1. Introduction
Biological signals can be gathered from a wide range of physiological sensors which are frequently used in different diagnostic and experimental settings. The digitized sensor outputs are time series data encoding information about the underlying biological processes and phenomena. It is, therefore, of interest to develop robust signal and data processing methods for the automatic extraction of relevant information from the collected data. In addition to various methods of classical statistical and causal inference, the regression and classification of time series data require their representation as feature vectors, such as the parameters of time series data models.
Time series data are often modeled as an autoregressive integrated moving average (ARIMA). A comprehensive overview of ARIMA modeling can be found in [
1]. The ARIMA-based clustering and classification of time series data is investigated, for example, in [
2,
3]. The order of ARIMA models can be determined by the Akaike information criterion (AIC) [
4,
5]. In this paper, we develop a new strategy for reducing the order of ARIMA models by introducing a novel technique referred to as signal folding. It should not be confused with other techniques referred to by the same term, which sometime appear in the literature, and which may refer, respectively, to frequency folding in signal sampling, simple time reversal of a signal, or to a technique for reducing the required number of elements in digital circuits. The signal folding method proposed in this paper partitions the signal into multiple fragments which are then linearly combined so that the signal variability is reduced. The fragment combining, including ordinary arithmetic averaging, requires the proper scaling and alignment of fragments to avoid the information aliasing. The proposed signal folding crucially exploits a cyclostationary property [
6] inherently present in many biological signals. The signal folding preserves the signal mean, whereas the signal variance is substantially reduced. More importantly, the folding of labeled signals for training machine learning models can greatly simplify the feature extraction since the required order of the underlying data models can be considerably reduced. It then leads to an order of magnitude speed-up in training the machine learning models.
The benefits of signal folding in this paper are demonstrated for a case of sleep apnea detection in a single-lead ECG signal. Sleep apnea is potentially a serious health condition marked by abnormal breathing activity in sleep, sleep fragmentation, intermittent hypoxia, systemic inflammation imbalance, and other serious health problems [
7,
8]. Sleep apnea in suspected patients can be detected by monitoring changes in their ECG patterns, although most patients remain undiagnosed. The ECG-based detection of sleep apnea can be administered outside specialized medical clinics, and it is often used prior to more complex but more indicative multi-channel ECG and EEG polysomnography. However, these methods require the overnight monitoring of patients, which hinders their widespread use. Sleep apnea detection from a single lead ECG signal can, therefore, become a viable alternative to polysomnography [
9].
Different features for detecting sleep apnea in ECG signals were investigated in [
10]. The R-peaks and the distances between R-peaks were found to be the most discriminating. The study of [
11] evaluated the accuracy of detecting sleep apnea in the ECG signal over different signal durations. It was found that there is an optimal ECG segment length, providing the most reliable apnea detection. The detection of arrhythmic conditions in ECG signals was studied in [
12]. The signal noise was suppressed in a frequency domain using the fast Fourier transform (FFT). The R-peaks and other waveforms of ECG signal were detected, and modeled as polynomials. The polynomial coefficients were then used as feature vectors for the random forest classifier. A singular value decomposition (SVD), empirical mode decomposition, and the FFT were used in [
13] to extract the features for ECG signal classification. The discrete wavelet transform following signal de-noising was studied in [
14] to identify the P, Q, R and S peaks, allowing to classify the ECG signals. The robustness of ECG classification to a sampling jitter was investigated in [
15] assuming the R-peak intervals, and other signal morphology and higher-order statistical features. The features for ECG signal classification in [
16] were defined by fitting mathematical expressions to the QRS waveforms. The R-peak intervals as well as amplitudes were exploited in [
9] to detect sleep apnea in a single lead ECG signal. In addition, it was shown in [
9] that assuming previous classification decisions can improve the per-segment classification accuracy from 80% to 86%.
The ARIMA model for low-frequency and high-frequency components of the ECG signal was used in [
17] to extrapolate future signal values. It was suggested to use smoothing of the ECG signal prior to further processing. A moving average filtering for the noise removal in ECG signals was analyzed in [
18]. A non-stationary ARIMA model of ECG signals with maximum-likelihood estimation of the model parameters was considered in [
19]. The ARIMA coefficients were then used to detect sleep apnea using different classifiers, including support vector machine (SVM), artificial neural network (ANN), quadratic and linear discriminant analysis, and the K-nearest neighbor (K-NN) classifier.
The detection of sleep apnea in ECG signal had 90% accuracy in [
10,
11]. Ref. [
19] reported an accuracy of 81% for an ARIMA model with 8 coefficients. The binary classification of arrhythmic conditions in the ECG signals reached 91% accuracy in [
12]. The R-peak based signal folding investigated in our numerical experiments can achieve 90% accuracy, but with significantly reduced complexity. In particular, this accuracy was achieved for an ARIMA model having only 3 coefficients enabled by the signal folding pre-processing. It leads to substantial, order-of-magnitude time savings in training the sleep apnea classifiers.
The rest of this paper is organized as follows. Signal folding and the associated folding operator are formally introduced, and their properties are analyzed in
Section 2.
Section 3 describes the classification procedure involving signal folding and the corresponding algorithms in more detail. The performance and sensitivity of signal folding for sleep apnea detection in a one-lead ECG signal are evaluated in
Section 4. The properties of signal folding are discussed in
Section 5 in light of other state-of-the-art methods used for ECG signal classification. The paper is concluded in
Section 6.
2. Mathematical Background
This section mathematically defines signal folding, and the associated folding operator, and explores their fundamental properties. The main objective is to understand why signal folding can improve the efficiency of signal classification.
A good model for random observations of a periodically occurring biological phenomenon can have a general mathematical form:
where
are random processes defined over a unit support interval, and
, and
are random variables. The
k-th summand in (
1) has the support,
, of length
. Using the expectation operator,
, the mean process is defined as
The variance of the random signal (
1) depends on the cross-covariances [
20],
Thus, the random process (
1) is generally non-stationary since both its mean and variance may be time dependent.
