Enrichment and Identification of the Most Abundant Zinc Binding Proteins in Developing Barley Grains by Zinc-IMAC Capture and Nano LC-MS/MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Osborne Protein Fractionation of the Microdissected Barley Tissues
2.3. SDS-PAGE, Coomassie, Zinc Blotting, and DTZ Staining Assay
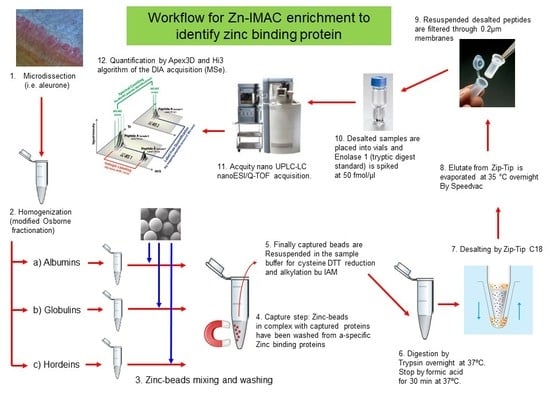
2.4. Zn-IMAC Chromatography of Osborne Fractionated Proteins of the Microdissected Barley Tissues
2.5. Sample Preparation for Proteomic and MS Analysis
2.6. Light Microscopy
2.7. Gel Fractionaction on Gelfree 8100 and Silver Staining
3. Results
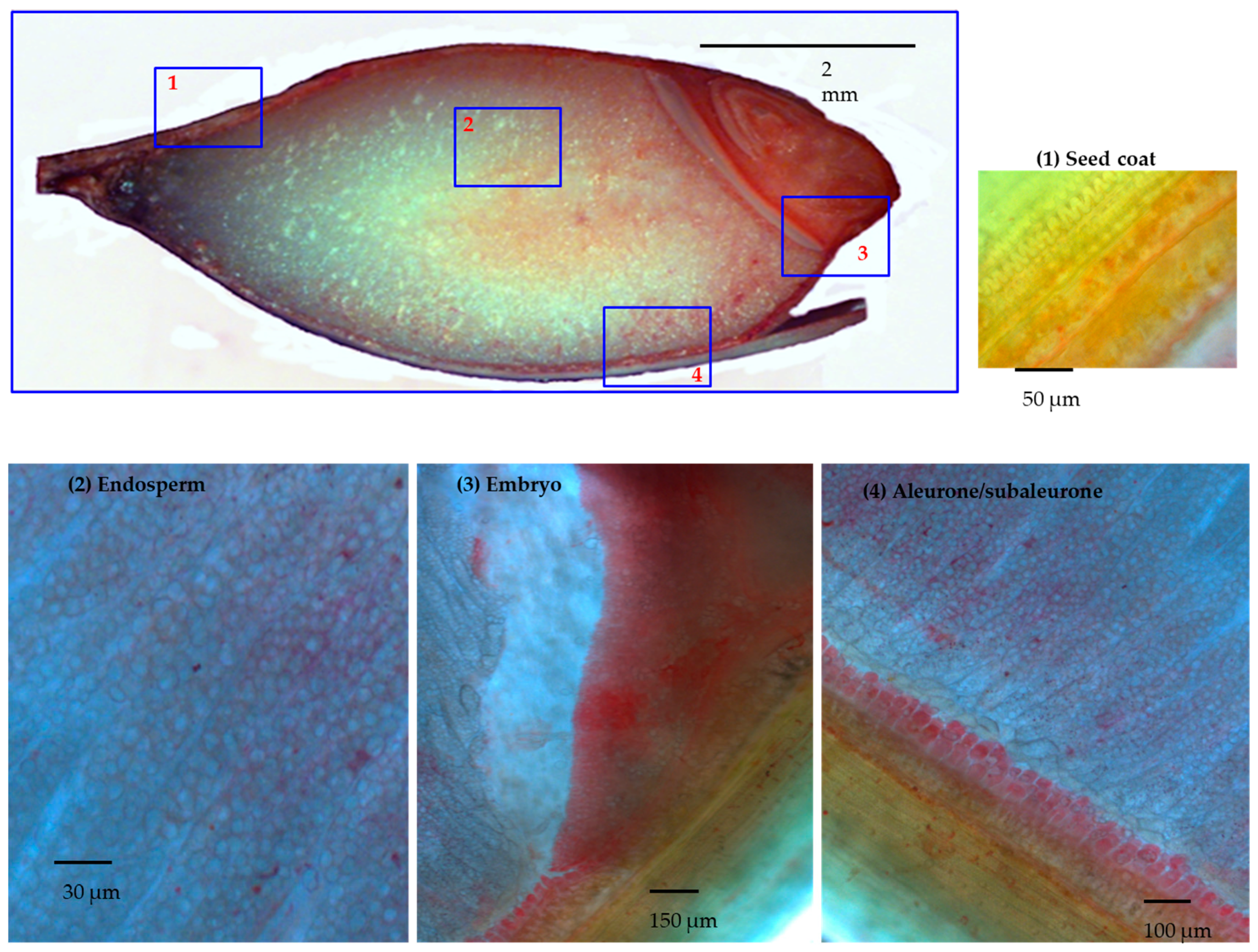
3.1. Manual Microdissection of Barley Developing Grain
3.2. DTZ Staining of the Longitudinal Section of Half Barley Grain
3.3. Osborne Protein Fractionation of the Microdissected Barley Tissues, Zn-IMAC Capture, and nanoLC MS/MS Identification
3.3.1. Seed Coat (Group 1)
3.3.2. Aleurone/Subaleurone (Group 2)
3.3.3. Embryo (Group 3)
3.3.4. Endosperm (Group 4)
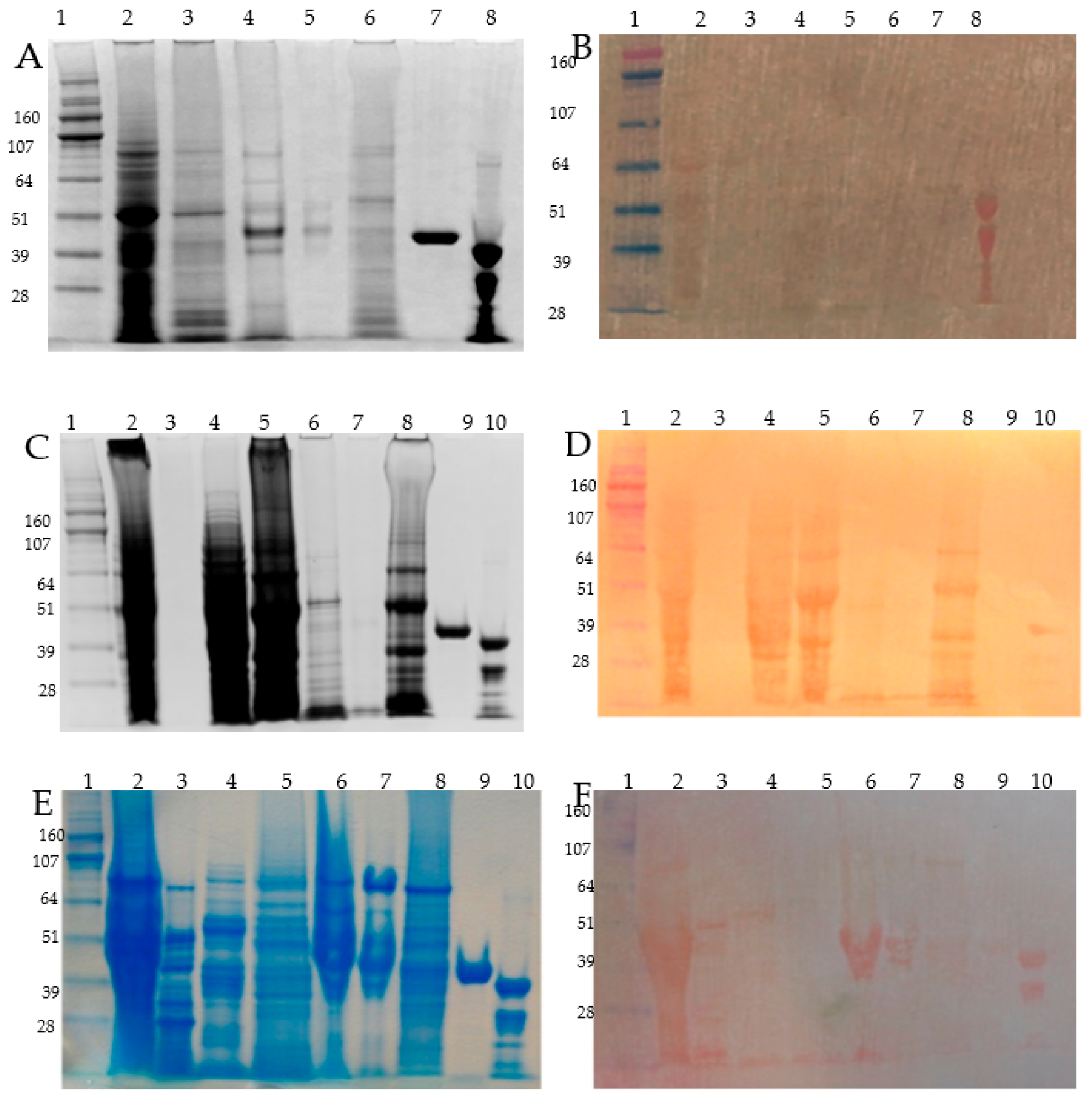
3.4. Detection of the Most Abundant Zn-IMAC Captured Proteins after DTT Pre-Treatment
3.5. Protein Size Fractionation and Electro-Elution by Gelfree 8100, Zinc DTZ Blotting, and MS/MS Identification of the High DTZ Stained Bands
4. Discussion
4.1. Zn Binding Proteins in Globulins
4.2. Zinc Binding Proteins in Prolamin Fractions
4.3. Other Groups of Zn Binding Proteins
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Broadley, M.R.; White, P.J.; Hammond, J.P.; Zelko, I.; Lux, A. Zinc in plants: Tansley review. New Phytol. 2007, 173, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.M.; Guerinot, M.L. Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 2009, 5, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Bertini, I.; Rosato, A. Metalloproteomes: A bioinformatic approach. Acc. Chem. Res. 2009, 42, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Bertini, I.; Cavallaro, G. Minimal functional sites allow a classification of zinc sites in proteins. PLoS ONE 2011, 6, e26325. [Google Scholar] [CrossRef] [PubMed]
- Maret, W. Zinc biochemistry: From a single zinc enzyme to a key element of life. Adv. Nutr. Int. Rev. J. 2013, 4, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Corbo, M.D.; Lam, J. Zinc deficiency and its management in the pediatric population: A literature review and proposed etiologic classification. J. Am. Acad. Dermatol. 2013, 69, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.J. Global impacts of human mineral malnutrition. Plant Soil 2010, 335, 133–154. [Google Scholar] [CrossRef]
- Osborne, T.B.; Voorhees, C.L. Proteids of the wheat kernel. J. Am. Chem. Soc. 1894, 16, 524–535. [Google Scholar] [CrossRef]
- Shewry, P.R.; Field, J.M.; Kirkman, M.A.; Faulks, A.J.; Miflin, B.J. The extraction, solubility, and characterization of 2 groups of barley storage polypeptides. J. Exp. Bot. 1980, 31, 393–407. [Google Scholar] [CrossRef]
