Abstract
The survival prediction of children undergoing hematopoietic stem-cell transplantation is essential for successful transplantation. However, the performance of current algorithms for predicting mortality in this patient group has not improved over recent decades. This paper proposes a new feature selection technique for survival prediction problems using the Mud Ring Algorithm (MRA). Experiments and tests were initially performed on 13 real datasets with varying occurrences to compare the suggested algorithm with other algorithms. After that, the constructed model classification performance was compared to other techniques using the bone marrow transplant children’s dataset. Modern techniques were used to acquire their classification results, which were then compared to the suggested outcomes using a variety of well-known metrics, graphical tools, and diagnostic analysis. This investigation has demonstrated that our suggested approach is comparable and outperformed other methods in terms of results. In addition, the results showed that the constructed model enhanced prediction accuracy by up to 82.6% for test cases.
Keywords:
hematopoietic stem-cell transplantation; children; Mud Ring Algorithm; feature selection; artificial intelligence MSC:
68U01
1. Introduction
Relapse to an underlying illness, which affects about 30% of the conditions on the rationale and stage of the disease, drastically reduces survival following heterologous hematopoietic stem-cell transplantation as a therapy for the blood malignancies hematopoietic system [1,2]. The graft-versus-leukemia (GvL) effect’s anti-neoplastic effectiveness of transplanted donor lymphocytes is constrained by tumor immune evasion and the restrictive preventive medicine required by the deadly graft-versus-host disease. Many germline-specific genes have also been shown to contribute to refusal and GvL, most notably minimal epitopes antigens, donor-recipient mismatches in common gene deletions, and donation polymorphisms outside the Haplotype in genes regulating, for example, immune response [3]. The alloimmunity capacity of the graft is primarily controlled by the genetic similarity of the human leukocyte antigen loci.
Mucosal inflammation brought on by cancer therapy can impact mucosa in the mouth, nose, and vagina. However, it is typically seen as inflammation and pain in the tongue, lips, and palate. Mucosal irritation brought on by treating cancer is caused by various pathobiology, dosage, medication, patient-related, and other unidentified variables [4,5]. One severe secondary effect of cancer therapy that necessitates hospitalization is severe oral mucositis, also known as ulcerative mucositis (UM). Reduced oral intake, bleeding, intense discomfort, and secondary dental problems are typical in cancer patients who develop UM; around half of them require adjustment of the cancer therapy, with a few resulting in treatment pauses. Up to 70% of people with cancer receiving high-dosage chemotherapy had an incidence of oral mucosal dermatitis, compared to 40% receiving a combination of conventional chemotherapy treatment therapy who are expected to experience this side effect. When receiving conditioning treatment before a bone marrow transplant, the UM incidence is abnormally high (70–80%). While concurrent chemotherapy and radiation to improve distant metastasis and central tumor control for head and neck malignancies, as well as continuous chemotherapy infusions for various cancers, result in severe UM in around 60–70% of patients [6,7].
Machine learning categorization enables decision-making in several facets of healthcare, including diagnosis, prediction, surveillance, treatment, and hospital administration. In classifiers for medical uses, accuracy is crucial [8,9]. The effectiveness of patients’ treatments may be improved by early illness diagnosis. Overcoming the curse of high dimensionality is one of the most crucial and essential challenges in any classifier, which serves as an incentive for utilizing an appropriate feature selection strategy. High dimensional datasets decrease the classification accuracy. The feature selection (FS) method is used to choose the most valuable characteristics from the provided medical datasets. It increases the predicted precision and decreases the calculation cost for illness diagnosis [10].
FS algorithms are data preprocessing techniques frequently used in classification or grouping tasks in data mining [11]. These techniques offer a condensed set of input attributes while maintaining the necessary discriminating data. High-dimensional datasets decrease the classification accuracy. The FS method chooses the most valuable characteristics from the medical datasets. It increases the predicted accuracy and decreases the calculation cost for illness diagnosis. FS algorithms are dimensionality reduction techniques frequently used in classification or clustering tasks in data mining. These techniques offer a condensed set of input attributes while maintaining the necessary discriminating data [12]. There are two types of FS: supervised and unsupervised. Unsupervised FS is suitable unless class labels for the data are given, in which case supervised feature selection is used. The filter, wrapper, and hybrid methods can be roughly grouped into three conventional FS methods [13].
Recently, there has been an emerging interest in finding a suitable alternative for patients with hematology where stem cells have been studied as a novel treatment. In addition, it has been shown that, in clinical application, it may be essential to identify patients suffering from unfavorable outcomes or who have a poor prognosis because they are genetically predisposed to developing acute graft-versus-host disease (GVHD) or transplant-related mortality. As part of an effort to promote the utilization of stem cells as a treatment for chronic and life-threatening diseases, in this paper, we proposed a new feature selection method using a new version of the Mud Ring Algorithm (MRA) for solving the survival prediction of children undergoing hematopoietic stem-cell transplantation. The proposed method used a modified MRA based on modifying the basic method’s exploration search ability. This paper proposes using the MRA as a feature selection method to select the optimal set of features from the bone marrow transplant; datasets have been collected from children to help improve the prediction models’ overall performance. MRA application for the feature selection problem was carried out by selecting the optimal subset of features from the training set, utilizing its improved optimization capability. The tests were initially carried out on 13 real datasets with varying occurrences and were allocated to evaluate the performance of the suggested approach categorization. The UCI machine learning repository provided the data that were chosen.
The rest of this paper is presented as follows. Section 2 presents the related work methods using optimization and machine learning techniques. Section 3 presents the proposed feature selection based on the Mud Ring Algorithm. Section 4 shows the experiments and results. Finally, the conclusions and future works are given in Section 5.