Provided that the parameters
can be accurately estimated, the random process (
1) can be folded as
In (
4), we introduced the folding operator
, so the folded process,
, is an arithmetic average of the constituting random processes
. The operator
F is deterministic, conditioned on
. The parameters
can be estimated by exploiting specific properties of the random signal (
1), for example, by minimizing a distance metric between subsequent fragments
and
, or between
and all the previous fragments
,
.
The signal folding (
4) is accomplished in two steps. The signal (
1) is first sliced into time-shifted fragments,
, which are then re-scaled by
. The re-scaled fragments are then time-aligned before computing their arithmetic average. If the variance of
is small, i.e., the probability,
, for
,
, the fragments,
, must still be optimally aligned, but they can be zero-padded to the same length instead of re-scaled; this is the case of near-cyclostationary signals, such as ECG or EEG.
Note also that the signal folding (
4) is a form of low-pass filtering since the higher frequency components in waves
are suppressed. It is different from classical low-pass signal filtering in which the signal samples are serially combined over time, whereas the signal folding proposed in this paper combines the samples in signal fragments in parallel. Consequently, we can assume and define the signal fragments to be cyclostationary, but mutually stationary [
6].
In practice, the folding order
K in (
4) is finite. For instance, some fragments
can be labeled and used as training data for learning a classifier in order to categorize other unseen fragments. Let
fragments,
, be partitioned into
L equal-size groups of
fragments each. The fragments in each group are folded, i.e., averaged as
Assuming mutual stationarity in Definition 1, the folded fragments,
, have the following properties.
Definition 1. The fragments, , , are (wide-sense) stationary, provided that the following conditions are satisfied:
- (1)
The mean, , for .
- (2)
The variance, , for .
- (3)
The cross-covariance, , for .
The expression (
6) shows that signal folding does not change the mean of signal fragments. The variance reduction in () was obtained assuming inequality,
. Note also that
For cyclostationary as well as generally non-stationary signals, the averaging (
9) can be further combined with a linear time-average operator,
. Finally, the general moments in () are bounded by the Jensen’s inequality. In addition, Jensen’s inequality for a 2D convex function
can be written as
2.1. ARIMA Modeling of Signal Fragments
The objective is to represent the folded average fragment,
, by an ARIMA model. In its general form, the model,
, is defined as a product of certain polynomials in a lag variable
, i.e., [
1]
The model (
11) relates the error term (input noise),
, to the output random process,
. The polynomials in (
11) are defined as follows [
1]:
Denote as
the vector of
ARIMA model parameters. The most likely parameter values
describing the observed fragments,
, maximize the likelihood,
. However, the model should also avoid overfitting the observed data. The AIC metric for model selection combines the model goodness-of-fit to the data and penalizes the model order, i.e., [
4,
5],
The model order is then a trade-off between the number of degrees-of-freedom to fit the observed data, and their likelihood in order to minimize the AIC metric (
13).
Assuming the property (), it is straightforward to show that signal folding reduces the variance of the model noise
in (
11). The reduced variance of
increases the likelihood,
, of the model parameters,
, given the observations,
. Consequently, if the parameters likelihood is increased due to the reduced variance of the model noise, maintaining the same AIC value allows for a larger number of parameters,
, to be assumed in the model. Consequently and equivalently, the number of parameters
can be reduced while maintaining the same model accuracy, provided that the variance of
is reduced. The reduced value of
directly translates into a substantial reduction of the computational complexity since the data model must be identified for possibly a large number of signal fragments.
2.2. Classification of Signal Fragments with and without Folding
In the last subsection, it was shown that signal folding allows for the reduced order ARIMA modeling of signal fragments. This has a major positive impact on the computational complexity of obtaining the feature representation of signal fragments. The feature vectors as the ARIMA model coefficients were considered for classification of time series, for example, in [
2,
3,
17,
19].
The task is to decide whether the observed fragment,
, indicates that a condition,
, or,
, has occurred. The optimum maximum a posteriori (MAP) rule is defined as [
21]
If there is no prior information on
C, both conditions can be assumed to be equally likely, i.e.,
. The decision rule (
14) represents a binary hypothesis testing problem to infer the condition,
C, from the observation,
.
Assume that groups of
K time-aligned fragments under the same condition
C are combined to create the average fragments,
The corresponding conditional distributions (likelihoods) are denoted as
,
,
, and
, where the first subscript indicates the number of averaged fragments. These distributions are sketched in
Figure 1, assuming the support,
. The cross-over points,
and
, in
Figure 1 partition the signal values into the regions where the likelihood of one condition is greater than the other. However, the actual decision threshold
is normally determined to set the desired probabilities of Type I and Type II errors. Since averaging (
15) reduces the variance whilst the mean is unaffected [cf. (
6) and ()], both of these error probabilities are reduced, i.e.,
Furthermore, averaging the groups of signal fragments reduces the required order of ARIMA models, but also the amount of available training samples. This greatly reduces the overall computational complexity of feature extraction; however, a smaller number of available training samples may also affect the classifier performance. In order to assess the effect of the smaller number of training samples having smaller variances due to averaging, let the total number of labeled signal fragments,
, be divided into
L groups. The
K fragments within each group are then averaged as in (
15). The fragments are generated under one of the conditions (classes),
C. The mean fragment in each class
C is denoted as
[cf. (
9)]. The classifier performance can be assessed by comparing the probabilities that the average fragment
representing the
l-th group,
, is closer in some sense to the mean fragment,
, of class 0 than to the mean fragment,
, of class 1, i.e.,
The actual norm
in (
17) must be derived for the specific features and decision rule assumed [cf. (
14)]. However, provided that the fragments are mutually independent and identically distributed (I.I.D.), and each fragment
is represented by a scalar feature
, we can compare the probabilities
and
of correctly classifying all
N fragments without folding and all
fragments with folding, respectively. In particular, denote
to be a feature vector of
K fragments in the fragment group
l, so
, for
. The corresponding probabilities of correct decisions are
where
denotes the
-norm of a vector, and the threshold
is the average of the mean features in each class, i.e.,
. Since for any vector
of
N elements, the norms,
, it is straightforward to show that
Consequently, the probability , so signal folding can not only significantly reduce the computational complexity of feature extraction, but also improve the decision accuracy of classifiers, effectively compensating the reduced number of training samples.