- Mills, E.N.C.; Jenkins, J.A.; Alcocer, M.J.C.; Shewry, P.R. Structural, biological, and evolutionary relationships of plant food allergens sensitizing via the gastrointestinal tract. Crit. Rev. Food Sci. Nutr. 2004, 44, 379–407. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R. Seed storage proteins: Structures and biosynthesis. Plant Cell Online 1995, 7, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Verdier, J.; Thompson, R.D. Networks of seed storage protein regulation in cereals and legumes at the dawn of the omics era. In Seed Development: OMICS Technologies toward Improvement of Seed Quality and Crop Yield; Springer: Dordrecht, The Netherlands, 2012; pp. 187–210. [Google Scholar]
- Verdier, J.; Thompson, R.D. Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol. 2008, 49, 1263–1271. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R. Plant storage proteins. Biol. Rev. 1995, 70, 375–426. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Kaczmarczyk, A.; Vincze, E. Effects of Zn fertilization on hordein transcripts at early developmental stage of barley grain and correlation with increased Zn concentration in the mature grain. PLoS ONE 2014, 9, e108546. [Google Scholar] [CrossRef] [PubMed]
- Cakmak, I.; Kalayci, M.; Kaya, Y.; Torun, A.; Aydin, N.; Wang, Y.; Arisoy, Z.; Erdem, H.; Yazici, A.; Gokmen, O. Biofortification and localization of zinc in wheat grain. J. Agric. Food Chem. 2010, 58, 9092–9102. [Google Scholar] [CrossRef] [PubMed]
- Kutman, U.B.; Kutman, B.Y.; Ceylan, Y.; Ova, E.A.; Cakmak, I. Contributions of root uptake and remobilization to grain zinc accumulation in wheat depending on post-anthesis zinc availability and nitrogen nutrition. Plant Soil 2012, 361, 177–187. [Google Scholar] [CrossRef]
- Persson, D.P.; Hansen, T.H.; Laursen, K.H.; Schjoerring, J.K.; Husted, S. Simultaneous iron, zinc, sulfur and phosphorus speciation analysis of barley grain tissues using SEC-ICP-MS and IP-ICP-MS. Metallomics 2009, 1, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Stomph, T.J.; Jiang, W.; Struik, P.C. Zinc biofortification of cereals: Rice differs from wheat and barley. Trends Plant Sci. 2009, 14, 123–124. [Google Scholar] [CrossRef] [PubMed]
- Lombi, E.; Smith, E.; Hansen, T.H.; Paterson, D.; De Jonge, M.D.; Howard, D.L.; Persson, D.P.; Husted, S.; Ryan, C.; Schjoerring, J.K. Megapixel imaging of (micro)nutrients in mature barley grains. J. Exp. Bot. 2011, 62, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Teklic, T.; Loncaric, Z.; Kovacevic, V.; Singh, B.R. Metallic trace elements in cereal grain—A review: How much metal do we eat? Food Energy Secur. 2013, 2, 81–95. [Google Scholar] [CrossRef]
- Myers, S.S.; Zanobetti, A.; Kloog, I.; Huybers, P.; Leakey, A.D.B.; Bloom, A.J.; Carlisle, E.; Dietterich, L.H.; Fitzgerald, G.; Hasegawa, T.; et al. Increasing CO2 threatens human nutrition. Nature 2014, 510, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, M.G.; Clemens, S.; Williams, L.E.; Krämer, U.; Borg, S.; Schjørring, J.K.; Sanders, D. Zinc biofortification of cereals: Problems and solutions. Trends Plant Sci. 2008, 13, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Hegelund, J.; Pedas, P.; Husted, S.; Schiller, M.; Schjoerring, J. Zinc fluxes into developing barley grains: Use of stable Zn isotopes to separate root uptake from remobilization in plants with contrasting Zn status. Plant Soil 2012, 361, 241–250. [Google Scholar] [CrossRef]
