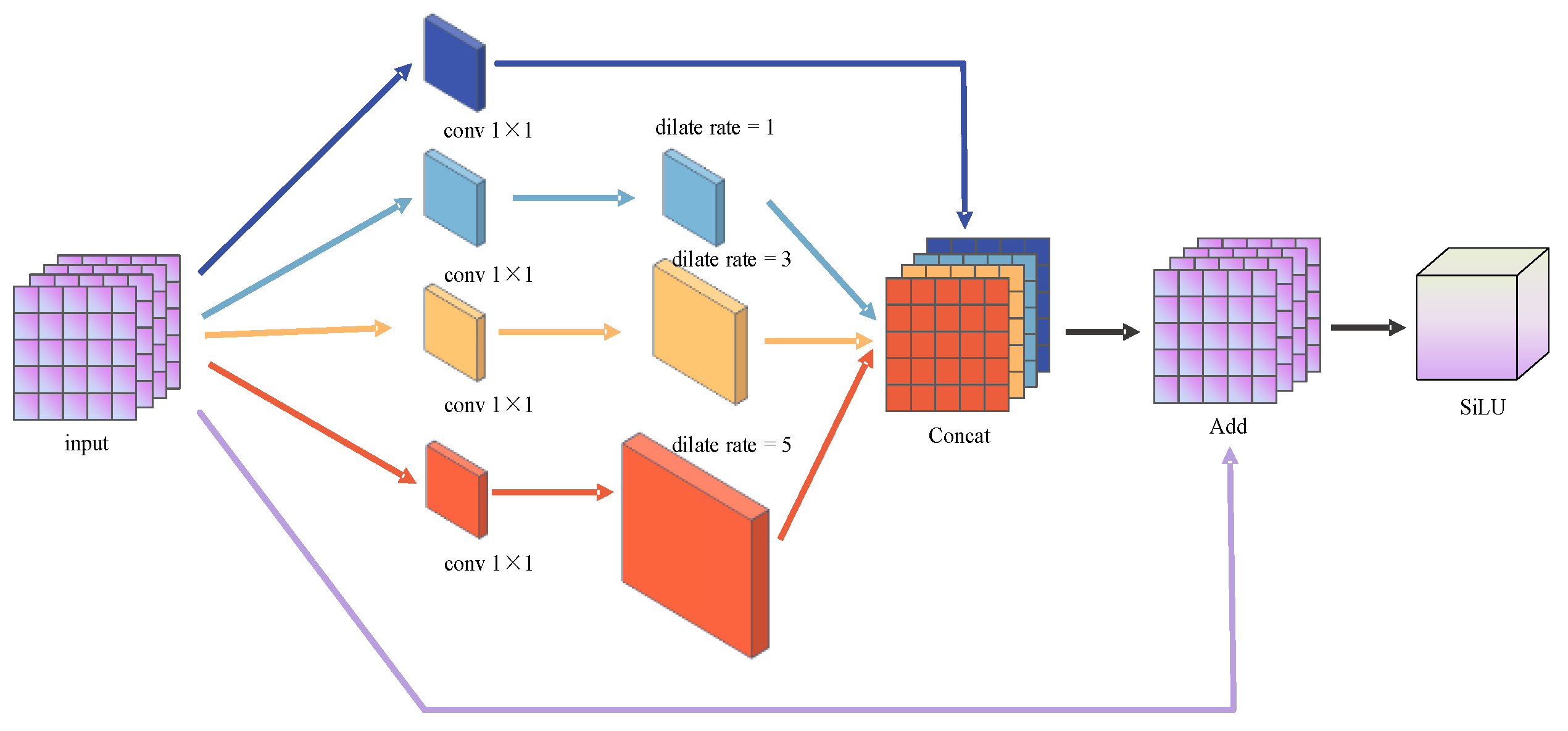
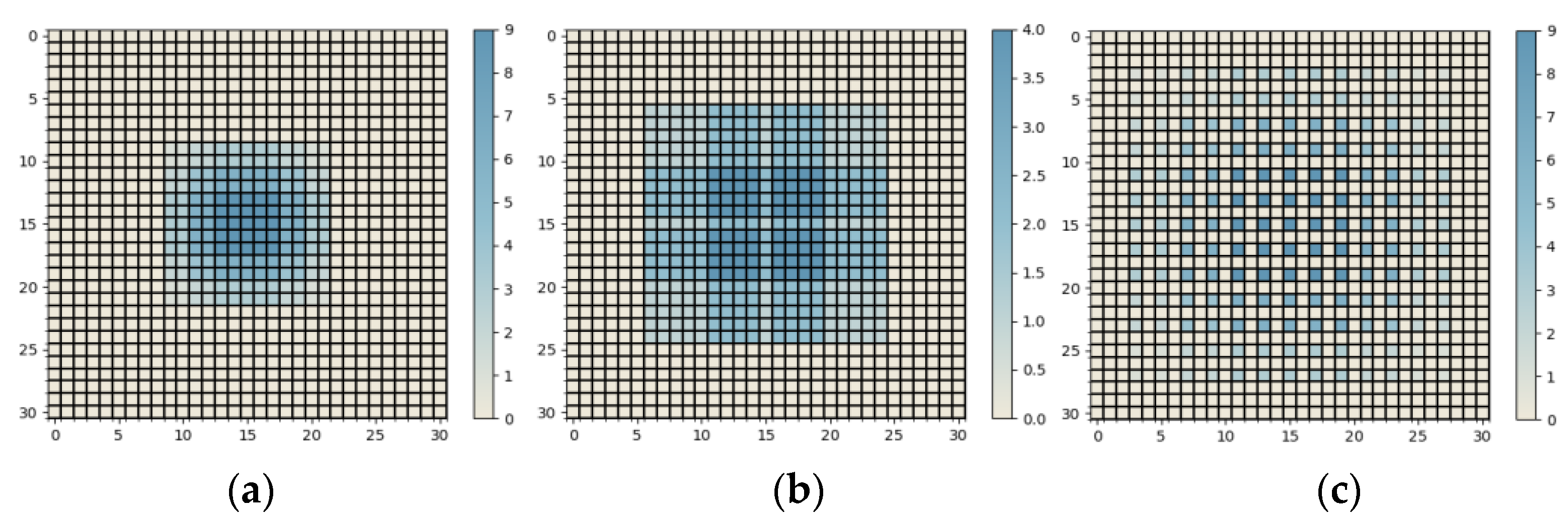
3.3.1. Series Receptive Field Enhancement Module (SRFEM)
3.3.2. Parallel Receptive Field Enhancement Module (PRFEM)
4. Experiments and Results
4.1. Dataset and Evaluation Metrics
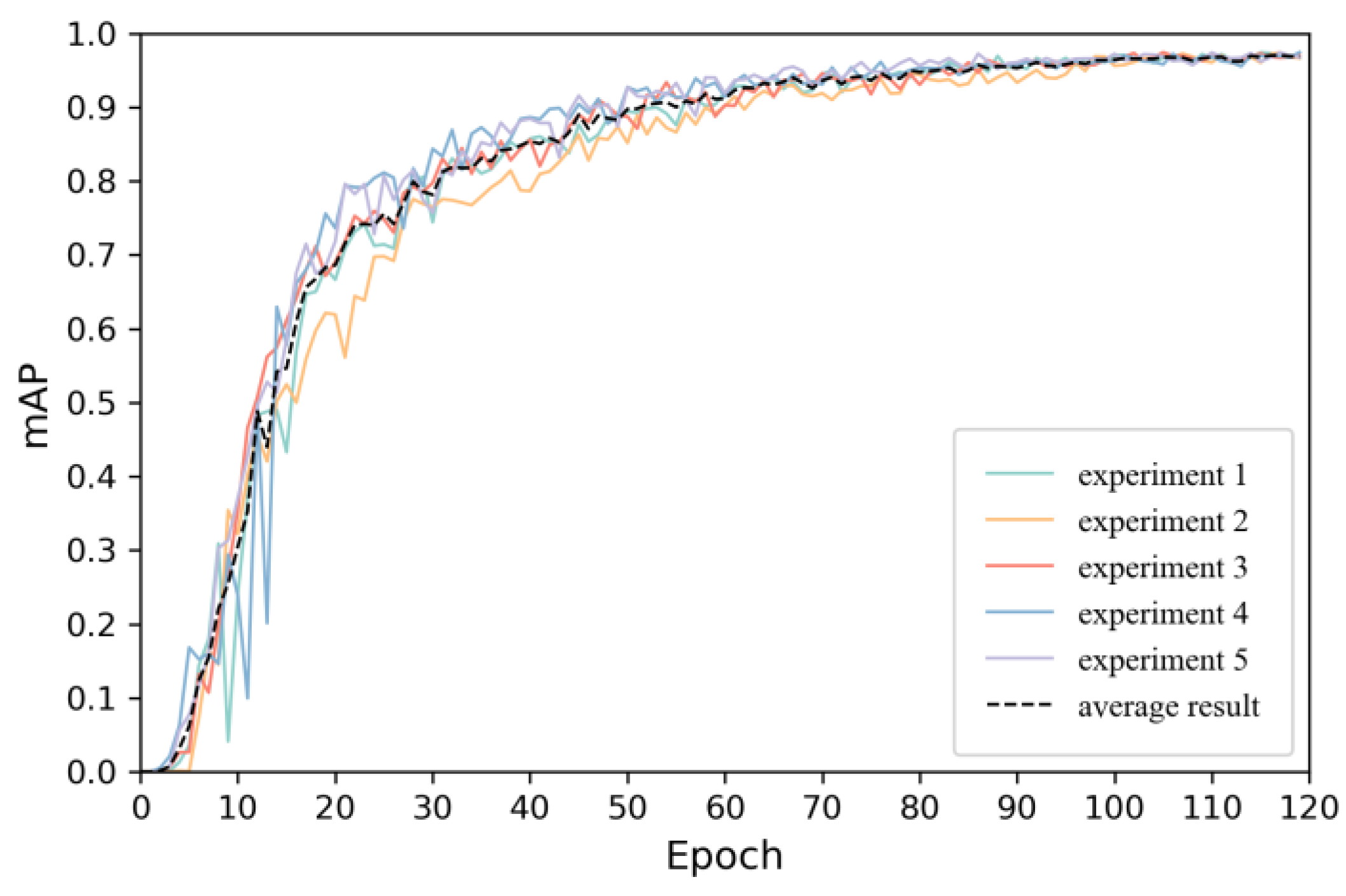
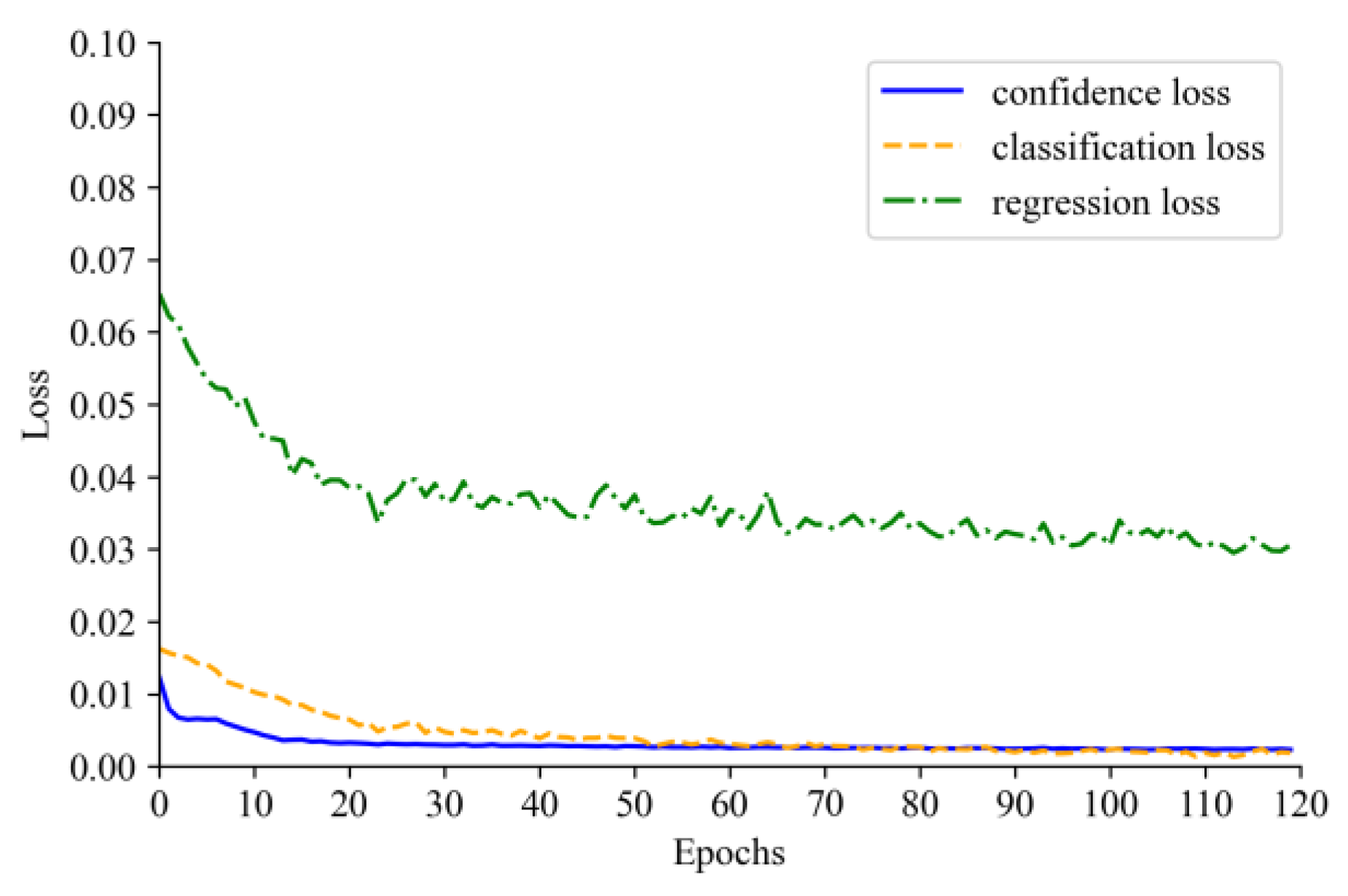
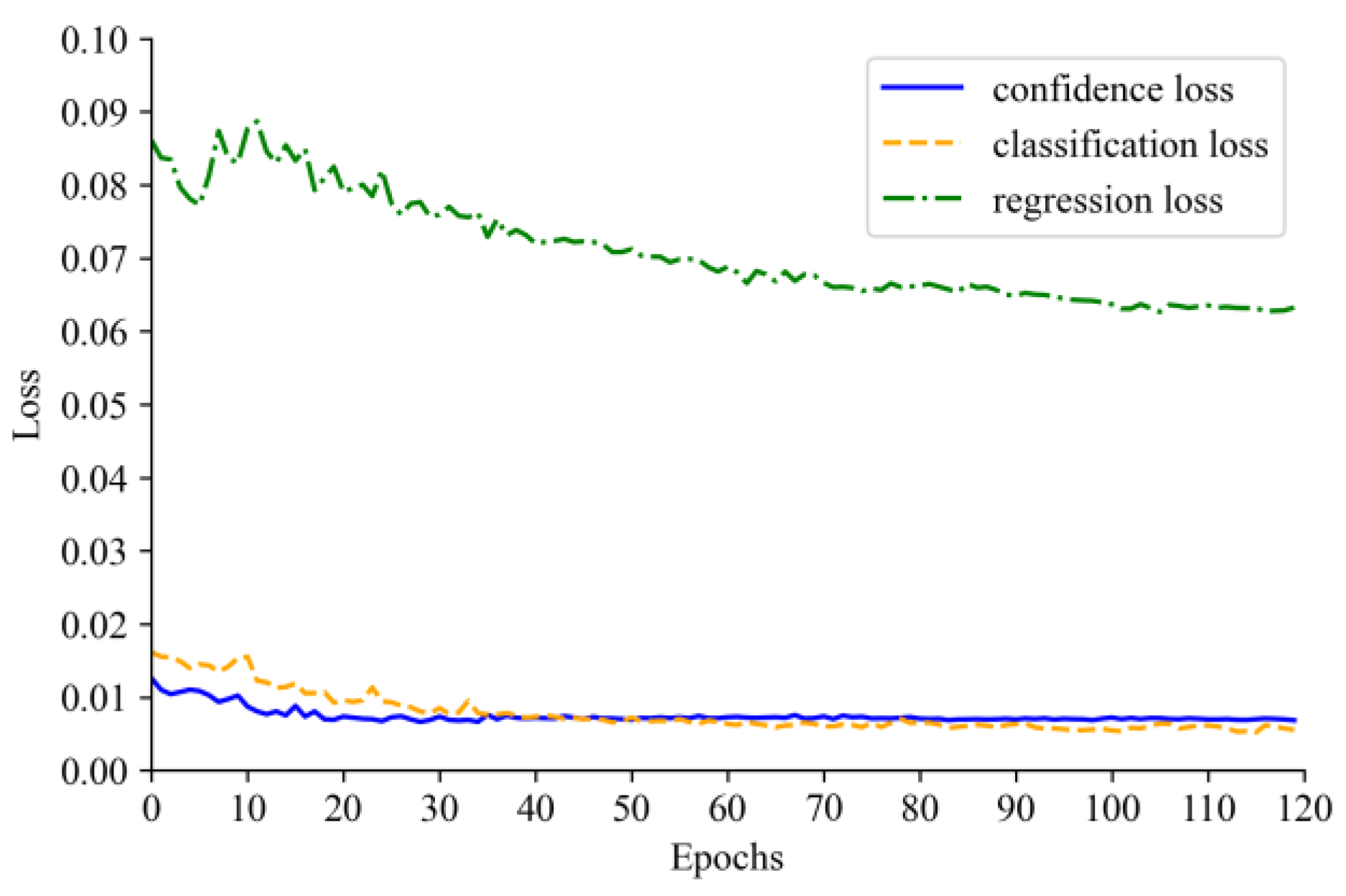
4.2. Model Training
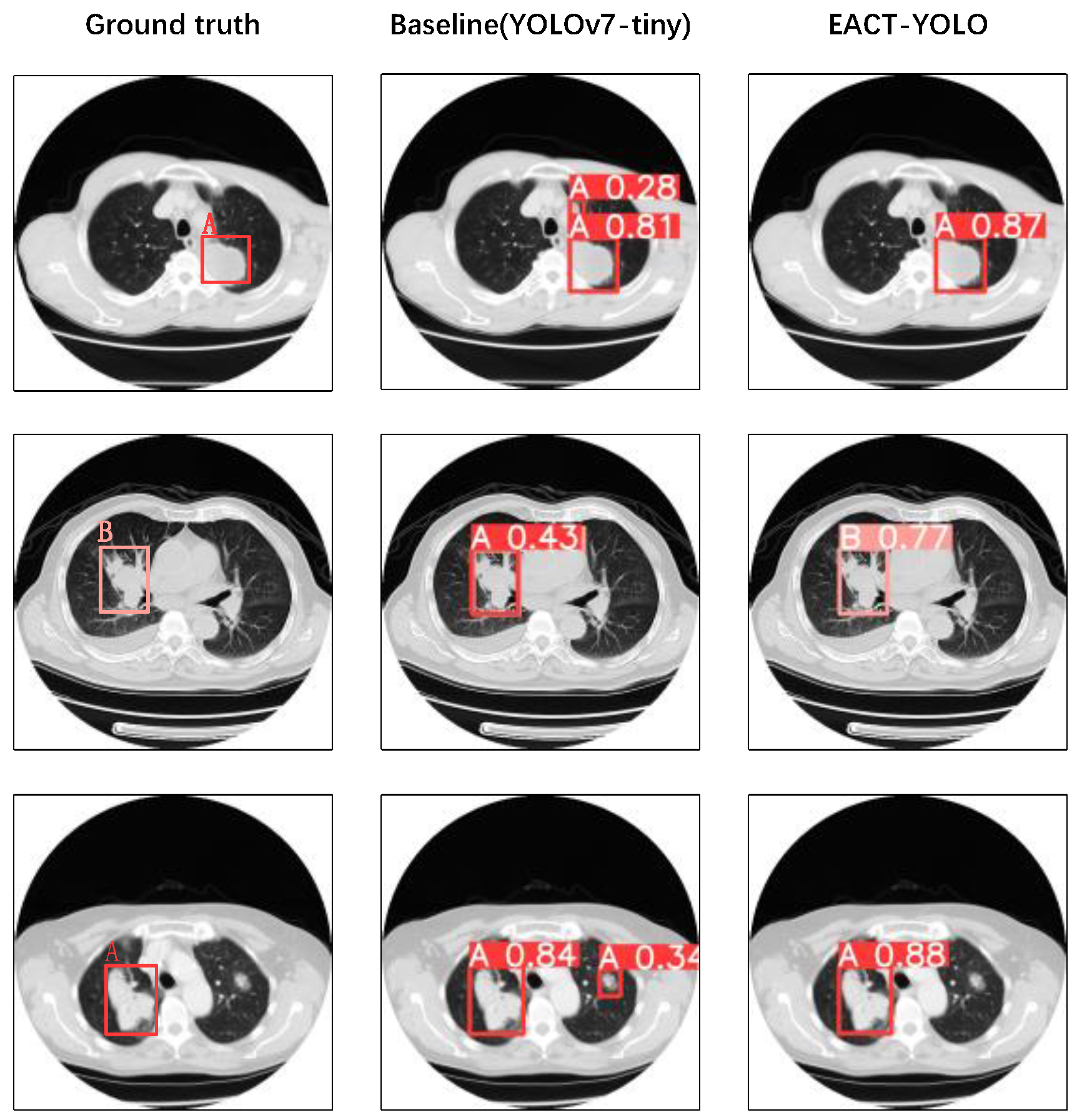
4.3. Comparison of ELCT-YOLO with State-of-the-Art Models
4.4. Ablation Study of ELCT-YOLO
4.5. CRS Study
4.6. DENeck Study
5. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Slatore, C.; Lareau, S.C.; Fahy, B. Staging of Lung Cancer. Am. J. Respir. Crit. Care Med. 2022, 205, P17–P19. [Google Scholar] [CrossRef] [PubMed]
- Nishino, M.; Schiebler, M.L. Advances in Thoracic Imaging: Key Developments in the Past Decade and Future Directions. Radiology 2023, 306, 222536. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, D.; Lu, M.T.; Raghu, V.K.; Park, C.M.; Goo, J.M.; Choi, S.H.; Kim, H. Deep learning to optimize candidate selection for lung cancer CT screening: Advancing the 2021 USPSTF recommendations. Radiology 2022, 305, 209–218. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, K.; Cui, H.; Jin, Q.; Cheng, P.; Nakaguchi, T.; Li, C.; Ning, Z.; Wang, L.; Xuan, P. Topological structure and global features enhanced graph reasoning model for non-small cell lung cancer segmentation from CT. Phys. Med. Biol. 2023, 68, 025007. [Google Scholar] [CrossRef]