3. Methodology
As shown in the previous section, signal folding exploits a cyclostationary property of signals in order to reduce their inherent variability and noise, but without completely removing important information embedded in the signals, which is crucial for their classification. An overall signal classification workflow involving signal folding as the first pre-processing step is shown in
Figure 2. The folding operation consists of first partitioning the signal segments into multiple fragments. The fragments can be re-scaled, but they must always be time aligned before they are averaged. The closer the input signal is to be more exactly cyclostationary, the more likely it is that the re-scaling step can be omitted, and replaced by padding the fragments with zero samples. In the second step, a mathematical model of the averaged fragment is identified, and the model parameters are used as a feature vector for classifying the original signal segment.
In this paper, the effect of signal folding on signal classification is investigated for the case of detecting sleep apnea in a one-lead ECG signal. The ECG data were retrieved from PhysioNet, a public repository of medical research data [
22,
23]. The dataset consists of 35 CSV (comma-separated values) files containing the ECG signals for men and women between ages 27 and 63 years over a duration of 7 to 10 h. The ECG signals are manually annotated once per every minute to indicate whether sleep apnea has occurred or not. The annotation marks are used to partition the signal into one minute segments of 6000 samples each, with the marks being in the segment centers. The segments of this length are deemed to be sufficient to reliably detect sleep apnea by classifying the extracted segment features [
11]. The R-waves in the ECG segments can be observed once per every 90 to 100 samples. In total, 6512 segments with sleep apnea and 10,483 normal segments, all having 6000 data points, are available in the dataset.
The feature extraction and subsequent classification can be made more robust by reducing the signal variations. It can be accomplished by a simple Butterworth or moving average filter in order to suppress high-frequency components in the signals [
18]. In this paper, signal folding is proposed to smooth out the signal variations. The signal folding can be succinctly described as storing the original long signal row by row into a matrix, taking the average signal across the matrix rows, and then classifying these shorter average signals. Such a procedure requires to properly slice the ECG segments into multiple fragments of possibly unequal sizes, and then to precisely scale and align the fragments before their averaging. The alignment is necessary, even if the fragments are of an equal length in order to avoid the information aliasing effects, which may suppress important information or create spurious features in the signal.
The following three methods were considered to slice the long ECG segments into shorter fragments. In all these methods, the length L of all fragments is constrained to ensure that they have at least samples, but no more than samples. The design parameters and are determined, so all fragments contain exactly one R-wave with very high probability; a small fraction of fragments which eventually do not satisfy this condition is discarded.
The first segment partitioning method simply slices the ECG signal into the equal-sized fragments, having between
and
samples. The procedure is outlined in Algorithm 1. In particular, the optimal fragment length
minimizes the cumulative squared Euclidean distance (ED) between all the pairs of fragments,
and
, having the equal length
L samples, i.e.,
and
| Algorithm 1 The signal fragmentation yielding equal size fragments |
|
1: functionCreate_Fragments_1()
|
# input signal and the fragments length limits
|
|
2: for all do | |
|
3: partition into disjoint segments of length L | |
|
4: |
# calculate the cumulative squared Euclidean
|
|
# distance between all pairs of segments
|
|
5: end for | |
|
6: return |
# return the optimum segment length
|
|
7: end function | |
The second segment partitioning method allows for non-equal sized fragments. The process is outlined in Algorithm 2. It is a greedy strategy which cuts out the next fragment, so its Euclidean distance to the previous fragment is minimized under the length constraint. The created fragments can be disjoint, or they are allowed to overlap. The averaging of fragments with unequal sizes requires their alignment and scaling prior to their averaging. For discrete-time signals, the re-scaling corresponds to digital interpolation or extrapolation. Alternatively, padding the shorter fragments with zeros can be used for near-cyclostationary signals instead of re-scaling.
| Algorithm 2 The signal fragmentation yielding non-equal size fragments |
|
1: functionCreate_Fragments_2()
|
# input signal and the fragments length limits
|
|
2: for all do | |
|
3: get |
# previous segment of length |
|
4: for all do | |
|
5: get |
# next segment of length |
|
6: |
# calculate their Euclidean distance
|
|
7: end for | |
|
8: |
# select the best |
|
9: |
# and store it
|
|
10: end for | |
|
11: return |
# return the fragment lengths
|
|
12: end function | |
The third strategy for slicing the ECG signal into fragments relies on detecting the R-peaks. We adopted the R-peak recognition algorithm presented in [
24]. This algorithm can be also used to estimate the inter-R-peak distances. The limits
and
can be then determined, so that most of the fragments contain exactly one R-peak. The histogram of the inter-R-peak distances,
, for one selected ECG segment of 6000 samples is shown in
Figure 3. For this segment, there are 70 R-peaks, so 69 inter-R-peak distances are reported in
Figure 3 with the mean value of about 87 samples. More importantly, the mid-points between the consecutive R-peaks can be assumed as the slicing points to create the fragments. This approach turned out to be more robust than the first two methods. This can be expected since Algorithms 1 and 2 are more general, and can be used for fragmenting any near-cyclostationary biological signals, whereas the fragmentation based on R-peak detection is specific to the ECG signals.