- Clemens, S. Evolution and function of phytochelatin synthases. J. Plant Physiol. 2006, 163, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Neal, A.L.; Geraki, K.; Borg, S.; Quinn, P.; Mosselmans, J.F.; Brinch-Pedersen, H.; Shewry, P.R. Iron and zinc complexation in wild-type and ferritin-expressing wheat grain: Implications for mineral transport into developing grain. J. Biol. Inorg. Chem. 2013, 18, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Erdal, I.; Kaya, M.; Küçükyumuk, Z. Effects of zinc and nitrogen fertilizations on grain yield and some parameters effecting zinc bioavailability in lentil seeds. Legume Res. 2014, 37, 55–61. [Google Scholar] [CrossRef]
- Erdal, I.; Yilmaz, A.; Taban, S.; Eker, S.; Torun, B.; Cakmak, I. Phytic acid and phosphorus concentrations in seeds of wheat cultivars grown with and without zinc fertilization. J. Plant Nutr. 2002, 25, 113–127. [Google Scholar] [CrossRef]
- Brejnholt, S.M.; Dionisio, G.; Glitsoe, V.; Skov, L.K.; Brinch-Pedersen, H. The degradation of phytate by microbial and wheat phytases is dependent on the phytate matrix and the phytase origin. J. Sci. Food Agric. 2011, 91, 1398–1405. [Google Scholar] [CrossRef] [PubMed]
- Lott, J.N.A.; Spitzer, E. X-ray-analysis studies of elements stored in protein body globoid crystals of triticum grains. Plant Physiol. 1980, 66, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, E.; Webber, M.; Lott, J.N.A. Elemental composition of globoid crystals in protein bodies of wheat-grain grown on soil treated with sewage-sludge. Can. J. Bot. 1981, 59, 403–409. [Google Scholar] [CrossRef]
- Tennstedt, P.; Peisker, D.; Böttcher, C.; Trampczynska, A.; Clemens, S. Phytochelatin synthesis is essential for the detoxification of excess zinc and contributes significantly to the accumulation of zinc. Plant Physiol. 2009, 149, 938–948. [Google Scholar] [CrossRef] [PubMed]
- Stomph, T.J.; Choi, E.Y.; Stangoulis, J.C.R. Temporal dynamics in wheat grain zinc distribution: Is sink limitation the key? Ann. Bot. 2011, 107, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Tauris, B.; Borg, S.; Gregersen, P.L.; Holm, P.B. A roadmap for zinc trafficking in the developing barley grain based on laser capture microdissection and gene expression profiling. J. Exp. Bot. 2009, 60, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Tiong, J.; McDonald, G.K.; Genc, Y.; Pedas, P.; Hayes, J.E.; Toubia, J.; Langridge, P.; Huang, C.Y. Hvzip7 mediates zinc accumulation in barley (Hordeum vulgare) at moderately high zinc supply. New Phytol. 2014, 201, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; Sankaran, R.P. Moving micronutrients from the soil to the seeds: Genes and physiological processes from a biofortification perspective. Plant Sci. 2011, 180, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Usuda, K.; Kobayashi, T.; Ishimaru, Y.; Kakei, Y.; Takahashi, M.; Higuchi, K.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Overexpression of the barley nicotianamine synthase gene hvnas1 increases iron and zinc concentrations in rice grains. Rice 2009, 2, 155–166. [Google Scholar] [CrossRef]
