Potential Alternative Strategy against Drug Resistant Tuberculosis: A Proteomics Prospect
Abstract
1. Introduction
1.1. Current Scenario
1.2. Cumulative Effort of Government Bodies and Public–Private Consortia
1.3. Slow Progress in Vaccines, Diagnostics and Drug Discovery
1.4. Causes of Drug Resistance
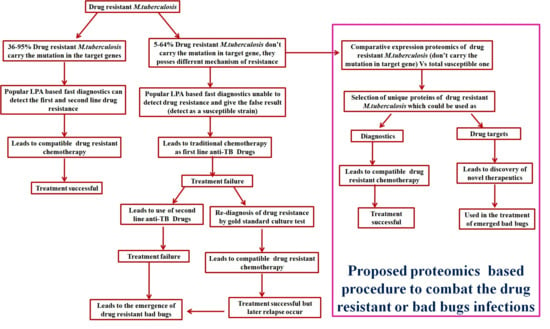
1.5. Alternative Strategies for Drug Resistance Tuberculosis
2. Proteomics and Bioinformatics Explored Potential Drug Targets and Newer Diagnostic Strategies against Drug-Resistant TB: A Future Perspective
3. Conclusions and Future Perspective
Author Contributions
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Global Tuberculosis Report 2016; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Revised National Tuberculosis Control Program (RNTCP). TB India Report 2016; Revised National Tuberculosis Control Program: New Delhi, India, 2016.
- World Health Organization (WHO). Global Tuberculosis Report 2015; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Andersen, P.; Doherty, T.M. The success and failure of BCG-implications for a novel tuberculosis vaccine. Nat. Rev. Microbiol. 2005, 3, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.E.; Weiner, J.; Reyn, C.F. Novel approaches to tuberculosis vaccine development. Int. J. Infect. Dis. 2017, 56, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Dhuriya, Y.K.; Deo, N. Repurposing and revival of the drugs: A new approach to combat the drug resistant tuberculosis. Front. Microbiol. 2017, 8, 2452. [Google Scholar] [CrossRef] [PubMed]
- Beauclerk, A.A.D.; Cundliffe, E. Site of action of two ribosomal RNA methylases responsible for resistance to aminoglycoside. J. Mol. Biol. 1987, 193, 661–671. [Google Scholar] [CrossRef]
- Welch, K.T.; Virga, K.G.; Whittemore, N.A.; Özen, C.; Wright, E.; Brown, C.L.; Lee, R.E.; Serpersu, E.H. Discovery of non-carbohydrate inhibitors of aminoglycoside-modifying enzymes. Bioorg. Med. Chem. 2005, 13, 6252–6363. [Google Scholar] [CrossRef] [PubMed]
- Magnet, S.; Courvalin, P.; Lambert, T. Resistance modulation cell division type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii BM4454. Antimicrob. Agents Chemother. 2001, 45, 3375–3380. [Google Scholar] [CrossRef] [PubMed]
- Magnet, S.; Smith, T.A.; Zheng, R.; Nordmann, P.; Blanchard, J.S. Aminoglycosides resistance resulting from tight drug binding to an altered aminoglycosides acetyl transferase. Antomicrob. Agents Chemother. 2003, 47, 1577–1583. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, B.; Gupta, Y.; Singhal, N.; Katoch, V.M.; Venkatesan, K.; Bisht, D. Proteomic analysis of streptomycin resistant and sensitive clinical isolates of Mycobacterium tuberculosis. Proteome Sci. 2010, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Sharma, D.; Sharma, P.; Katoch, V.M.; Venkatesan, K.; Bisht, D. Proteomic analysis of Mycobacterium tuberculosis isolates resistant to kanamycin and amikacin. J. Proteom. 2013, 94, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Kumar, B.; Lata, M.; Oshi, B.; Venkatesan, K.; Shukla, S.; Bisht, D. Comparative proteomic analysis of aminoglycosides resistant and susceptible Mycobacterium tuberculosis clinical isolates for exploring potential drug targets. PLoS ONE 2015, 10, e0139414. [Google Scholar] [CrossRef] [PubMed]