The histograms as the one in
Figure 3 were obtained for 20 randomly selected ECG segments. The fragment length bounds
and
samples were inferred from the observed statistics of the inter-R-peak distances. The inferred bounds
and
can be used in Algorithms 1 and 2. The third fragmentation method can assume the fixed-length fragments of
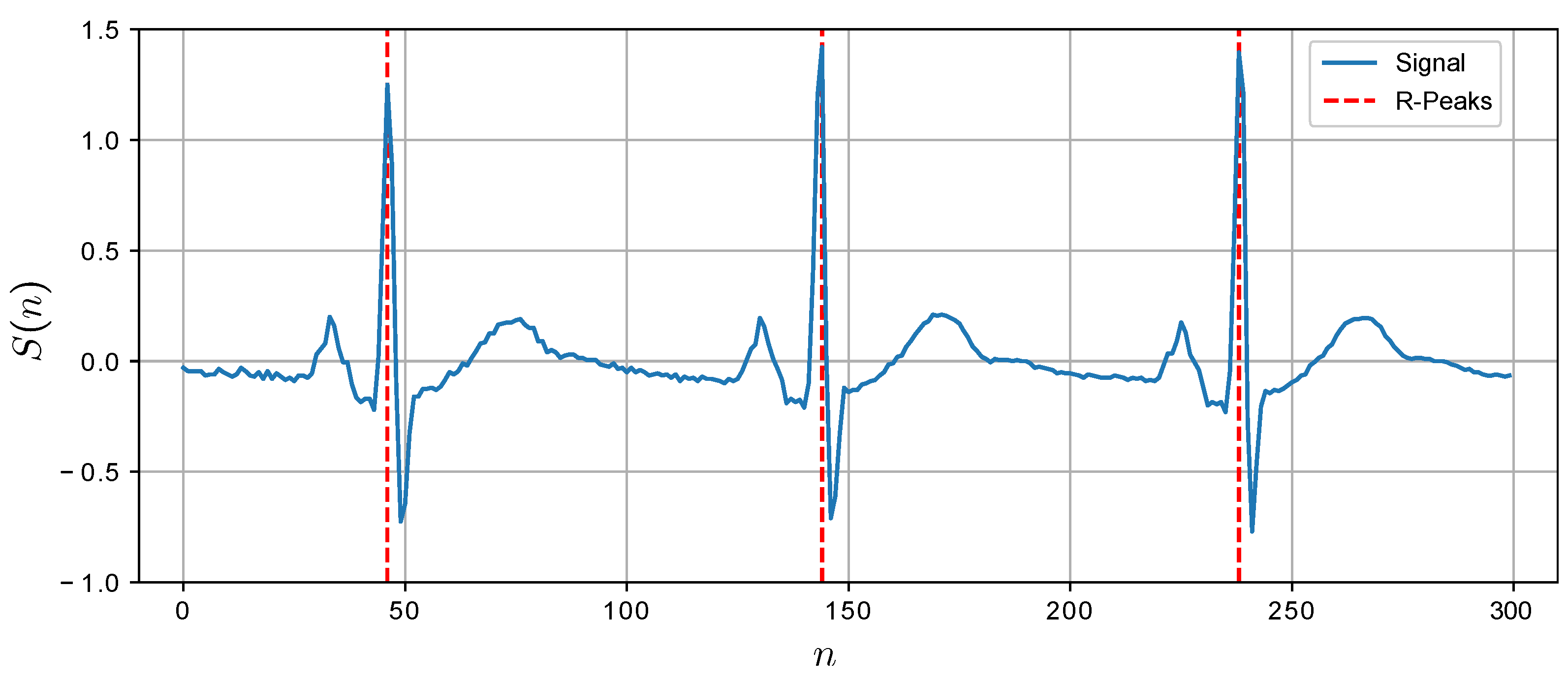
samples with the fragment centers placed at the detected R-peaks. This causes some fragments to overlap whilst there may be gaps in between other fragments. However, it is guaranteed that each fragment contains exactly one R-peak. The presence of a single R-peak within the fragments also enables their robust alignment. A snipset of the ECG segment with automatically identified R-peaks is shown in
Figure 4.