- Moreno-Moyano, L.T.; Bonneau, J.P.; Sanchez-Palacios, J.T.; Tohme, J.; Johnson, A.A.T. Association of increased grain iron and zinc concentrations with agro-morphological traits of biofortified rice. Front. Plant Sci. 2016, 7, 1463. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Persson, D.P.; Hansen, T.H.; Husted, S.; Schjoerring, J.K.; Kim, Y.S.; Jeon, U.S.; Kim, Y.K.; Kakei, Y.; Masuda, H.; et al. Bio-available zinc in rice seeds is increased by activation tagging of nicotianamine synthase. Plant Biotechnol. J. 2011, 9, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Mir, G.; Domènech, J.; Huguet, G.; Guo, W.J.; Goldsbrough, P.; Atrian, S.; Molinas, M. A plant type 2 metallothionein (mt) from cork tissue responds to oxidative stress. J. Exp. Bot. 2004, 55, 2483–2493. [Google Scholar] [CrossRef] [PubMed]
- Roosens, N.H.; Leplae, R.; Bernard, C.; Verbruggen, N. Variations in plant metallothioneins: The heavy metal hyperaccumulator thlaspi caerulescens as a study case. Planta 2005, 222, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Ruttkay-Nedecky, B.; Nejdl, L.; Gumulec, J.; Zitka, O.; Masarik, M.; Eckschlager, T.; Stiborova, M.; Adam, V.; Kizek, R. The role of metallothionein in oxidative stress. Int. J. Mol. Sci. 2013, 14, 6044–6066. [Google Scholar] [CrossRef] [PubMed]
- Schiller, M.; Hegelund, J.N.; Pedas, P.; Kichey, T.; Laursen, K.H.; Husted, S.; Schjoerring, J.K. Barley metallothioneins differ in ontogenetic pattern and response to metals. Plant Cell Environ. 2014, 37, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.F.; Eagling, T.; He, J.B.; Zou, C.Q.; McGrath, S.P.; Shewry, P.R.; Zhao, F.J. Effects of nitrogen on the distribution and chemical speciation of iron and zinc in pearling fractions of wheat grain. J. Agric. Food Chem. 2014, 62, 4738–4746. [Google Scholar] [CrossRef] [PubMed]
- Regvar, M.; Eichert, D.; Kaulich, B.; Gianoncelli, A.; Pongrac, P.; Vogel-Mikus, K.; Kreft, I. New insights into globoids of protein storage vacuoles in wheat aleurone using synchrotron soft X-ray microscopy. J. Exp. Bot. 2011, 62, 3929–3939. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Vincze, E.; Wieser, H.; Schjoerring, J.K.; Holm, P.B. Suppression of C-hordein synthesis in barley by antisense constructs results in a more balanced amino acid composition. J. Agric. Food Chem. 2007, 55, 6074–6081. [Google Scholar] [CrossRef] [PubMed]
- Lookhart, G.; Bean, S. Separation and characterization of wheat protein fractions by high-performance capillary electrophoresis. Cereal Chem. 1995, 72, 527–532. [Google Scholar]
- Uddin, M.N.; Nielsen, A.L.L.; Vincze, E. Zinc blotting assay for detection of zinc-binding prolamin in barley (hordeum vulgare) grain. Cereal Chem. 2014, 91, 228–232. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.Z.; Vissers, J.P.C.; Geromanos, S.J. Absolute quantification of proteins by LCMSE—A virtue of parallel MS acquisition. Mol. Cell. Proteom. 2006, 5, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Ullmann-Zeunert, L.; Muck, A.; Wielsch, N.; Hufsky, F.; Stanton, M.A.; Bartram, S.; Bocker, S.; Baldwin, I.T.; Groten, K.; Svatos, A. Determination of N-15-incorporation into plant proteins and their absolute quantitation: A new tool to study nitrogen flux dynamics and protein pool sizes elicited by plant-herbivore interactions. J. Proteome Res. 2012, 11, 4947–4960. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Label-free protein quantitation using weighted spectral counting. Methods Mol. Biol. 2012, 893, 321–341. [Google Scholar] [PubMed]