- Lin, J.; Yu, Y.; Zhang, X.; Wang, Z.; Li, S. Classification of Histological Types and Stages in Non-small Cell Lung Cancer Using Radiomic Features Based on CT Images. J. Digit. Imaging 2023, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, H.; Yatabe, Y.; Watanabe, H.; Akai, H.; Abe, O.; Watanabe, S.-I.; Kusumoto, M. Radiological precursor lesions of lung squamous cell carcinoma: Early progression patterns and divergent volume doubling time between hilar and peripheral zones. Lung Cancer 2023, 176, 31–37. [Google Scholar] [CrossRef]
- Halder, A.; Dey, D.; Sadhu, A.K. Lung nodule detection from feature engineering to deep learning in thoracic CT images: A comprehensive review. J. Digit. Imaging 2020, 33, 655–677. [Google Scholar] [CrossRef]
- Huang, S.; Yang, J.; Shen, N.; Xu, Q.; Zhao, Q. Artificial intelligence in lung cancer diagnosis and prognosis: Current application and future perspective. Semin. Cancer Biol. 2023, 89, 30–37. [Google Scholar] [CrossRef]
- Mousavi, Z.; Rezaii, T.Y.; Sheykhivand, S.; Farzamnia, A.; Razavi, S. Deep convolutional neural network for classification of sleep stages from single-channel EEG signals. J. Neurosci. Methods 2019, 324, 108312. [Google Scholar] [CrossRef]
- Gong, J.; Liu, J.; Hao, W.; Nie, S.; Zheng, B.; Wang, S.; Peng, W. A deep residual learning network for predicting lung adenocarcinoma manifesting as ground-glass nodule on CT images. Eur. Radiol. 2020, 30, 1847–1855. [Google Scholar] [CrossRef]
- Mei, J.; Cheng, M.M.; Xu, G.; Wan, L.R.; Zhang, H. SANet: A Slice-Aware Network for Pulmonary Nodule Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 4374–4387. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Liu, Z.; Luo, Y.; Hu, H.; Shen, L.; Du, B.; Kuang, K.; Yang, J. SGDA: Towards 3D Universal Pulmonary Nodule Detection via Slice Grouped Domain Attention. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 1–13. [Google Scholar] [CrossRef]