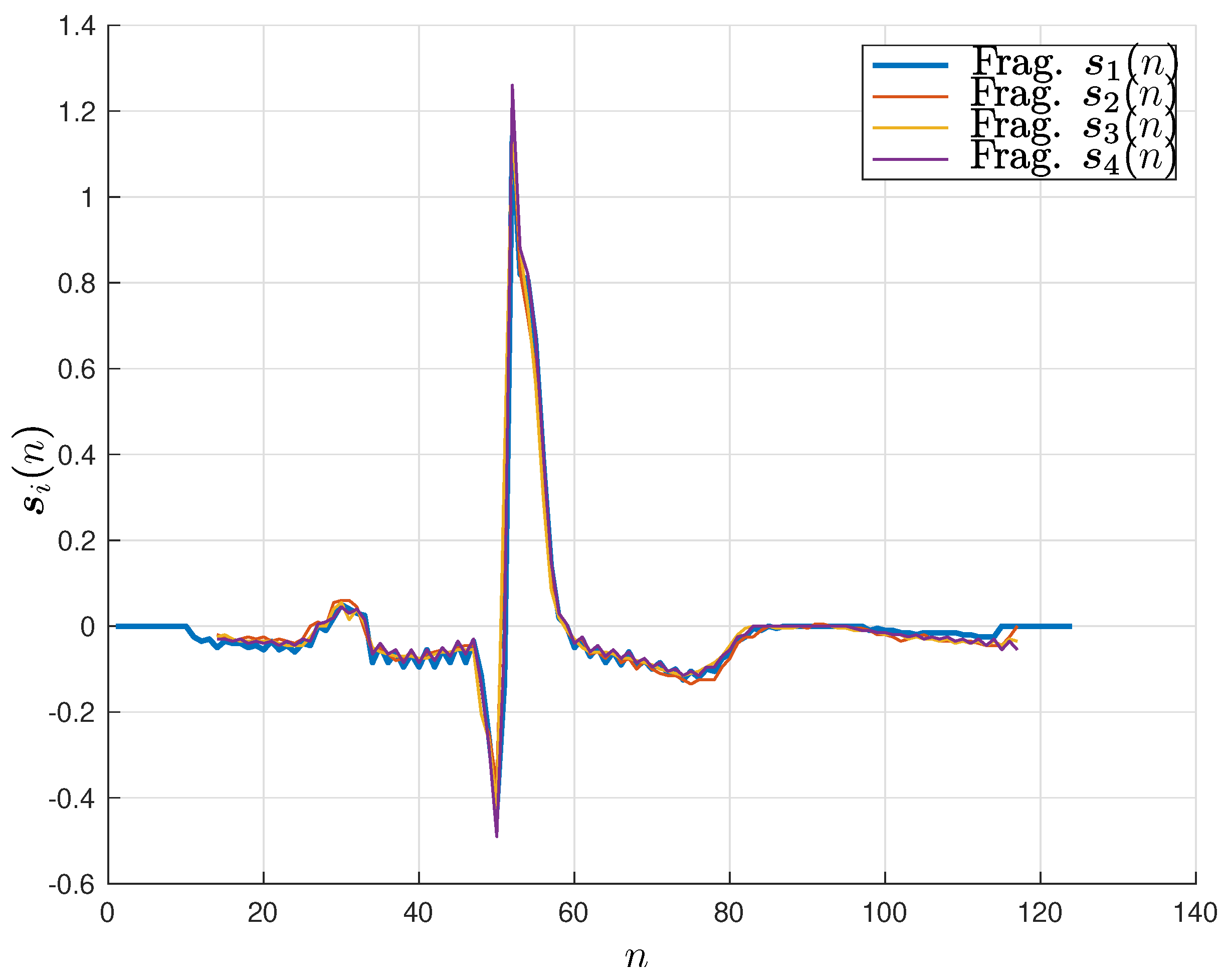
Figure 5 illustrates the alignment of four fragments of different lengths at their R-peak values.
The fragment alignment for the other two methods can be achieved by finding the fragment shifts, minimizing the Euclidean distance between the fragments. Once the fragments are aligned, their averaging is straightforward, although the zero-padding samples should be discounted.
Sleep Apnea Detection in the ECG Signal
Automated classification of time series data by classical machine learning classifiers requires to manually define the features upon which the classification decisions are made. In this study, the ECG signal segments are fitted to ARIMA models, and the ARIMA model coefficients are then used as the feature vectors for subsequent ECG signal classification.
The order of ARIMA model can be estimated by the Akaike information criterion (AIC). However, finding the coefficients of the ARIMA model of a given order must be performed repeatedly many times for all ECG segments or fragments in order to obtain the labeled feature vectors for training the classifiers. Since fitting the model to data has an associated computational cost; these costs quickly accumulate for larger training datasets.
In order to assess the computational cost of extracting the feature vectors from the whole ECG dataset, we measured the time
to fit the ARIMA model of a given order to 100 ECG fragments. Thus, the time
is assumed as a proxy for inferring the actual computational complexity. Recall that our ECG dataset consists of 16,995 segments, and each segment can be further subdivided into 60 fragments on average. The estimated time for fitting the whole dataset is then equal to
(hours). The measured and estimated times
and
, respectively, are reported in
Table 1. The ARMA model order
was assumed in [
19], and the ARMA model order
was determined by minimizing the AIC values for fitting the whole ECG segments (see below).
Our numerical experiments indicate that the time required for fitting the ARIMA models to time series data is strongly dependent on the model order and much less dependent on the length of the data sequences. In general, the ECG signal requires higher order ARIMA models to maintain good classification accuracy, even though such models incur the significant time penalty as shown in
Table 1. Moreover, fitting a single ARIMA model of a given order to the whole ECG segment can incur a substantial loss in the classifier accuracy in comparison when the same model order is used for the individual ECG fragments. More importantly, we observed that employing the segment folding to represent each ECG segment by an average fragment provides sufficient information for the classifier to maintain its good accuracy. Even though the segment fragmentation, alignment and averaging consume a certain amount of time, the average fragments can be represented by the lower order ARMA models. This yields a good trade-off between the time required for training the classifier and the achievable classifier accuracy. Specifically, the ECG segment folding reduces the time required for feature extraction by an order of magnitude while maintaining the same classifier accuracy as when the higher order ARMA models are fitted to all the available (16,995 × 60) ECG fragments.
The detection of sleep apnea in ECG segments was performed assuming the following eight classifiers: logistic regression, SVM, decision tree, random forest, naïve Bayes, K-NN, extreme gradient boosting (XGBoost), and ANN. The ANN has two fully connected layers with 16 neurons in each layer. One output neuron gives predicted values between 0 and 1, and the threshold is assumed to make the final binary decisions. The random forest classifier is configured to contain 100 decision trees, and its performance is validated by a so-called out-of-bag score.
The 70% of labeled ECG fragments are used for the classifier training, and the remaining 30% of data are used for testing and evaluating the classifier performance. The training data are selected by stratified sampling in order to reflect the ratio of the available fragments with and without sleep apnea, respectively.
The performance of binary classifiers is normally evaluated by various metrics involving the true positive (TP), true negative (TN), false positive (FP) and false negative (FN) rates. Specifically, in this paper, the performance of classifiers is compared, assuming the accuracy, specificity and sensitivity. The accuracy reports the percentage of correctly identified segment types, i.e., it is the fraction, . The specificity is determined as a percentage of correctly identified normal segments (without sleep apnea) among the segments that were identified as being normal, i.e., it is the fraction, . The sensitivity is the percentage of segments correctly identified as being affected by sleep apnea, i.e., it is the fraction, . The classifiers are also compared by the times required for their training. Note that we do not evaluate the goodness-of-fit of ARIMA models to ECG signal segments since the ultimate metric of interest is the classification performance.