2. Related Work
A new meta-heuristic optimization technique called the Mud Ring Algorithm (MRA) is presented, which imitates the feeding behavior of bottlenose dolphins along Florida’s Atlantic coast [14]. MRA is primarily inspired by the mud ring feeding method and foraging behavior of bottlenose dolphins. By rapidly moving his tail in the sand and swimming around the school of fish, a single dolphin may use this technique to catch fish. When the fish gets lost, they jump above the surface and land in the dolphins’ waiting mouths. The MRA optimization method mimics this feeding approach physically and demonstrates the effectiveness of its optimization through a thorough comparison with other meta-heuristic methods. The statistical comparisons and outcomes demonstrated that the MRA outperforms previous meta-heuristic optimizers in handling various optimization problems and may produce the best solutions.
Due to its limited therapeutic range and considerable pharmacokinetic variability, Busulfan must be dosed correctly to minimize systemic toxicity from overexposure and graft failure from underexposure [15]. Biomarkers for predicting busulfan exposure are researched using global metagenomics. Fifty-nine pediatric patients were separated into three groups based on the area under the busulfan concentration-time curve (AUC), i.e., low-, medium-, and high-AUC groups [16]. They had urine samples collected prior to receiving busulfan. Deferoxamine metabolites were found in the high-AUC group. Compared to the low-AUC group, phenylacetylglutamine and two acylcarnitines were considerably lower in the high-AUC group. The high-AUC group had individuals with high ferritin levels because deferoxamine, an iron-chelating drug that reduces serum ferritin levels, was found in their blood. Therefore, they supported our theory that there is a significant negative relationship between serum ferritin level and busulfan absorption (dose/AUC) in a retrospective analysis of 130 pediatric patients.
This research [17] presents a new approach to feature selection and microarray data categorization. The initial step is to choose the essential genes (features) from the original collection of genes using a statistical filter called ANOVA. Then, a suggested evolutionary wrapper-based method to identify the ideal set of genes from the previously chosen genes uses the concepts of the improved Jaya (EJaya) method and forest optimization algorithm (FOA). Tuning the two crucial FOA parameters, mainly local seeding changes and global seeding adjustments, is the primary goal of utilizing EJaya. The suggested approach is examined on binary-class and multi-class microarray datasets to conduct extensive experimental research. It has been determined through a thorough review of the results that the suggested approach delivers more excellent classification performance with a fewer number of attributes than the benchmark system.
This work aims to collect information on DNA viruses and breast tumors to develop an accurate diagnosis model for fibroadenoma and breast cancer [17]. Research has consistently shown that the support vector machine (SVM) is more capable of making accurate diagnoses. In order to distinguish between breast cancer and fibroadenoma and to identify the critical risk factor for breast cancer, this study develops a hybrid SVM-based technique with feature selection. The SVM-based classifier’s categorical performance is marginally higher than LDA’s in terms of overall hit ratio, negative hit ratio, and positive hit ratio.
As a result of technological advancements in several industries, enormous amounts of data are produced. Medical datasets can have a high degree of complexity and small sample numbers. The classification of this high-dimensional data presents a challenge for researchers. In this study [18], they introduced a unique optimization strategy for better feature selection in the categorization of medical data. On 15 illness datasets with various feature sizes collected from UCI, the statistical findings of cmSAR were either the same or better than those of well-known metaheuristic algorithms and the original SAR method. All of the medical datasets were performed better by the suggested cmSAR.
This article presented an enhanced whale optimization algorithm, the Chaotic Multi-Squadron Whale Optimizer (CMWOA), which combines chaotic and multi-swarm techniques to conduct parameter optimization and feature selection concurrently for SVM [19]. The suggested SVM model was matched with numerous competitive SVM models based on other algorithms, using several clinical diagnosis issues of breast cancer, diabetes, and erythematous-squamous. The experimental findings show that the proposed method was considerably superior to all of its rivals in terms of classification accuracy and subset size.
The WOA is a well-known algorithm frequently used to resolve NP-hard issues, such as feature selection. However, it, and the majority of its versions, are plagued by a weak search technique and a population with little variety. Therefore, implementing effective tactics to address the feature selection issue and other fundamental WOA flaws is crucial. Because there are three efficient search strategies—migrating, selective picking, and enriched encircling prey—this study is devoted to developing an improved whale optimization algorithm called E-WOA [20]. To resolve global optimization issues, the effectiveness of E-WOA is assessed and contrasted with standard WOA versions. The data gathered demonstrated that the E-WOA performs better than the WOA versions. After E-WOA had shown comparable stability, a binary E-WOA called BE-WOA was proposed to pick beneficial characteristics, notably from medical datasets. In terms of fitness, accuracy, sensitivity, precision, and quantity of features, the BE-WOA is evaluated with the most recent high-performing optimization algorithms and verified using datasets related to medical conditions. The BE-WOA is also used to identify COVID-19 illness.
This study suggests a wrapper feature selection (FS) using an improved moth flame optimization (MFO) method. Its primary goal is to enhance the categorization tasks used in medical applications [21]. This study used a freshly created approach to find a nearly ideal feature subset for accurate illness detection. The moth movement style inspired the methodology. The two steps of enhancement are the foundation of the suggested modification technique. The proposed methods are validated using 23 medical datasets collected from UCI, Keel, and Kaggle data sources. The Levy flight operator and transfer functions have a striking impact on the functionality of MFO, as shown by the empirical results and numerous comparisons. Table 1 shows an overview of the selected related works.

Table 1.
An overview of the selected related works.