- Su, A.; PP, F.R.; Abraham, A.; Stephen, D. Deep Learning-Based BoVW–CRNN Model for Lung Tumor Detection in Nano-Segmented CT Images. Electronics 2023, 12, 14. [Google Scholar] [CrossRef]
- Mousavi, Z.; Shahini, N.; Sheykhivand, S.; Mojtahedi, S.; Arshadi, A. COVID-19 detection using chest X-ray images based on a developed deep neural network. SLAS Technol. 2022, 27, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Jiang, H.; Ma, L. YOLO-lung: A Practical Detector Based on Imporved YOLOv4 for Pulmonary Nodule Detection. In Proceedings of the 2021 14th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 23–25 October 2021; pp. 1–6. [Google Scholar]
- Causey, J.; Li, K.; Chen, X.; Dong, W.; Huang, X. Spatial Pyramid Pooling with 3D Convolution Improves Lung Cancer Detection. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 19, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhao, L.; Yuan, J.; Yu, H. MSANet: Multiscale Aggregation Network Integrating Spatial and Channel Information for Lung Nodule Detection. IEEE J. Biomed. Health Inform. 2022, 26, 2547–2558. [Google Scholar] [CrossRef]
- Guo, N.; Bai, Z. Multi-scale Pulmonary Nodule Detection by Fusion of Cascade R-CNN and FPN. In Proceedings of the 2021 International Conference on Computer Communication and Artificial Intelligence (CCAI), Guangzhou, China, 7–9 May 2021; pp. 15–19. [Google Scholar]
- Yan, C.-M.; Wang, C. Automatic Detection and Localization of Pulmonary Nodules in CT Images Based on YOLOv5. J. Comput. 2022, 33, 113–123. [Google Scholar] [CrossRef]