4. Results
The segments extracted from the original ECG dataset were folded into fragments as described in Methods. There are 60 fragments on average in every ECG segment containing exactly one R-peak. The fragments were then aligned and averaged to yield the short ECG signals of about 100 samples each with much less variation than the original signals. The order of ARIMA models to represent the average ECG fragments was determined using the AIC. The AIC values were calculated for 1000 randomly selected fragments. The maximum order of both the AR and MA model parts was limited to 5 in order to limit the overall computation times required for determining the model order. It should be noted that without fragment averaging, the required order of ARIMA models would be at least doubled.
The results of the ARIMA model order selection using the AIC are given in
Table 2. The seasonality was set to
corresponding to an ordinary ARMA model, and to
, respectively. Assuming the values in
Table 2, the ARMA model
without seasonality was selected for all the subsequent numerical experiments since it provides the best trade-off between the model likelihood and the model complexity.
The ARIMA coefficients are used as feature vectors to classify the ECG fragments and indicate whether sleep apnea is present or absent. The performance of all eight classifiers is compared in
Table 3 in terms of the accuracy, specificity, and sensitivity. The first accuracy values are given for the general fragmentation method described by Algorithm 2. The second accuracy values assume the R-peak-based ECG segment folding. The training times reported in
Table 3 are the average times per one instance of training data. The performance reported in
Table 3 is the maximum achievable values over 25 independent training runs to suppress the random effects observed in the classifier training and testing.
Overall, the results in
Table 3 suggest that the best performing classifier is random forest. The R-peak-based segment folding outperforms the general segment folding method for decision trees, random forest, K-NN, ANN and XGBoost classifiers. The better performance is likely due to exploiting the specific structure of ECG signals containing distinct R-peaks, which can be reliably detected. It is interesting that this feature is implicitly exploited by some but not all classifier types. The unit training times of all classifiers are relatively small, except for the ANN, which requires a much longer time to learn the classification model from training data.
Sensitivity Analysis
It is desirable to gain an insight into how the key parameters of the proposed segment folding affect the classification performance. In particular, we carry out numerical experiments to evaluate the effect of the number of averaged fragments, and how the fragments are selected for averaging. We also evaluate the effect of fragment misalignment prior to their averaging. Finally, we investigate the consequences of performing the averaging of ECG segments prior to their fragmentation. The numerical results are obtained only for the random forest classifier since it has the best performance as shown in the last subsection. In this subsection, the general fragmentation method in Algorithm 2 is used for the segment folding.
Consider first the problem of choosing a limited number of available fragments from the ECG segment for averaging. The fragments are optimally aligned by their R-peaks. We compare two fragment selection strategies. The first strategy is to deterministically select the fragments consecutively, as they were created from the original ECG segment. For example, the first fragments are chosen, and the remaining fragments in the ECG segment are discarded. The second strategy selects the same number of fragments; however, now the fragments are selected from the ECG segments at random with an equal probability. The selected fragments are aligned, averaged, and then used for training and testing the classifier.
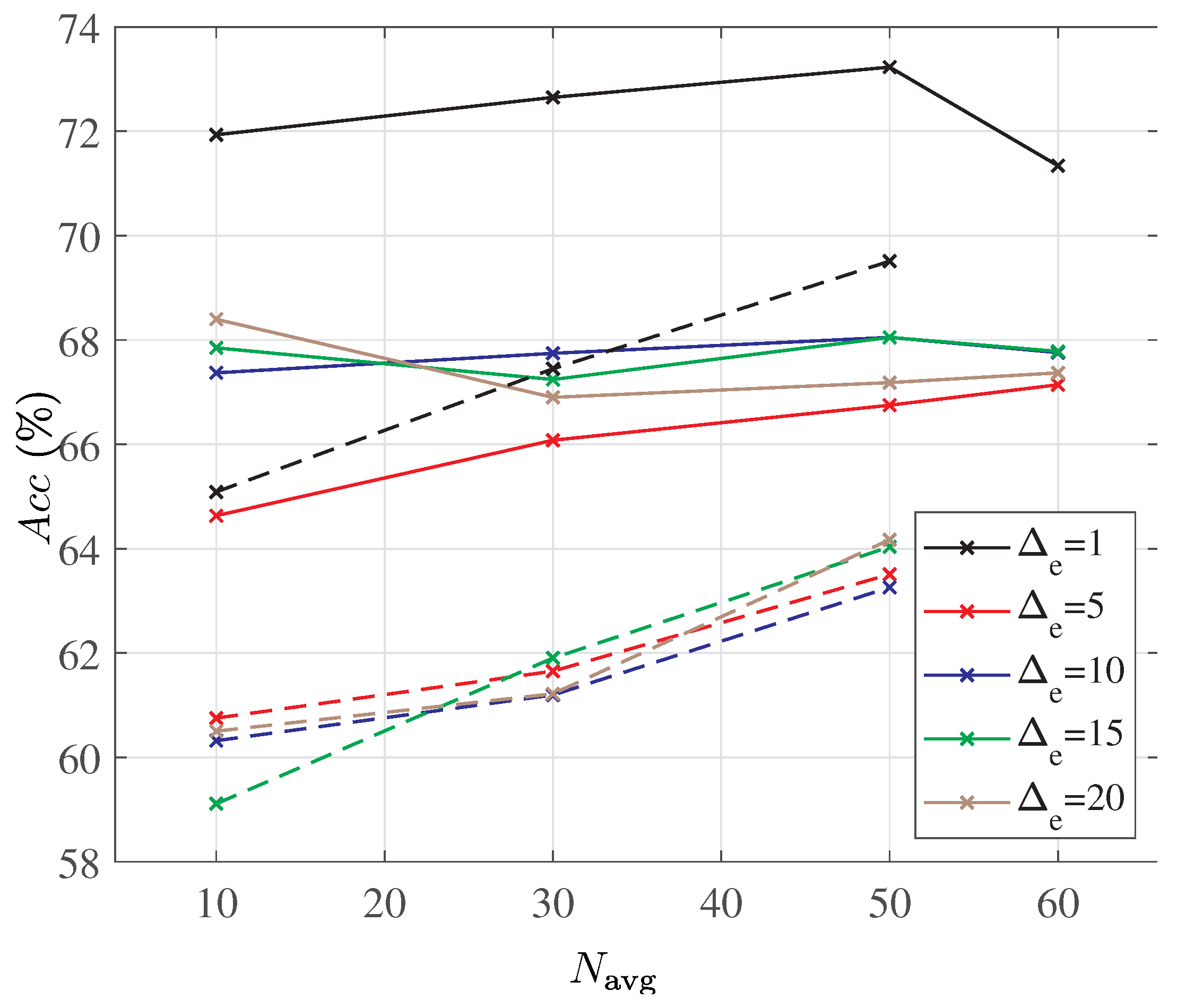
Assuming that
fragments are selected from the 60 fragments available in each ECG segment, the accuracy of the random forest classifier with the deterministic and the random fragment selection is compared in
Figure 6. In order to eliminate randomness in training the classifier, a ten-fold cross-validation is assumed, and the classifier is trained five times on each training dataset. The best achieved accuracy is recorded for each value of
and for both fragment selection strategies.