3. Methodology
Dolphins use different strategies to hunt and feed on schools of fish, such as shrimp, squid, and mollusk. They work together to maximize their harvest of a large number of species. Group hunting includes a whacking strategy [22], strand feeding [23], and mud ring feeding [24]. Mud ring fishing is a unique hunting strategy used by dolphins to encircle a school of fish. In this process, the dolphins create circular waves with their tail, which disorientates the position of the school of fish, trapping them against a sandbar or reef, making them easier to pick up by mouth. This unique strategy used by bottlenose dolphins to search and trap prey inspired the Mud Ring Algorithm (MRA) optimizer technique. The Mud Ring Algorithm (MRA) is a simulation that recreates the hunting behavior of bottlenose dolphins and models their mud ring formation as they seek prey utilizing echolocation.
The Mud Ring Algorithm identifies the paths where the dolphins’ swarm gets closer to prey each time, shown with the K parameter. The K parameter shows that, as the hunting process progresses, the dolphin swarm draws closer and closer to its victim and the volume of the swarm’s sounds decreases as it gets closer to the target. Consequently, it regulates the transition between the exploration and exploitation stages of looking for prey (mud rings). The MRA algorithm explores worldwide and looks for better solutions in the exploitation process during the exploration phase. Exploration occurs in the search space when this parameter is large, while it tends to exploit when this value is low.
3.1. Exploration and Hunting for Prey Phase
Let us idealize some of the characteristics of dolphins’ echolocation; we operate under the following rules: Dolphins use a combination of velocities, , and positions, , as well as sound loudness, , to search for prey using echolocation. All dolphins are capable of automatically adjusting the volume of the sounds they produce in response to the proximity of their prey; even though the volume could change in a variety of ways, we hypothesize that the volume changes in response to the time step and the pulse rate, ‘r’, which can range from 0 to 1, where 0 indicates that there are no emission pulses and 1 indicates a highest pulse emission rate. The following are the computations for the vector :
where is a random vector ranging from 0 to 1, and:
Virtual dolphins are organically employed as search agents to find prey (exploring). Dolphins search in a completely random position within a parameter space with d-dimensions. The dolphins’ relative positions define their position to one another. Therefore, in order to encourage the dolphins to part ways with one another and search for the fittest prey, we need a variable called that either has a value greater than 1 or a value that is less than 1; thus, a dolphin chosen at random is selected rather than the best dolphin in the group. This selection mechanism, in conjunction with 1, encourages exploration and makes it possible for the MRA algorithm to conduct a global search. The mathematical derivation of the MRA algorithm is presented in the following.
It is essential to provide up-to-date criteria for periodically refreshing the positions and velocities. The following provides the workability () depending on the velocity () at time step t by:
where is a random velocity that is initialized. Each dolphin is initially assigned a random velocity in the range of based on the challenge size [14].
3.2. Feeding and Exploitation Phase of the Mud Ring
Dolphins can locate their prey and surroundings after they have detected them. However, the optimal design’s location in the search space is unknown a priori; hence, the MRA technique considers the target prey (optimal or a solution that is close to it) as the current best solution. As a result, once the best search agent has been identified, the other dolphins will seek to update their positions by the best dolphin position. These equations describe this behavior:
where are coefficient vectors and is the dolphin position vector, while is the position vector of the best dolphin position thus far; t denotes the current time step and l is a random number.
Every time step, , positions should be adjusted if there is a better position available. To make a plume, the best dolphin circles its target while waving its tail quickly in the sand, creating a sine waveform, while the rest of the dolphins continue to circle the prey. The following formula is used to calculate the vector, :
By finding the random vector , any position inside the search area can be located. Thus, Equation (5) acts as a simulation of the prey encircling, and it assists any dolphin in justifying its location as being near the current optimal position.
The MRA’s search process begins with a population of randomly selected solutions (dolphin positions). Dolphins justify their locations at each time step based on either the optimum site available so far or a randomly selected dolphin. As a result, the parameter depends on the time step taken to move from exploration to use. When , the optimal position for the dolphins is chosen, whereas at , a dolphin is elected at random to justify dolphin positions. Finally, it is worth noting that the MRA algorithm has only two basic parameters that can be modified (C and K).
This paper proposes using the MRA as a feature selection method to select the optimal set of features from the bone marrow transplant; datasets have been collected from children to help improve the prediction models’ overall performance. MRA application for the feature selection problem is carried out by selecting the optimal subset of features from the training set, utilizing the improved optimization capability of the previously published MRA algorithm [14], which begins its search with randomly generated search individuals. The binary encoding type is used as the suggested algorithm’s representation scheme to pick the most optimal features. In this type of encoding (binarization), each search is represented as its distinct entry in a binary array, and the features are regarded as either being present (1) or absent (0). The 1s stand for the traits that were kept from the training set, whereas the zeroes denote eliminated elements. As a measure of an individual’s overall fitness, the F-Meas of search individuals in MRA is calculated and analyzed, as shown in Equation (11).
The MRA feature selection method is depicted in Pseudocode, as given in Algorithm 1, and its application to real-world datasets is discussed in the subsequent section. The initial dataset is then segmented into training, validation, and testing subsets. The proposed MRA looks for the best possible feature subset from the training set and then puts that subset through its paces on the validation set. The best search individual is presumed to have converged once the termination criteria have been met. Finally, this search individual with the highest possible fitness value is decoded to find the solution, which consists of a feature set with fewer elements.
| Algorithm 1: MRA Algorithm |
| 1. Initialize the population of dolphins randomly, Di, i ∈ [1, 2, ..., n] and velocity vi 2. Evaluate the fitness function of each dolphin 3. Find the best dolphin position (features set) D* 4. while ( < ) 5. for i = 1 to n 6. Modify , , , and 7. if |K| >= 1 then Generate new solutions by modifying velocity vi using Equation (3) 8. Else /* Forming the mud ring*/ Update the current dolphin location using Equation (5) 9. end if 10. end for 11. Update the bounds for dolphins outside the search space 12. Attain the dolphin’s fitness functions 13. Update D* in case of a better position (features) 14. Set 15. end while Return D* (the best subset of features) |
The flow diagram of the implication of the MRA method in the feature selection issue for the datasets of real-world is depicted in Figure 1. As the diagram shows, the initial dataset is first divided into three sub-sets: train, test, and validation. The proposed MRA aims basically to find the best sub-set of features from the set of training data, which is then tested using the set of validation data. After satisfying the termination criteria, the optimal search agent is considered as converged, and the search agent with the best fitness value is then decoded to be the solution that presents the reduced sub-set of features.