- Zhong, G.; Ding, W.; Chen, L.; Wang, Y.; Yu, Y.F. Multi-Scale Attention Generative Adversarial Network for Medical Image Enhancement. IEEE Trans. Emerg. Top. Comput. Intell. 2023, 1–13. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H.-Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar]
- Ghiasi, G.; Lin, T.-Y.; Le, Q.V. Nas-fpn: Learning scalable feature pyramid architecture for object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 7036–7045. [Google Scholar]
- Tan, M.; Pang, R.; Le, Q.V. Efficientdet: Scalable and efficient object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 14–19 June 2020; pp. 10781–10790. [Google Scholar]
- Wang, P.; Chen, P.; Yuan, Y.; Liu, D.; Huang, Z.; Hou, X.; Cottrell, G. Understanding convolution for semantic segmentation. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 1451–1460. [Google Scholar]
- Jocher, G.; Chaurasia, A.; Stoken, A.; Borovec, J.; Kwon, Y.; Michael, K.; Fang, J.; Yifu, Z.; Wong, C.; Montes, D.; et al. ultralytics/yolov5: v7.0—YOLOv5 SOTA Realtime Instance Segmentation; Zenodo: Geneva, Switzerland, 2022. [Google Scholar]
- Alsaedi, D.; El Badawe, M.; Ramahi, O.M. A Breast Cancer Detection System Using Metasurfaces With a Convolution Neural Network: A Feasibility Study. IEEE Trans. Microw. Theory Tech. 2022, 70, 3566–3576. [Google Scholar] [CrossRef]
- Fang, H.; Li, F.; Fu, H.; Sun, X.; Cao, X.; Lin, F.; Son, J.; Kim, S.; Quellec, G.; Matta, S.; et al. ADAM Challenge: Detecting Age-Related Macular Degeneration From Fundus Images. IEEE Trans. Med. Imaging 2022, 41, 2828–2847. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Wang, S.; Yin, Y. Explainable Multitask Shapley Explanation Networks for Real-time Polyp Diagnosis in Videos. IEEE Trans. Ind. Inform. 2022, 1–10. [Google Scholar] [CrossRef]