- Lata, M.; Sharma, D.; Deo, N.; Tiwari, P.K.; Bisht, D.; Venkatesan, K. Proteomic analysis of ofloxacin-mono resistant Mycobacterium tuberculosis isolates. J. Proteom. 2015, 127, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Gopinath, K.; Sharma, P.; Bisht, D.; Sharma, P.; Singh, N.; Singh, S. Comparative proteomic analysis of sequential isolates of Mycobacterium tuberculosis from a patient with pulmonary tuberculosis turning from drug sensitive to multidrug resistant. Indian J. Med. Res. 2015, 141, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Lata, M.; Singh, R.; Deo, N.; Venkatesan, K.; Bisht, D. Cytosolic proteome profiling of aminoglycosides resistant Mycobacterium tuberculosis clinical isolates using MALDI-TOF/MS. Front. Microbiol. 2016, 7, 1816. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Bisht, D. Secretory proteome analysis of streptomycin resistant Mycobacterium tuberculosis clinical isolates. SLAS Discov. 2017, 22, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Hasse, B.; Walker, A.S.; Fehr, J.; Furrer, H.; Hoffmann, M.; Battegay, M.; Calmy, A.; Fellay, J.; Di Benedetto, C.; Weber, R.; et al. Cotrimoxazole prophylaxis is associated with reduced risk of incident tuberculosis in participants in the Swiss HIV Cohort Study. Antimicrob. Agents Chemother. 2014, 58, 2363–2368. [Google Scholar] [CrossRef] [PubMed]
- Jaspard, M.; Elefant-Amoura, E.; Melonio, I.; De Montgolfier, I.; Veziris, N.; Caumes, E. Bedaquiline and linezolid for extensively drug-resistant tuberculosis in pregnant woman. Emerg. Infect. Dis. 2017, 23, 1731–1732. [Google Scholar] [CrossRef] [PubMed]
- Tobin, D.M. Host-directed therapies for tuberculosis. Cold Spring Harb. Perspect. Med. 2015, 1, 417–432. [Google Scholar]
- Wallis, R.S.; Hafner, R. Advancing host-directed therapy for tuberculosis. Nat. Rev. Immunol. 2015, 15, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Coussens, A.K.; Wilkinson, R.J.; Hanifa, Y.; Nikolayevskyy, V.; Elkington, P.T.; Islam, K.; Timms, P.M.; Venton, T.R.; Bothamley, G.H.; Packe, G.E.; et al. Vitamin D accelerates resolution of inflammatory responses during tuberculosis treatment. Proc. Natl. Acad. Sci. USA 2012, 109, 15449–15454. [Google Scholar] [CrossRef] [PubMed]
- Katoch, K.; Singh, P.; Adhikari, T.; Benara, S.K.; Singh, H.B.; Chauhan, D.S.; Sharma, V.D.; Lavania, M.; Sachan, A.S.; Katoch, V.M. Potential of Mw as a prophylactic vaccine against pulmonary tuberculosis. Vaccine 2008, 26, 1228–1234. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Lata, M.; Faheem, M.; Ullah Khan, A.; Joshi, B.; Venkatesan, K.; Shukla, S.; Bisht, D. Cloning, expression and correlation of Rv0148 to amikacin & kanamycin resistance. Curr. Proteom. 2015, 12, 96–100. [Google Scholar]
- Sharma, D.; Lata, M.; Faheem, M.; Khan, A.U.; Joshi, B.; Venkatesan, K.; Shukla, S.; Bisht, D. M. tuberculosis ferritin (Rv3841): Potential involvement in Amikacin (AK) & Kanamycin (KM) resistance. Biochem. Biophys. Res. Commun. 2016, 478, 908–912. [Google Scholar] [PubMed]
- Sharma, D.; Bisht, D. M. tuberculosis hypothetical proteins and proteins of unknown function: Hope for exploring novel resistance mechanisms as well as future target of drug resistance. Front. Microbiol. 2017, 8, 465. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Bisht, D. Role of bacterioferritin & ferritin in M. tuberculosis pathogenesis and drug resistance: A future perspective by interactomic approach. Front. Cell. Infect. Microbiol. 2017, 7, 240. [Google Scholar] [PubMed]
- Sharma, D.; Shankar, H.; Lata, M.; Joshi, B.; Venkatesan, K.; Bisht, D. Culture filtrate proteome analysis of aminoglycoside resistant clinical isolates of Mycobacterium tuberculosis. BMC Infect. Dis. 2014, 14 (Suppl. S3), 60. [Google Scholar] [CrossRef]
- Sharma, D.; Bisht, D. An efficient and rapid lipophilic proteins extraction from Mycobacterium tuberculosis H37Rv for two dimensional gel electrophoresis. Electrophoresis 2016, 37, 1187–1190. [Google Scholar] [CrossRef] [PubMed]
- Lata, M.; Sharma, D.; Kumar, B.; Deo, N.; Tiwari, P.K.; Bisht, D.; Venkatesan, K. Proteome analysis of ofloxacin and moxifloxacin induced Mycobacterium tuberculosis isolates by proteomic approach. Protein Pept. Lett. 2015, 22, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Deo, N.; Bisht, D. Proteomics and Bioinformatics: A Modern Way to Elucidate the Resistome in Mycobacterium tuberculosis. J. Proteom. Bioinform. 2017, 10, e33. [Google Scholar] [CrossRef]