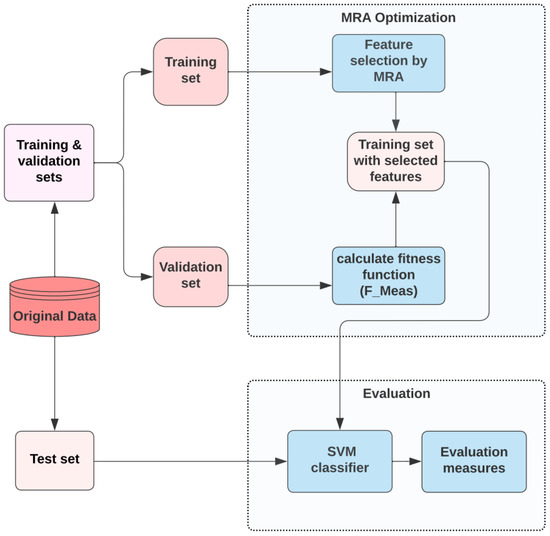
Figure 1.
Flow diagram of MRA for feature selection method in classification.
The experiments are performed to assess the MRA feature selection method performance, and the modern feature selection method results are used in comparison with the MRA results. In this experiment, the Support Vector Machine classifier (SVM) [25] is the classifier used to test the classification performance of the reduced sub-set of data. Various evaluation metrics are used, and all are defined in Equations (7)–(11).
For evaluation metrics, if , , and represent the true negatives, the true positives, the false negatives, and the false positives, respectively [26,27], then:
4. Results and Discussion
Firstly, the experiments were performed on thirteen different real-world datasets with different numbers of instances and attributes to assess the proposed method’s performance. These data were chosen from the UCI machine learning repository [28]. Their results of classification were obtained from comparison between the modern methods (Archimedes optimization algorithm (AOA) [22], Jellyfish Search (JS) [29], Ant Lion Optimizer (ALO) [23], Whale Optimization Algorithm (WOA) [24,30], Heap-based Optimizer (HBO) [31], and Equilibrium Optimizer (EO) [32]) and the proposed method. Table 2 shows the characteristics of the real-world datasets.

Table 2.
Characteristics of the real-world datasets.
Each dataset instance was randomly divided into three subsets in all experiments: train, validation, and test. For each dataset, the feature selection was performed using the proposed MRA feature selection method and the modern methods, while the SVM classifier was used as an evaluator for the selected features. To measure the performance of our proposal, 30 runs were performed independently, and the ACC, F-Meas, G-Mean, and Time metrics for each dataset were recorded for all used algorithms; then, each metric average was calculated. In addition, the selected feature number (# features) was recorded for each method. Table 3 shows the performances of all feature selection methods used in the comparison of real-world datasets. As reference, the first row is preserved for the original dataset (Or) results that consists of full features.

Table 3.
Performance measures for real-world datasets.
The results in Table 3 show that the MRA is better than the modern compared methods in terms of classification accuracy on nine datasets out of thirteen data, where the HBO method reaped the second-best order of ACC for six data only. Furthermore, for the G-Mean and F-Meas metrics, our proposal also occupies the best values for seven datasets out of the thirteen used data, which proves the MRA feature selection method’s stability against other modern methods. On the other hand, it can be noticed from Table 3 and Figure 2 that, although decreasing the number of features as in “yeast-2_vs_4” and “Segment (BRICKFACE-others) “data, the MRA method can keep the same level of performance gained by all feature sets, while for all other datasets, the MRA approach obtained an improved performance compared with the performance obtained by all of the feature sets. Based on these previous results, one can conclude that the proposed method for feature selection (MRA) can obtain a better performance in terms of computational complexity than the other algorithms because it has fewer features, which yields lower runtime and less space while guaranteeing the best performance.
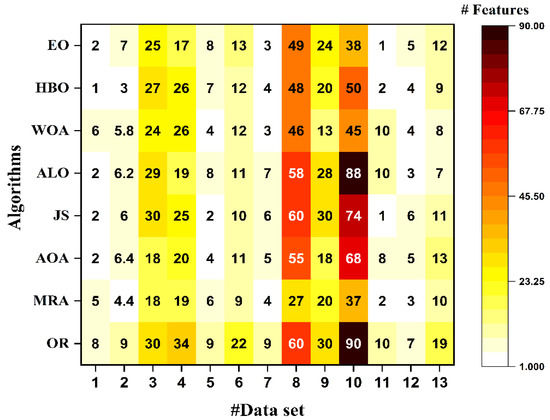
Figure 2.
Comparison between the feature selection values of the proposed algorithm and other algorithms.
The next step involved applying the proposed method to the bone marrow transplant children’s dataset to select the best feature (see Table A1), which helps improve the survival prediction performance of children undergoing hematopoietic stem-cell transplantation. The classification performance results of 30 independent runs on the bone marrow transplant children’s dataset are depicted in Figure 3, along with a comparison of the classification performance obtained using the AOA, JS, ALO, WOA, HBO, and EO methods. Malignant illnesses (such as acute lymphoblastic leukemia, acute myelogenous leukaemia, chronic myelogenous leukaemia, and myelodysplastic syndrome) and nonmalignant instances are all described in this dataset for children with hematologic diseases (i.e., severe aplastic anemia, Fanconi anemia, with X-linked adrenoleukodystrophy). Unrelated donor hematopoietic stem-cell transplantation was performed on all patients without any manipulation.