- Ahmed, I.; Chehri, A.; Jeon, G.; Piccialli, F. Automated pulmonary nodule classification and detection using deep learning architectures. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhao, Z.; Zhong, J.; Wang, W.; Wen, Z.; Qin, J. Polypseg+: A lightweight context-aware network for real-time polyp segmentation. IEEE Trans. Cybern. 2022, 53, 2610–2621. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Sahoo, D.; Hoi, S.C. Recent advances in deep learning for object detection. Neurocomputing 2020, 396, 39–64. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2016; pp. 7263–7271. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Li, C.; Li, L.; Jiang, H.; Weng, K.; Geng, Y.; Li, L.; Ke, Z.; Li, Q.; Cheng, M.; Nie, W. YOLOv6: A single-stage object detection framework for industrial applications. arXiv 2022, arXiv:2209.02976. [Google Scholar]
- Gevorgyan, Z. SIoU loss: More powerful learning for bounding box regression. arXiv 2022, arXiv:2205.12740. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path aggregation network for instance segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8759–8768. [Google Scholar] [CrossRef]
- Qiao, S.; Chen, L.-C.; Yuille, A. Detectors: Detecting objects with recursive feature pyramid and switchable atrous convolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021; pp. 10213–10224. [Google Scholar]
- Liu, Y.; Li, H.; Cheng, J.; Chen, X. MSCAF-Net: A General Framework for Camouflaged Object Detection via Learning Multi-Scale Context-Aware Features. IEEE Trans. Circuits Syst. Video Technol. 2023, 1. [Google Scholar] [CrossRef]