The accuracy results in
Figure 6 offer several interesting observations. A random selection of fragments is clearly inferior to the consecutive fragment selection, especially when only a small fraction of fragments is considered. However, whereas the accuracy of the random fragment selection monotonically and steadily increases with the number of selected fragments, the accuracy of the consecutive fragment selection reaches a maximum when about half of the fragments are chosen for averaging. The dependency of the classifier accuracy with the consecutive fragment selection on the actual number of fragments selected is also less pronounced. Furthermore, selecting only a subset of consecutive fragments for averaging appears to always outperform the case when all the fragments are averaged (denoted as the horizontal green line in
Figure 6). This is a rather interesting outcome which may indicate that there are limits on how much information can be removed from the signal by the proposed signal folding.
Next, we investigate the robustness of the random forest classifier to the fragment misalignment prior to the fragment averaging. The misalignment may be less of an issue for the ECG signals due to the availability of the R-peaks in each ECG fragment, which allows for their accurate alignment. However, the alignment problem can, in general, be more severe for other types of biological signals. The misalignment error can be systematic, i.e., constant, or vary at random for different fragments. Since the systematic misalignment error is more likely to be identified and corrected, here, we only study the case when the misalignment error
(in samples) is chosen at random with an equal probability from the interval:
The procedure for evaluating the performance of the random forest classifier is the same as above. Thus, the classifier is trained independently five times on each training dataset, and the maximum accuracy among 25 runs is recorded. Both the consecutive and the random fragment selection strategies are considered.
Figure 7 and
Figure 8 show the accuracy results from the same set of experiments.
Figure 7 shows the achieved accuracy as the number of averaged fragments
for different misalignment limits
.
Figure 8 then shows the achieved accuracy as the maximum misalignment
for the different number of averaged fragments
. Note that the value,
, corresponds to the case of averaging all the available fragments. We can observe from
Figure 7 that the misalignment error has larger effect on the consecutive fragment selection than on the random fragment selection. For the consecutive selection with a larger misalignment of fragments, the accuracy becomes largely independent of the number of fragments being averaged. On the other hand, the classification accuracy improves with the number of fragments for the random selection, even if the fragment misalignment is large.
The last experiment assumes that the averaging is performed directly on the ECG segments prior to their fragmentation. In this case, the alignment must be done for the whole segments, which may be more problematic than for the individual fragments containing only a single R-peak. In order to have a fair comparison with the previous results, the ECG segments with the same label (sleep apnea or normal) are averaged in batches of 60. This allows to create 282 averaged ECG segments from the original pool of 16,995 segments. The averaged ECG segments are then sliced into equal-length fragments of 100 samples each. The fragments are used to train the random forest classifier using the coefficients of ARIMA models as the feature vectors. The best performance from 25 independent training runs is 85%, which is better than the segment folding method, using Algorithm 2, having only 78.6% accuracy, but worse then the R-peak-based segment folding, having 89.5% accuracy as shown in
Table 3. For comparison, Ref. [
19] reported the best accuracy of 83% for the SVM classifier assuming the
ARMA model with the time-varying variance of the error term fitted to the whole ECG segment.
5. Discussion
In the literature, the classification of ECG signals seems to be mainly concerned with detecting heart arrhythmia, although other diagnostic objectives are also considered such as detecting stages of myocardial infarction [
25], stages of sleep [
26], and changes in ECG due to hypertension [
27]. The detection of abnormal conditions and their classification are usually performed assuming signal segments [
13], their frequency sub-bands [
27], and possibly multiple time scales [
28]. The raw ECG signal normally contains additive measurement noise and power-line interference, but it may be also subject to other distortions, such as baseline wander, and muscle and electrode motion artifacts. The noise and other distortions can be suppressed by a wide range of techniques, including Fourier and wavelet transforms and other signal decompositions, machine learning autoenconders, and statistical filtering methods [
29]. Consequently, the performance of classifiers is strongly dependent on the quality and quantity of the ECG data, i.e., the actual dataset being considered [
30].