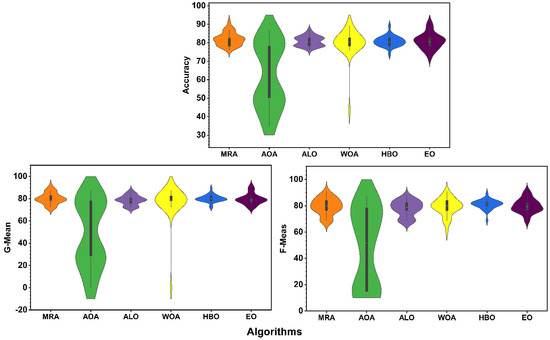
Figure 3.
A violin plot illustrating a full performance distribution of classifiers.
The violin plots illustrated in Figure 3 show the ACC, G-Mean, and F-Meas for the proposed MRA method and all compared methods. A violin plot is a hybrid of box and kernel density plots, which presents peaks in the data. These plots are commonly used to visualize the numerical data distribution and show summary statistics with the density of each variable. The distribution shape is extremely wide in the middle and skinny at the two ends for the MRA, ALO, HBO, and EO methods for the three used measures. In addition, ACC, G-Mean, and F-Meas for the MRA method are highly concentrated around the median, which directly indicates that the MRA algorithm occupies the first order in achieving the best performance in terms of ACC, G-Mean, and F-Meas and is then followed by HBO, EO, and ALO.
The violin plot elements in Figure 3 show that the median ACC, G-Mean, and F-Meas for the AOA method is lower than other compared methods. Additionally, the WOA method shows outlier points in ACC and G-Mean; the comparison leads us to remove both from the next comparison figure (Figure 4).
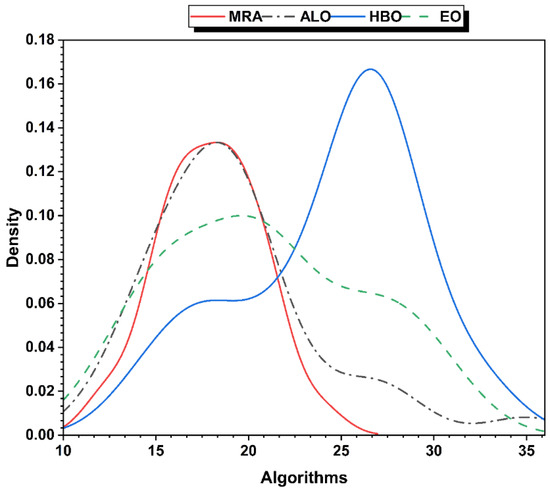
Figure 4.
FS distributions of selected algorithms.
For more validation to our results, a statistical significance analysis is applied. A paired difference, two-sided signed-rank test “Wilcoxon signed-rank test”, which is a non-parametric statistical hypothesis [33] is used here for the thirteen real-world datasets beside the bone marrow transplant children’s dataset to fairly derive strong conclusions. The proposed MRA is compared with the three selected methods for each dataset. From the smallest, 1, to the largest, 14, all the ranked differences were recorded for each two compared methods; r and r− were assigned to the positive and the negative ranks after summing up separately. The significance level alpha “α” equals 0.05, used in comparison with the “T” value, has a critical value, which equals 21 for 14 datasets, while T = min {r, r−}. The null hypothesis—all differences in the performance between any two compared methods may be found by chance—was rejected only if the “T” value was less than or equal to 21 (critical value).
Table 4, Table 5 and Table 6 present the significance test results; note that the NaN values were replaced by the value −1 in the three tables, in order for them to be excluded by being at the bottom of the ranked list.

Table 4.
Wilcoxon signed-rank for test ACC.

Table 5.
Wilcoxon signed-rank for test F-Meas.

Table 6.
Wilcoxon signed-rank for test G-mean.
The results of the significance test of average ACC for MRA vs. ALO, WOA, HBO, and EO are presented in Table 4. In the case of MRA vs. EO, MRA has a positive difference for 13 datasets; hence, it is better than this algorithm, while EO has a negative difference and is better than MRA for only one dataset. The sum of all the positive ranks, R, is 86, and the sum of negative ranks, R−, is 18 for MRA vs. EO, respectively. Therefore, it can be concluded that MRA statistically outperforms EO. In addition, in the case of MRA vs. ALO and HBO, MRA statistically outperforms these algorithms where the values of T are 0 and 17. On the other hand, the results of the significance test of average F-Meas and G-mean for MRA vs. ALO, HBO, and EO are presented in Table 5 and Table 6. In the case of MRA vs. HBO and EO, MRA has a positive difference for 12 datasets; hence, it is better than these algorithms, while HBO and EO have a negative difference and are better than MRA for only two datasets. Based on the sum of all the positive and negative ranks (R and R−), it is clear that MRA performs better statistically than ALO and HBO, but not better than EO.
The feature selection distributions of the selected algorithms (MRA, ALO, HBO, and EO) are presented in Figure 4. MRA shows the most uniform distribution curve for the density of selected features than other compared algorithms while keeping the lower number of features (less than 30) through different iterations. The HBO algorithm shows the highest density of selected features with non-uniform curve, so it will be removed from the next comparison.
Figure 5 presents the bar chart for the selected feature numbers and time measures of the selected algorithms. Regarding the number of selected features, the proposed MRA shows the lowest number and is first in the order of the three selected algorithms. The ALO algorithm comes after the MRA in comparing the number of selected features, while the EO algorithm holds the last order in the comparison.