- Liu, J.-J.; Hou, Q.; Cheng, M.-M.; Feng, J.; Jiang, J. A simple pooling-based design for real-time salient object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 3917–3926. [Google Scholar]
- Xiang, W.; Mao, H.; Athitsos, V. ThunderNet: A turbo unified network for real-time semantic segmentation. In Proceedings of the 2019 IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa Village, HI, USA, 7–11 January 2019; pp. 1789–1796. [Google Scholar]
- Xu, L.; Xue, H.; Bennamoun, M.; Boussaid, F.; Sohel, F. Atrous convolutional feature network for weakly supervised semantic segmentation. Neurocomputing 2021, 421, 115–126. [Google Scholar] [CrossRef]
- Liu, J.; Yang, D.; Hu, F. Multiscale object detection in remote sensing images combined with multi-receptive-field features and relation-connected attention. Remote Sens. 2022, 14, 427. [Google Scholar] [CrossRef]
- Guo, C.; Fan, B.; Zhang, Q.; Xiang, S.; Pan, C. Augfpn: Improving multi-scale feature learning for object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 14–19 June 2020; pp. 12595–12604. [Google Scholar]
- Bhattacharjee, A.; Murugan, R.; Goel, T.; Mirjalili, S. Pulmonary nodule segmentation framework based on fine-tuned and pre-trained deep neural network using CT images. IEEE Trans. Radiat. Plasma Med. Sci. 2023, 7, 394–409. [Google Scholar] [CrossRef]
- Ezhilraja, K.; Shanmugavadivu, P. Contrast Enhancement of Lung CT Scan Images using Multi-Level Modified Dualistic Sub-Image Histogram Equalization. In Proceedings of the 2022 International Conference on Automation, Computing and Renewable Systems (ICACRS), Pudukkottai, India, 13–15 December 2022; pp. 1009–1014. [Google Scholar]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Li, P.; Wang, S.; Li, T.; Lu, J.; HuangFu, Y.; Wang, D. A large-scale CT and PET/CT dataset for lung cancer diagnosis [dataset]. Cancer Imaging Arch. 2020. [Google Scholar] [CrossRef]