- Kumar, G.; Shankar, H.; Sharma, D.; Sharma, P.; Bisht, D.; Katoch, V.M.; Joshi, B. Proteomics of culture filtrate of prevalent Mycobacterium tuberculosis strains: 2D-PAGE map and MALDI-TOF/MS analysis. SLAS Discov. 2017, 22, 1142–1149. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Singh, R.; Deo, N.; Bisht, D. Interactome analysis of Rv0148 to predict potential targets and their pathways linked to aminoglycosides drug resistance: An insilico approach. Microb. Pathog. 2018, in press. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Leitner, A.; Aebersold, R. Mass spectrometry applied to bottom-up proteomics: Entering the high-throughput era for hypothesis testing. Annu. Rev. Anal. Chem. 2016, 9, 449–472. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.S.; Calder, B.; Gonnelli, G.; Degroeve, S.; Rajaonarifara, E.; Mulder, N.; Soares, N.C.; Martens, L.; Blackburn, J.M. Identification of quantitative proteomic differences between Mycobacterium tuberculosis lineages with altered virulence. Front. Microbiol. 2016, 7, 813. [Google Scholar] [CrossRef] [PubMed]
- Malen, H.; De Souza, G.A.; Pathak, S.; Søfteland, T.; Wiker, H.G. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiol. 2011, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Jhingan, G.D.; Kumari, S.; Jamwal, S.V.; Kalam, H.; Arora, D.; Jain, N.; Kumaar, L.K.; Samal, A.; Rao, K.V.S.; Kumar, D.; et al. Comparative proteomic analyses of avirulent, virulent, and clinical strains of Mycobacterium tuberculosis identify strain-specific patterns. J. Biol. Chem. 2016, 291, 14257–14273. [Google Scholar] [CrossRef] [PubMed]
- De Keijzer, J.; de Haas, P.E.; de Ru, A.H.; van Veelen, P.A.; van Soolingen, D. Disclosure of selective advantages in the “modern” sublineage of the Mycobacterium tuberculosis Beijing genotype family by quantitative proteomics. Mol. Cell. Proteom. 2014, 13, 2632–2645. [Google Scholar] [CrossRef] [PubMed]
- De Keijzer, J.; Mulder, A.; de Haas, P.E.W.; de Ru, A.H.; Heerkens, E.M.; Amaral, L.; van Soolingen, D.; van Veelen, P.A. Thioridazine alters the cell-envelope permeability of Mycobacterium tuberculosis. J. Proteome Res. 2016, 15, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Schubert, O.T.; Mouritsen, J.; Ludwig, C.; Röst, H.L.; Rosenberger, G.; Arthur, P.K.; Claassen, M.; Campbell, D.S.; Sun, Z.; Farrah, T.; et al. The Mtb proteome library: A resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 2013, 13, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Schubert, O.T.; Ludwig, C.; Kogadeeva, M.; Zimmermann, M.; Rosenberger, G.; Gengenbacher, M.; Gillet, L.C.; Collins, B.C.; Rost, H.L.; Kaufmann, S.H.E.; et al. Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 2015, 18, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Sharma, D.; Bisht, D. Comparative protein profiling of intraphagosomal expressed proteins of Mycobacterium bovis BCG. Protein Pept. Lett. 2016, 23, 51–54. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, D.; Bisht, D.; Khan, A.U. Potential Alternative Strategy against Drug Resistant Tuberculosis: A Proteomics Prospect. Proteomes 2018, 6, 26. https://doi.org/10.3390/proteomes6020026
Sharma D, Bisht D, Khan AU. Potential Alternative Strategy against Drug Resistant Tuberculosis: A Proteomics Prospect. Proteomes. 2018; 6(2):26. https://doi.org/10.3390/proteomes6020026
Chicago/Turabian StyleSharma, Divakar, Deepa Bisht, and Asad U. Khan. 2018. "Potential Alternative Strategy against Drug Resistant Tuberculosis: A Proteomics Prospect" Proteomes 6, no. 2: 26. https://doi.org/10.3390/proteomes6020026
APA StyleSharma, D., Bisht, D., & Khan, A. U. (2018). Potential Alternative Strategy against Drug Resistant Tuberculosis: A Proteomics Prospect. Proteomes, 6(2), 26. https://doi.org/10.3390/proteomes6020026