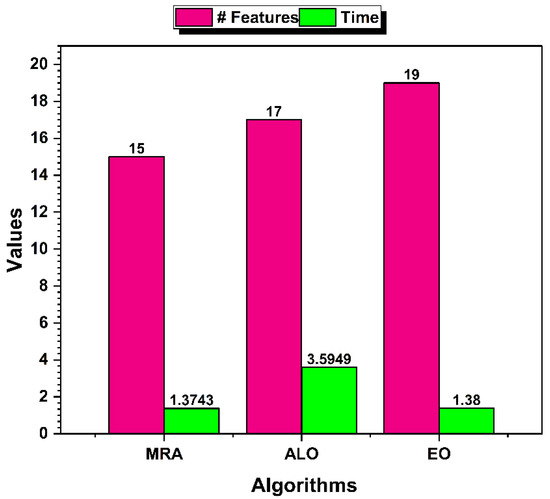
Figure 5.
Selected feature numbers and Time measures of selected algorithms.
Regarding the run time, the MRA algorithm shows the lower run time and comes first in the order of the three selected algorithms. The EO algorithm comes after the MRA in comparing the run time, while the ALO algorithm keeps the last order in this comparison. The results and attributes chosen by the best three algorithms are displayed in Table 7. It is clear that the suggested MRA is superior to the other compared algorithms, even though all three selected the same seven features (underlined for emphasis).

Table 7.
The performance metrics and selected features by the selected algorithms.
Finally, all the results of the preceding statistical tests prove that the MRA feature selection method can statistically outperform other algorithms and that it has strong competitiveness in terms of all the performance measures utilized. However, the proposed algorithm is plagued mostly by the drawback of having a randomized nature, because it is based on using an optimization algorithm that is performed by hundreds of iterations and is started with initial random solutions.
5. Conclusions
This research proposes the MRA algorithm applied to the SVM classifier as a feature selection algorithm to find the optimal features. Thirteen real-world datasets are taken into account to assess the performance of our proposal. The tested datasets were chosen from the UCI Machine Learning repository. Each investigated dataset is divided randomly into three training, validation, and testing sub-sets. To measure the performance of each dataset, the ACC, F-Meas, G-Mean, and the # features for each algorithm were recorded. As a result, the MRA shows superior and comparable results in term of feature selection and classification performance when compared to the AOA, JS, ALO, WOA, HBO, and EO algorithms. After proving its validity, the proposed MRA feature selection algorithm is applied to the bone marrow transplant children’s dataset to select the best features that can be used to achieve the best prediction performance.
Compared to the state-of-the-art methods, the MRA feature selection method showed superiority over other methods in the survival prediction of children undergoing hematopoietic stem-cell transplantation. The constructed model generated a higher numerical range of the significance test values, corresponding to maximum F-Meas values of 83.33 for bone marrow transplant data. Moreover, the accuracy of the results obtained using the constructed model for all test cases used in the present work exceeded 82%. It has been proven to be quicker than other algorithms, with high accuracy.
In future studies, multi-objective versions of MRA can be developed. Furthermore, it can address various issues in renewable energy and other disciplines. Another promising direction to explore is comparing various constraint-handling strategies for solving real-world constrained problems.
Author Contributions
Conceptualization, L.M.E.B. and N.B.; methodology and software validation L.M.E.B., A.S.D., and S.U.; formal analysis and writing—original draft, A.S.D., L.M.E.B., N.B., and L.A.; supervision, N.B.; writing—review and editing, N.B., L.A., and S.U.; visualization, S.U., L.M.E.B., N.B., and L.A.; funding acquisition, S.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R79), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Data Availability Statement
Not applicable.
Acknowledgments
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R79), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no-conflict of interest.
Appendix A

Table A1.
Dataset attributes.
Table A1.
Dataset attributes.
| Attribute | Information |
|---|---|
| Donor_age | Age of the donor at the time of hematopoietic stem cells apheresis |
| donor_age_below_35 | Is donor age less than 35 (yes, no) |
| donor_ABO | ABO blood group of the donor of hematopoietic stem cells (0, A, B, AB) |
| donor_CMV | Presence of cytomegalovirus infection in the donor of hematopoietic stem cells prior to transplantation (present, absent) |
| recipient_age | Age of the recipient of hematopoietic stem cells at the time of transplantation |
| recipient_age_below_10 | Is recipient age below 10 (yes, no) |
| recipient_age_int | Age of the recipient discretized to intervals (0,5], (5, 10], (10, 20] |
| recipient_gender | Gender of the recipient (female, male) |
| recipient_body_mass | Body mass of the recipient of hematopoietic stem cells at the time of the transplantation |
| recipient_ABO | ABO blood group of the recipient of hematopoietic stem cells (0, A, B, AB) |
| recipient_rh | Presence of the Rh factor on recipient red blood cells (plus, minus) |
| recipient_CMV | Presence of cytomegalovirus infection in the donor of hematopoietic stem cells prior to transplantation (present, absent) |
| disease | Type of disease (ALL, AML, chronic, nonmalignant, lymphoma) |