- Mustafa, B.; Loh, A.; Freyberg, J.; MacWilliams, P.; Wilson, M.; McKinney, S.M.; Sieniek, M.; Winkens, J.; Liu, Y.; Bui, P. Supervised transfer learning at scale for medical imaging. arXiv 2021, arXiv:2101.05913. [Google Scholar]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU loss: Faster and better learning for bounding box regression. In Proceedings of the AAAI Conference on Artificial Intelligence, vol.34, New York, NY, USA, 7–12 February 2020; pp. 12993–13000. [Google Scholar] [CrossRef]
- Wang, C.; Sun, S.; Zhao, C.; Mao, Z.; Wu, H.; Teng, G. A Detection Model for Cucumber Root-Knot Nematodes Based on Modified YOLOv5-CMS. Agronomy 2022, 12, 2555. [Google Scholar] [CrossRef]
- Alamro, W.; Seet, B.-C.; Wang, L.; Parthiban, P. Early-Stage Lung Tumor Detection based on Super-Wideband Microwave Reflectometry. Electronics 2023, 12, 36. [Google Scholar] [CrossRef]
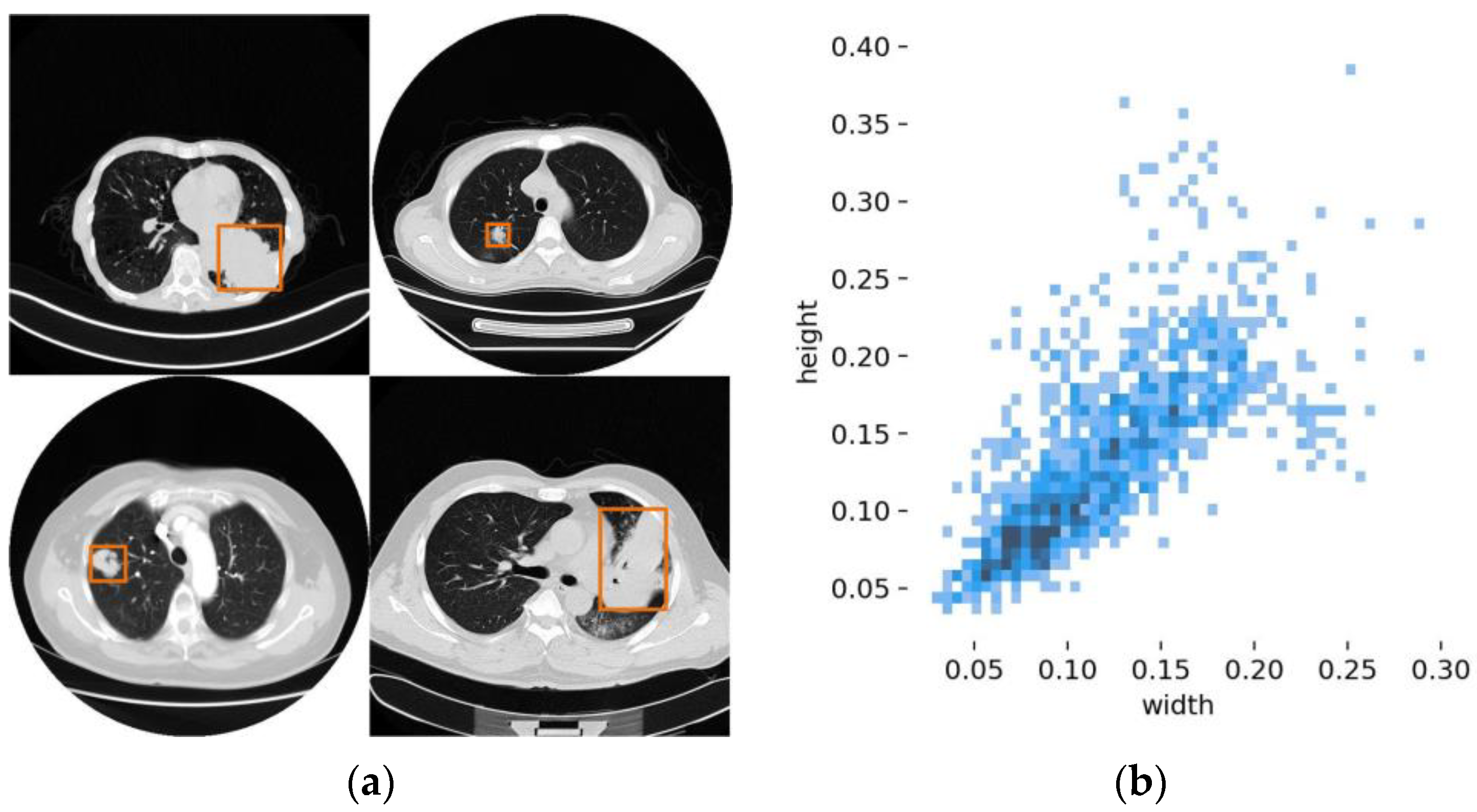

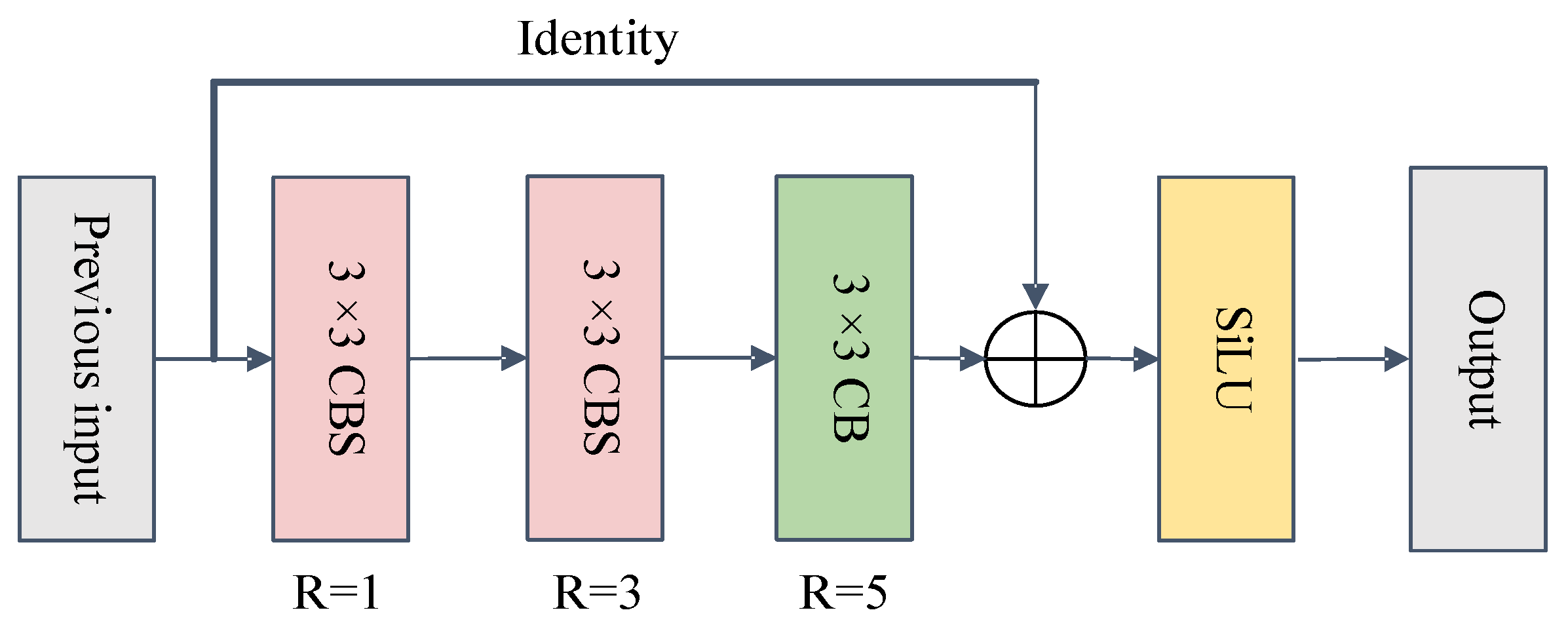
| Model | P | R | mAP | FPS | Size (MB) |
|---|---|---|---|---|---|
| YOLOv3 | 0.930 | 0.925 | 0.952 | 63 | 117.0 |
| YOLOv5 | 0.949 | 0.923 | 0.961 | 125 | 17.6 |
| YOLOv7-tiny | 0.901 | 0.934 | 0.951 | 161 | 11.8 |
| YOLOv8 | 0.967 | 0.939 | 0.977 | 115 | 21.4 |
| SSD | 0.878 | 0.833 | 0.906 | 95 | 182.0 |
| Faster R-CNN | 0.903 | 0.891 | 0.925 | 28 | 315.0 |
| ELCT-YOLO | 0.923 | 0.951 | 0.974 | 159 | 11.4 |
| No. | SPPF | DENeck | CRS | mAP | Size (MB) |
|---|---|---|---|---|---|
| 1 | 0.955 | 12.0 | |||
| 2 | ✓ | 0.958 | 11.2 | ||
| 3 | ✓ | 0.968 | 11.2 | ||
| 4 | ✓ | 0.966 | 12.6 | ||