- Mueller, L.N.; Brusniak, M.Y.; Mani, D.R.; Aebersold, R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 2008, 7, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Hibbard, P.L. Estimation of copper, zinc and cobalt (with nickel) in soil extracts—Dithizone methods particularly adapted to examination of soils. Ind. Eng. Chem. 1938, 10, 0615–0618. [Google Scholar] [CrossRef]
- Wu, X.S.; Bishopric, N.H.; Discher, D.J.; Murphy, B.J.; Webster, K.A. Physical and functional sensitivity of zinc finger transcription factors to redox change. Mol. Cell. Biol. 1996, 16, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Koziol, A.G.; Loit, E.; McNulty, M.; MacFarlane, A.J.; Scott, F.W.; Altosaar, I. Seed storage proteins of the globulin family are cleaved post-translationally in wheat embryos. BMC Res. Notes 2012, 5, 385. [Google Scholar] [CrossRef] [PubMed]
- Vanderschuren, H.; Lentz, E.; Zainuddin, I.; Gruissem, W. Proteomics of model and crop plant species: Status, current limitations and strategic advances for crop improvement. J. Proteom. 2013, 93, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Dalcorso, G.; Fasani, E.; Furini, A. Recent advances in the analysis of metal hyperaccumulation and hypertolerance in plants using proteomics. Front. Plant Sci. 2013, 4, 280. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Persson, D.P.; Husted, S.; Schellenberg, M.; Gehrig, P.; Lee, Y.; Martinoia, E.; Schjoerring, J.K.; Meyer, S. A proteomics approach to investigate the process of Zn hyperaccumulation in Noccaea caerulescens (J and C. Presl) F.K. Meyer. Plant J. 2013, 73, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Visioli, G.; Marmiroli, N. The proteomics of heavy metal hyperaccumulation by plants. J. Proteom. 2013, 79, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Martinoia, E.; Massonneau, A.; Frangne, N. Transport processes of solutes across the vacuolar membrane of higher plants. Plant Cell Physiol. 2000, 41, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Tsukamoto, T.; Inoue, H.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Deoxymugineic acid increases Zn translocation in Zn-deficient rice plants. Plant Mol. Biol. 2008, 66, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, A.B.; Landberg, T.; Berglund, T.; Greger, M. Increased metal tolerance in salix by nicotinamide and nicotinic acid. Plant Physiol. Biochem. 2008, 46, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Persson, D.P.; de Bang, T.C.; Pedas, P.R.; Kutman, U.B.; Cakmak, I.; Andersen, B.; Finnie, C.; Schjoerring, J.K.; Husted, S. Molecular speciation and tissue compartmentation of zinc in durum wheat grains with contrasting nutritional status. New Phytol. 2016, 211, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S. Zinc coordination sphere in biochemical zinc sites. In Zinc Biochemistry, Physiology, and Homeostasis; Springer: Berlin, Germany, 2001; pp. 85–127. [Google Scholar]