| disease_group | Type of disease (malignant, nonmalignant) |
| gender_match | Compatibility of the donor and recipient according to their gender (female to male, other) |
| ABO_match | Compatibility of the donor and the recipient of hematopoietic stem cells according to ABO blood group (matched, mismatched) |
| CMV_status | Serological compatibility of the donor and the recipient of hematopoietic stem cells according to cytomegalovirus infection prior to transplantation (the higher the value, the lower the compatibility) |
| HLA_match | Compatibility of antigens of the main histocompatibility complex of the donor and the recipient of hematopoietic stem cells (10/10, 9/10, 8/10, 7/10) |
| HLA_mismatch | HLA matched or mismatched |
| antigen | In how many antigens there is a difference between the donor and the recipient (0–3) |
| allel | In how many allele there is a difference between the donor and the recipient (0–4) |
| HLA_group_1 | The difference type between the donor and the recipient (HLA matched, one antigen, one allel, DRB1 cell, two allele or allel+antigen, two antigenes+allel, mismatched) |
| risk_group | Risk group (high, low) |
| stem_cell_source | Source of hematopoietic stem cells (peripheral blood, bone marrow) |
| tx_post_relapse | The second bone marrow transplantation after relapse (yes ,no) |
| CD34_x1e6_per_kg | CD34kgx10d6-CD34+ cell dose per kg of recipient body weight (106/kg) |
| CD3_x1e8_per_kg | CD3+ cell dose per kg of recipient body weight (108/kg) |
| CD3_to_CD34_ratio | CD3+ cell to CD34+ cell ratio |
| ANC_recovery | Neutrophils recovery defined as neutrophils count > 0.5 × 109/L (yes, no) |
| time_to_ANC_recovery | Time in days to neutrophils recovery |
| PLT_recovery | Platelet recovery defined as platelet count > 50,000/mm3 (yes, no) |
| time_to_PLT_recovery | Time in days to platelet recovery |
| acute_GvHD_II_III_IV | Development of acute graft versus host disease stage II or III or IV (yes, no) |
| acute_GvHD_III_IV | Development of acute graft versus host disease stage III or IV (yes, no) |
| time_to_acute_GvHD_III_IV | Time in days to development of acute graft versus host disease stage III or IV |
| extensive_chronic_GvHD | Development of extensive chronic graft versus host disease (yes, no) |
| relapse | Relapse of the disease (yes, no) |
| survival_time | Time of observation (if alive) or time to event (if dead) in days |
| survival_status | Survival status (0—alive, 1—dead) |
References
- Ritari, J.; Hyvärinen, K.; Koskela, S.; Itälä-Remes, M.; Niittyvuopio, R.; Nihtinen, A.; Salmenniemi, U.; Putkonen, M.; Volin, L.; Kwan, T.; et al. Genomic Prediction of Relapse in Recipients of Allogeneic Haematopoietic Stem Cell Transplantation. Leukemia 2019, 33, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-J.; Davies, S.M.; Camitta, B.M.; Logan, B.; Tiedemann, K.; Eapen, M.; Thiel, E.L. Comparison of Outcomes after HLA-Matched Sibling and Unrelated Donor Transplantation for Children with High-Risk Acute Lymphoblastic Leukemia. Biol. Blood Marrow Transpl. 2012, 18, 1204–1210. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fürst, D.; Müller, C.; Vucinic, V.; Bunjes, D.; Herr, W.; Gramatzki, M.; Schwerdtfeger, R.; Arnold, R.; Einsele, H.; Wulf, G.; et al. High-Resolution HLA Matching in Hematopoietic Stem Cell Transplantation: A Retrospective Collaborative Analysis. Blood 2013, 122, 3220–3229. [Google Scholar] [CrossRef] [PubMed]
- Satheeshkumar, P.S.; El-Dallal, M.; Mohan, M.P. Feature Selection and Predicting Chemotherapy-Induced Ulcerative Mucositis Using Machine Learning Methods. Int. J. Med. Inform. 2021, 154, 104563. [Google Scholar] [CrossRef] [PubMed]
- Lalla, R.V.; Sonis, S.T.; Peterson, D.E. Management of Oral Mucositis in Patients Who Have Cancer. Dent. Clin. N. Am. 2008, 52, 61–77. [Google Scholar] [CrossRef]
- Berger, K.; Schopohl, D.; Bollig, A.; Strobach, D.; Rieger, C.; Rublee, D.; Ostermann, H. Burden of Oral Mucositis: A Systematic Review and Implications for Future Research. Oncol. Res. Treat. 2018, 41, 399–405. [Google Scholar] [CrossRef]
- Blijlevens, N.; Schwenkglenks, M.; Bacon, P.; D’Addio, A.; Einsele, H.; Maertens, J.; Niederwieser, D.; Rabitsch, W.; Roosaar, A.; Ruutu, T.; et al. Prospective Oral Mucositis Audit: Oral Mucositis in Patients Receiving High-Dose Melphalan or BEAM Conditioning Chemotherapy—European Blood and Marrow Transplantation Mucositis Advisory Group. J. Clin. Oncol. 2008, 26, 1519–1525. [Google Scholar] [CrossRef]
- Abualigah, L.M.Q. Krill Herd Algorithm. In Feature Selection and Enhanced Krill Herd Algorithm for Text Document Clustering; Springer: Cham, Switzerland, 2018; pp. 11–19. [Google Scholar]
- Elaziz, M.A.; Abualigah, L.; Yousri, D.; Oliva, D.; Al-Qaness, M.A.A.; Nadimi-Shahraki, M.H.; Ewees, A.A.; Lu, S.; Ali Ibrahim, R. Boosting Atomic Orbit Search Using Dynamic-Based Learning for Feature Selection. Mathematics 2021, 9, 2786. [Google Scholar] [CrossRef]