| 5 | ✓ | ✓ | ✓ | 0.974 | 11.4 |
| Scheme | P | R | mAP |
|---|---|---|---|
| PPP | 0.961 | 0.929 | 0.965 |
| SPP | 0.977 | 0.893 | 0.969 |
| SSP | 0.958 | 0.924 | 0.969 |
| SSS | 0.921 | 0.957 | 0.974 |
| Dilation Rate | P | R | mAP |
|---|---|---|---|
| (1,2,3) | 0.938 | 0.923 | 0.958 |
| (1,3,5) | 0.969 | 0.933 | 0.967 |
| (2,4,6) | 0.954 | 0.921 | 0.961 |
| Method | mAP | Size (MB) |
|---|---|---|
| FPN | 0.957 | 14.6 |
| PANet | 0.963 | 11.7 |
| BiFPN | 0.967 | 11.2 |
| DENeck | 0.971 | 11.3 |
| Network | mAP | Size (MB) |
|---|---|---|
| YOLOv7-tiny’s | 0.965 | 11.4 |
| YOLOv7’s | 0.967 | 70.4 |
| YOLOv7x’s | 0.971 | 128.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Z.; Zhao, J.; Liu, J.; Zeng, X.; Zhang, H.; Zhang, X.; Ganchev, I. ELCT-YOLO: An Efficient One-Stage Model for Automatic Lung Tumor Detection Based on CT Images. Mathematics 2023, 11, 2344. https://doi.org/10.3390/math11102344
Ji Z, Zhao J, Liu J, Zeng X, Zhang H, Zhang X, Ganchev I. ELCT-YOLO: An Efficient One-Stage Model for Automatic Lung Tumor Detection Based on CT Images. Mathematics. 2023; 11(10):2344. https://doi.org/10.3390/math11102344
Chicago/Turabian StyleJi, Zhanlin, Jianyong Zhao, Jinyun Liu, Xinyi Zeng, Haiyang Zhang, Xueji Zhang, and Ivan Ganchev. 2023. "ELCT-YOLO: An Efficient One-Stage Model for Automatic Lung Tumor Detection Based on CT Images" Mathematics 11, no. 10: 2344. https://doi.org/10.3390/math11102344
APA StyleJi, Z., Zhao, J., Liu, J., Zeng, X., Zhang, H., Zhang, X., & Ganchev, I. (2023). ELCT-YOLO: An Efficient One-Stage Model for Automatic Lung Tumor Detection Based on CT Images. Mathematics, 11(10), 2344. https://doi.org/10.3390/math11102344