- McCall, K.A.; Huang, C.-C.; Fierke, C.A. Function and mechanism of zinc metalloenzymes. J. Nutr. 2000, 130, 1437S–1446S. [Google Scholar] [PubMed]
- Peroza, E.A.; Kaabi, A.A.; Meyer-Klaucke, W.; Wellenreuther, G.; Freisinger, E. The two distinctive metal ion binding domains of the wheat metallothionein Ec-1. J. Inorg. Biochem. 2009, 103, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Khuri, S.; Bakker, F.T.; Dunwell, J.M. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol. Biol. Evol. 2001, 18, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Cunsolo, V.; Muccilli, V.; Saletti, R.; Foti, S. Mass spectrometry in the proteome analysis of mature cereal kernels. Mass Spectrom. Rev. 2012, 31, 448–465. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.M.; Davidson, R.M.; Liu, B.; Zhu, X.; Hulbert, S.H.; Leung, H.; Leach, J.E. A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol. 2009, 149, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M.; Purvis, A.; Khuri, S. Cupins: The most functionally diverse protein superfamily? Phytochemistry 2004, 65, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Breiteneder, H.; Radauer, C. A classification of plant food allergens. J. Allergy Clin. Immunol. 2004, 113, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Nosworthy, N.; Caldwell, R.A. The zinc (ii) binding sites of soya bean glycinin. J. Sci. Food Agric. 1987, 41, 55–63. [Google Scholar] [CrossRef]
- Nosworthy, N.; Caldwell, R.A. The interaction of zinc (ii) and phytic acid with soya bean glycinin. J. Sci. Food Agric. 1988, 44, 143–150. [Google Scholar]
- Wang, Y.; Li, H.; Qiu, Y.F.; Li, N.; Sun, W.H.; Shan, Z.H. Identification of copper-binding proteins in soybean seeds by immobilized metal affinity chromatography and mass spectrometry. Plant Biosyst. 2014, 148, 88–95. [Google Scholar] [CrossRef]
- Zheng, H.G.; Yang, X.Q.; Ahmad, I.; Min, W.; Zhu, J.H.; Yuan, D.B. Soybean beta-conglycinin constituent subunits: Isolation, solubility and amino acid composition. Food Res. Int. 2009, 42, 998–1003. [Google Scholar] [CrossRef]
- Ralet, M.-C.; Fouques, D.; Leonil, J.; Molle, D.; Meunier, J.-C. Soybean β-conglycinin α subunit is phosphorylated on two distinct serines by protein kinase ck2 in vitro. J. Protein Chem. 1999, 18, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, B.; Ao, J. Separation and identification of zinc-chelating peptides from sesame protein hydrolysate using IMAC-Zn2+ and LC–MS/MS. Food Chem. 2012, 134, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Shewry, P. Identification of homologous globulins from embryos of wheat, barley, rye and oats. J. Exp. Bot. 1986, 37, 1863–1871. [Google Scholar] [CrossRef]
- Bell, S.G.; Vallee, B.L. The metallothionein/thionein system: An oxidoreductive metabolic zinc link. Chembiochem 2009, 10, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Florack, D.; Stiekema, W. Thionins: Properties, possible biological roles and mechanisms of action. Plant Mol. Biol. 1994, 26, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Kreis, M.; Forde, B.; Rahman, S.; Miflin, B.; Shewry, P. Molecular evolution of the seed storage proteins of barley, rye and wheat. J. Mol. Biol. 1985, 183, 499–502. [Google Scholar] [CrossRef]