- Mostafa, R.R.; Ewees, A.A.; Ghoniem, R.M.; Abualigah, L.; Hashim, F.A. Boosting Chameleon Swarm Algorithm with Consumption AEO Operator for Global Optimization and Feature Selection. Knowl.-Based Syst. 2022, 246, 108743. [Google Scholar] [CrossRef]
- Wu, D.; Jia, H.; Abualigah, L.; Xing, Z.; Zheng, R.; Wang, H.; Altalhi, M. Enhance Teaching-Learning-Based Optimization for Tsallis-Entropy-Based Feature Selection Classification Approach. Processes 2022, 10, 360. [Google Scholar] [CrossRef]
- Abualigah, L.; Diabat, A. Chaotic Binary Group Search Optimizer for Feature Selection. Expert Syst. Appl. 2022, 192, 116368. [Google Scholar] [CrossRef]
- Jiang, Y.; Luo, Q.; Wei, Y.; Abualigah, L.; Zhou, Y. An Efficient Binary Gradient-Based Optimizer for Feature Selection. Math. Biosci. Eng. 2021, 18, 3813–3854. [Google Scholar] [CrossRef]
- Desuky, A.S.; Cifci, M.A.; Kausar, S.; Hussain, S.; El Bakrawy, L.M. Mud Ring Algorithm: A New Meta-Heuristic Optimization Algorithm for Solving Mathematical and Engineering Challenges. IEEE Access 2022, 10, 50448–50466. [Google Scholar] [CrossRef]
- Shehab, M.; Abualigah, L.; Shambour, Q.; Abu-Hashem, M.A.; Shambour, M.K.Y.; Alsalibi, A.I.; Gandomi, A.H. Machine Learning in Medical Applications: A Review of State-of-the-Art Methods. Comput. Biol. Med. 2022, 145, 105458. [Google Scholar] [CrossRef]
- Kim, B.; Lee, J.W.; Hong, K.T.; Yu, K.-S.; Jang, I.-J.; Park, K.D.; Shin, H.Y.; Ahn, H.S.; Cho, J.-Y.; Kang, H.J. Pharmacometabolomics for Predicting Variable Busulfan Exposure in Paediatric Haematopoietic Stem Cell Transplantation Patients. Sci. Rep. 2017, 7, 1711. [Google Scholar] [CrossRef]
- Baliarsingh, S.K.; Vipsita, S.; Dash, B. A New Optimal Gene Selection Approach for Cancer Classification Using Enhanced Jaya-Based Forest Optimization Algorithm. Neural Comput. Appl. 2019, 32, 8599–8616. [Google Scholar] [CrossRef]
- Houssein, E.H.; Saber, E.; Ali, A.A.; Wazery, Y.M. Centroid Mutation-Based Search and Rescue Optimization Algorithm for Feature Selection and Classification. Expert Syst. Appl. 2022, 191, 116235. [Google Scholar] [CrossRef]
- Wang, M.; Chen, H. Chaotic Multi-Swarm Whale Optimizer Boosted Support Vector Machine for Medical Diagnosis. Appl. Soft Comput. 2020, 88, 105946. [Google Scholar] [CrossRef]
- Nadimi-Shahraki, M.H.; Zamani, H.; Mirjalili, S. Enhanced Whale Optimization Algorithm for Medical Feature Selection: A COVID-19 Case Study. Comput. Biol. Med. 2022, 148, 105858. [Google Scholar] [CrossRef]
- Abu Khurmaa, R.; Aljarah, I.; Sharieh, A. An Intelligent Feature Selection Approach Based on Moth Flame Optimization for Medical Diagnosis. Neural Comput. Appl. 2020, 33, 7165–7204. [Google Scholar] [CrossRef]
- Hashim, F.A.; Hussain, K.; Houssein, E.H.; Mabrouk, M.S.; Al-Atabany, W. Archimedes Optimization Algorithm: A New Metaheuristic Algorithm for Solving Optimization Problems. Appl. Intell. 2020, 51, 1531–1551. [Google Scholar] [CrossRef]
- Mirjalili, S. The Ant Lion Optimizer. Adv. Eng. Softw. 2015, 83, 80–98. [Google Scholar] [CrossRef]
- Mirjalili, S.; Lewis, A. The Whale Optimization Algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- El-Kenawy, E.-S.M.; Zerouali, B.; Bailek, N.; Bouchouich, K.; Hassan, M.A.; Almorox, J.; Kuriqi, A.; Eid, M. Improved Weighted Ensemble Learning for Predicting the Daily Reference Evapotranspiration under the Semi-Arid Climate Conditions. Environ. Sci. Pollut. Res. 2022, 29, 81279–81299. [Google Scholar] [CrossRef] [PubMed]
- Desuky, A.S.; Hussain, S.; Kausar, S.; Islam, M.A.; El Bakrawy, L.M. EAOA: An Enhanced Archimedes Optimization Algorithm for Feature Selection in Classification. IEEE Access 2021, 9, 120795–120814. [Google Scholar] [CrossRef]
- Gong, M. A Novel Performance Measure for Machine Learning Classification. Int. J. Manag. Inf. Technol. 2021, 13, 11–19. [Google Scholar] [CrossRef]
- Asuncion, A.; Newman, D. UCI Machine Learning Repository; UCI: San Diego, CA, USA, 2007. [Google Scholar]
- Chou, J.-S.; Truong, D.-N. A Novel Metaheuristic Optimizer Inspired by Behavior of Jellyfish in Ocean. Appl. Math. Comput. 2021, 389, 125535. [Google Scholar] [CrossRef]
- El-kenawy, E.-S.M.; Ibrahim, A.; Bailek, N.; Bouchouicha, K.; Hassan, M.A.; Jamei, M.; Al-Ansari, N. Sunshine Duration Measurements and Predictions in Saharan Algeria Region: An Improved Ensemble Learning Approach. Theor. Appl. Climatol. 2022, 147, 1015–1031. [Google Scholar] [CrossRef]
- Askari, Q.; Saeed, M.; Younas, I. Heap-Based Optimizer Inspired by Corporate Rank Hierarchy for Global Optimization. Expert Syst. Appl. 2020, 161, 113702. [Google Scholar] [CrossRef]
- Faramarzi, A.; Heidarinejad, M.; Stephens, B.; Mirjalili, S. Equilibrium Optimizer: A Novel Optimization Algorithm. Knowl.-Based Syst. 2020, 191, 105190. [Google Scholar] [CrossRef]
- Sheskin, D.J. Handbook of Parametric and Nonparametric Statistical Procedures; Chapman and Hall/CRC: London, UK, 2003; ISBN 0429186169. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).