A brief survey of state-of-the-art techniques for classifying ECG signals is given in
Table 4. It is clear that most of the recent papers focus on different types of neural network classifiers including convolutional (CNN), recurrent, and long short-term memory (LSTM) neural networks. The signal pre-processing aims to suppress the undesired distortions and inherent noises. This is usually achieved by a simple low-pass filtering [
26,
27,
29]. The ECG signal can be transformed into other domains using SVD, discrete Fourier transform (DFT), and wavelet transform. The signal features can be learned by the NN classifier [
25,
31,
32,
33], or they can be defined explicitly [
15,
34,
35]. Furthermore, as indicated in
Table 4, typical classification accuracy of ECG signals reported in the literature is often close to perfect 100%. An exception is Ref. [
33], reporting an accuracy which is in a good agreement with the values observed in our numerical experiments. The better performance of the NN classifiers is to be expected at the cost of much higher computational complexity and the required larger training datasets.
The main difference in ECG signal classification in this paper and the previous studies is the introduction of signal folding as the key pre-processing step for classifying near-cyclostationary biological signals. Signal folding reduces the signal variability, so the models with lower order can be assumed. Such models often have much lower computational complexity, and also the model overfitting is easier to detect. In the literature, the signal variability is normally reduced by smoothing the signal by low-pass filtering. However, signal folding requires to slice the signal into separate fragments, scale the fragments to the same length, and then align them, so they can be averaged. The fragment scaling and alignment can be combined into one step as a simple linear transformation, even though the parameters of the linear transformation must be determined individually for each fragment. For near-cyclostationary signals, the scaling can be replaced with padding the shorter fragments with zero samples as was assumed for the case of ECG signals in our numerical experiments. In addition, a simple averaging could be replaced with the weighted averaging in order to emphasize the signal fragments, which carry more important information, or are less noisy.
More importantly, even though signal folding removes some information by suppressing the signal variations, the classifier accuracy to detect sleep apnea in one-lead ECG segments was not affected. This is a rather surprising and unexpected finding. Moreover, as discussed in Methodology, there is a trade-off in reducing the number of training samples by their folding, and reducing the signal model order to learn the classifier more efficiently. A simple slicing of the signal into equal sizes fragments is easy to implement. However, the equal-sized fragments may be more difficult or even impossible to properly align. The scaling and alignment errors may create information aliasing, which can either obscure useful information, or create spurious information. Both phenomena have the detrimental effect on knowledge extraction and drawing valid conclusions from data. A better strategy for signal slicing is to create fragments that have good statistical similarity. The similarity between fragments can be evaluated, for example, by their Euclidean distance, or time warping [
13]. In such a case, the segment slicing can be combined with the fragment scaling and alignment. Although this method is rather general, and it can be used for any cyclostationary time series data, the disadvantage may be the larger required numerical complexity. The third approach to signal slicing can exploit specific unique signal features. In the case of ECG signals, there are near-periodically occurring R-peaks. The mid-points between the detected consecutive R-peaks then define the fragment boundaries. Moreover, the R-peaks can be used to align the fragments, although, in our numerical experiments, the scaling was replaced with zero padding at both ends of the fragments to have the same length. The R-peak-based fragmentation provided more reliable feature representation, and more accurate subsequent classification.
Our numerical results demonstrated that signal folding can greatly reduce the computational complexity of feature extraction. The numerical savings come from the reduced modeling order required to faithfully describe the folded segments rather than from making the fragments shorter, since identifying the model parameters in the smaller number of dimensions is much faster. Moreover, it was shown that folded segments have smaller variability, whereas their means are preserved. The smaller variability of labeled fragments improves the quality of training samples for learning the classifier since unimportant variations are suppressed, whereas the important variations are preserved for making the correct classification decisions. This is also crucial as the number of training data is reduced in proportion to the folding order. In our numerical experiments, we observed that there is an optimal number of combined fragments to achieve the best classification performance. Moreover, combining consecutive fragments always outperforms the case when the fragments are selected at random, which may be expected as indicated in [
9].
Mathematically, a new folding operator was introduced to succinctly describe the process of creating and averaging the signal fragments. The order of folding and expectation operations can be interchanged. The expectation is performed over the distribution of given random variables, whereas the signal folding requires defining the set of signal slicing points. The slicing points are used to split the signal into many fragments followed by their re-scaling, alignment, and arithmetic averaging. This allows creating multiple averaged fragments from signals of long duration. The folding order, i.e., how many fragments are averaged, is an important design parameter affecting the performance of subsequent data processing, and it deserves further investigation.
In our case of sleep apnea detection in ECG signals, the ECG data were already partitioned into one-minute segments. The ECG segments were completely folded, so there was exactly one R-peak contained in each fragment. Such folding exploits the cyclostationary property often present in many biological signals. The scaling and accurate alignment of the fragments is necessary to avoid information aliasing, which can otherwise be caused by an ordinary low-pass filtering. However, the low-pass pre-filtering can be combined with the signal folding, which is an interesting research problem to investigate.
The signal folding investigated in our numerical experiments appears to be robust since it exploits the unique patterns and cyclostationarity of the ECG signal. The more general greedy algorithm introduced in this paper relies on signal similarity, such as the Euclidean distance of the newly created fragment with all the previous fragments, but the subsequent classification has somewhat inferior (as much as 10% lower) performance compared to the R-peak-based signal fragmentation. One strategy for improving the signal folding robustness is to create the average fragments cumulatively, or to employ more rigorous methods of statistical inference to optimally estimate the key parameters of signal folding.
In summary, signal folding reduces the signal variability while preserving the important signal features. The feature extraction and signal classification can be then performed an order of magnitude faster assuming both linear and non-linear signal models. Signal folding can be readily combined with other signal processing techniques and incorporated into signal processing workflows. It can play a vital role in machine learning classification and regression. However, signal folding appears to be very sensitive to the selection of proper slicing instances and accurately scaling and aligning the created signal fragments in order to avoid information aliasing.