- Holding, D.; Messing, J. Evolution, structure, and function of prolamin storage proteins. In Seed Genomics; John Wiley & Sons: New York, NY, USA, 2013; pp. 139–158. [Google Scholar]
- Shewry, P.R.; Tatham, A.S. The prolamin storage proteins of cereal seeds: Structure and evolution. Biochem. J. 1990, 267, 1. [Google Scholar] [CrossRef] [PubMed]
- Thaimattam, R.; Tykarska, E.; Bierzynski, A.; Sheldrick, G.; Jaskolski, M. Atomic resolution structure of squash trypsin inhibitor. Acta Crystallogr. Sect. D 2002, 58, 1448–1461. [Google Scholar] [CrossRef] [PubMed]
- Starks, T.L.; Johnson, P.E. Techniques for intrinsically labeling wheat with zinc-65. J. Agric. Food Chem. 1985, 33, 691–698. [Google Scholar] [CrossRef]
- Kutman, U.B.; Yildiz, B.; Ozturk, L.; Cakmak, I. Biofortification of durum wheat with zinc through soil and foliar applications of nitrogen. Cereal Chem. 2010, 87, 1–9. [Google Scholar] [CrossRef]
- Katayama, A.; Tsujii, A.; Wada, A.; Nishino, T.; Ishihama, A. Systematic search for zinc-binding proteins in escherichia coli. Eur. J. Biochem. 2002, 269, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.; Moore, P.; Steitz, T. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004, 340, 141–177. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Nissen, P.; Hansen, J.; Moore, P.B.; Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Härd, T.; Rak, A.; Allard, P.; Kloo, L.; Garber, M. The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold. J. Mol. Biol. 2000, 296, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Katz, B.A.; Elrod, K.; Luong, C.; Rice, M.J.; Mackman, R.L.; Sprengeler, P.A.; Spencer, J.; Hataye, J.; Janc, J.; Link, J. A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site. J. Mol. Biol. 2001, 307, 1451–1486. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.; Lipscomb, W. Refined crystal structure of the potato inhibitor complex of carboxypeptidase A at 2.5 Å resolution. J. Mol. Biol. 1982, 160, 475–498. [Google Scholar] [CrossRef]
- Hara, M.; Fujinaga, M.; Kuboi, T. Metal binding by citrus dehydrin with histidine-rich domains. J. Exp. Bot. 2005, 56, 2695–2703. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.; Xing, X.; Sun, L.; Pan, J.; Kong, X.; Zhang, M.; Li, D. Zmlea3, a multifunctional group 3 lea protein from maize (Zea mays L.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 2013, 54, 944–959. [Google Scholar] [CrossRef] [PubMed]
- Krüger, C.; Berkowitz, O.; Stephan, U.W.; Hell, R. A metal-binding member of the late embryogenesis abundant protein family transports iron in the phloem Ofricinus communis L. J. Biol. Chem. 2002, 277, 25062–25069. [Google Scholar] [CrossRef] [PubMed]
- Heyen, B.J.; Alsheikh, M.K.; Smith, E.A.; Torvik, C.F.; Seals, D.F.; Randall, S.K. The calcium-binding activity of a vacuole-associated, dehydrin-like protein is regulated by phosphorylation. Plant Physiol. 2002, 130, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.W.; Zhu, B.; Close, T.J. The barley (Hordeum vulgare L.) dehydrin multigene family: Sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor. Appl. Genet. 1999, 98, 1234–1247. [Google Scholar] [CrossRef]
- Alsheikh, M.K.; Heyen, B.J.; Randall, S.K. Ion binding properties of the dehydrin erd14 are dependent upon phosphorylation. J. Biol. Chem. 2003, 278, 40882–40889. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, M.; Hincha, D.K. Lea (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Hincha, D.; Thalhammer, A. Lea proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem. Soc. Trans. 2012, 40, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using targetp, signalp and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Dure, L.; Crouch, M.; Harada, J.; Ho, T.H.D.; Mundy, J.; Quatrano, R.; Thomas, T.; Sung, Z.R. Common amino-acid sequence domains among the lea proteins of higher-plants. Plant Mol. Biol. 1989, 12, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Rohrig, H.; Schmidt, J.; Colby, T.; Brautigam, A.; Hufnagel, P.; Bartels, D. Desiccation of the resurrection plant Craterostigma plantagineum induces dynamic changes in protein phosphorylation. Plant Cell Environ. 2006, 29, 1606–1617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.J.; Cascio, D.; Eisenberg, D. Crystal-structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition-state analog, 2-carboxy-d-arabinitol 1,5-bisphosphate. Protein Sci. 1994, 3, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Xiao, W.; Hao, H.; Xiaoqing, L.; Chao, L.; Lei, Z.; Fashui, H. Effect of Mg2+ on the structure and function of ribulose-1,5-bisphosphate carboxylase/oxygenase. Biol. Trace Elem. Res. 2008, 121, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Cléry, A.; Blatter, M.; Allain, F.H. RNA recognition motifs: Boring? Not quite. Curr. Opin. Struct. Biol. 2008, 18, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Kimball, S.R. Eukaryotic initiation factor eif2. Int. J. Biochem. Cell Biol. 1999, 31, 25–29. [Google Scholar] [CrossRef]
- Shen, Y.-G.; Zhang, W.-K.; He, S.-J.; Zhang, J.-S.; Liu, Q.; Chen, S.-Y. An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and aba stress. Theor. Appl. Genet. 2003, 106, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Rorat, T. Plant dehydrins—Tissue location, structure and function. Cell. Mol. Biol. Lett. 2006, 11, 536–556. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dionisio, G.; Uddin, M.N.; Vincze, E. Enrichment and Identification of the Most Abundant Zinc Binding Proteins in Developing Barley Grains by Zinc-IMAC Capture and Nano LC-MS/MS. Proteomes 2018, 6, 3. https://doi.org/10.3390/proteomes6010003
Dionisio G, Uddin MN, Vincze E. Enrichment and Identification of the Most Abundant Zinc Binding Proteins in Developing Barley Grains by Zinc-IMAC Capture and Nano LC-MS/MS. Proteomes. 2018; 6(1):3. https://doi.org/10.3390/proteomes6010003
Chicago/Turabian StyleDionisio, Giuseppe, Mohammad Nasir Uddin, and Eva Vincze. 2018. "Enrichment and Identification of the Most Abundant Zinc Binding Proteins in Developing Barley Grains by Zinc-IMAC Capture and Nano LC-MS/MS" Proteomes 6, no. 1: 3. https://doi.org/10.3390/proteomes6010003
APA StyleDionisio, G., Uddin, M. N., & Vincze, E. (2018). Enrichment and Identification of the Most Abundant Zinc Binding Proteins in Developing Barley Grains by Zinc-IMAC Capture and Nano LC-MS/MS. Proteomes, 6(1), 3. https://doi.org/10.3390/proteomes6010003