Protein Oxidation in the Lungs of C57BL/6J Mice Following X-Irradiation
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Animals
2.3. Thoracic Irradiation
2.4. Protein Oxidation Detection by OxyBlot
2.5. Two Dimensional (2-D) Gel Electrophoresis
2.6. Peptide Mass Fingerprinting for Protein Identification
2.7. DNP-Labeled Protein Confirmation by Immunopercipitation and Western Blotting
2.8. Statistics
3. Results and Discussion
3.1. Survival Studies: 17 Gy Thoracic Irradiation Induces Lung Fibrosis within 180 Days
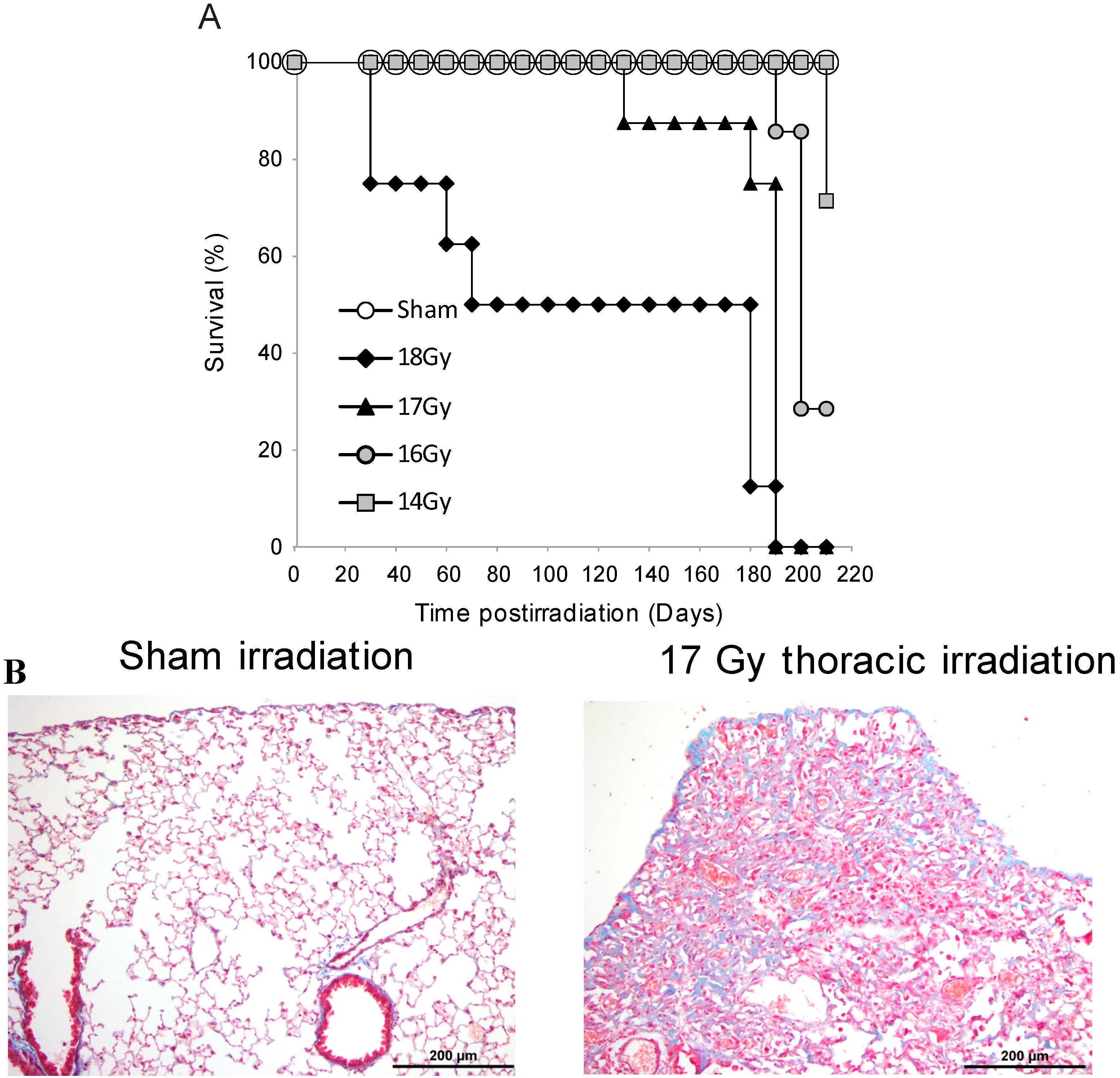
3.2. Radiation-Induced Protein Carbonylation in Lung Tissue
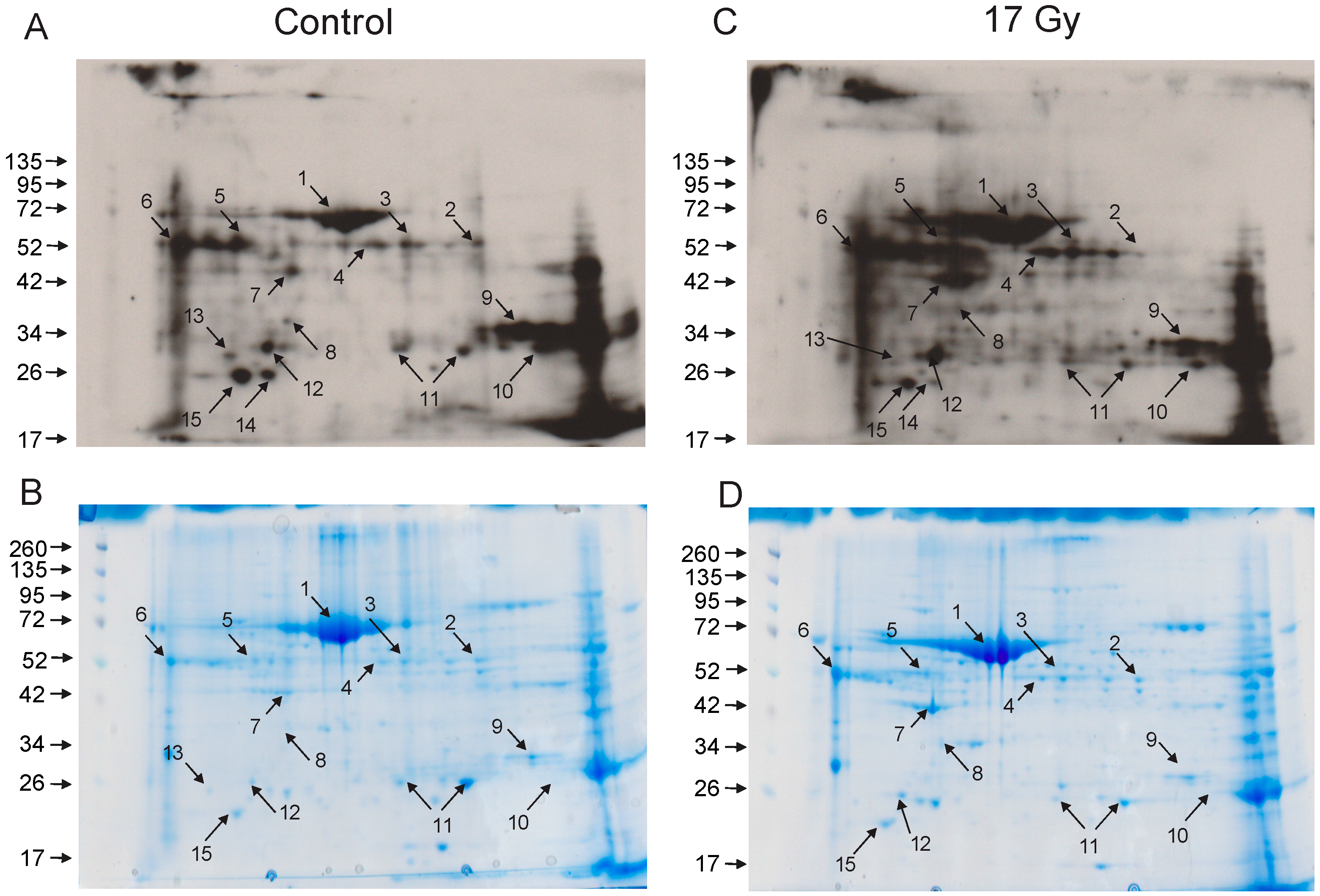
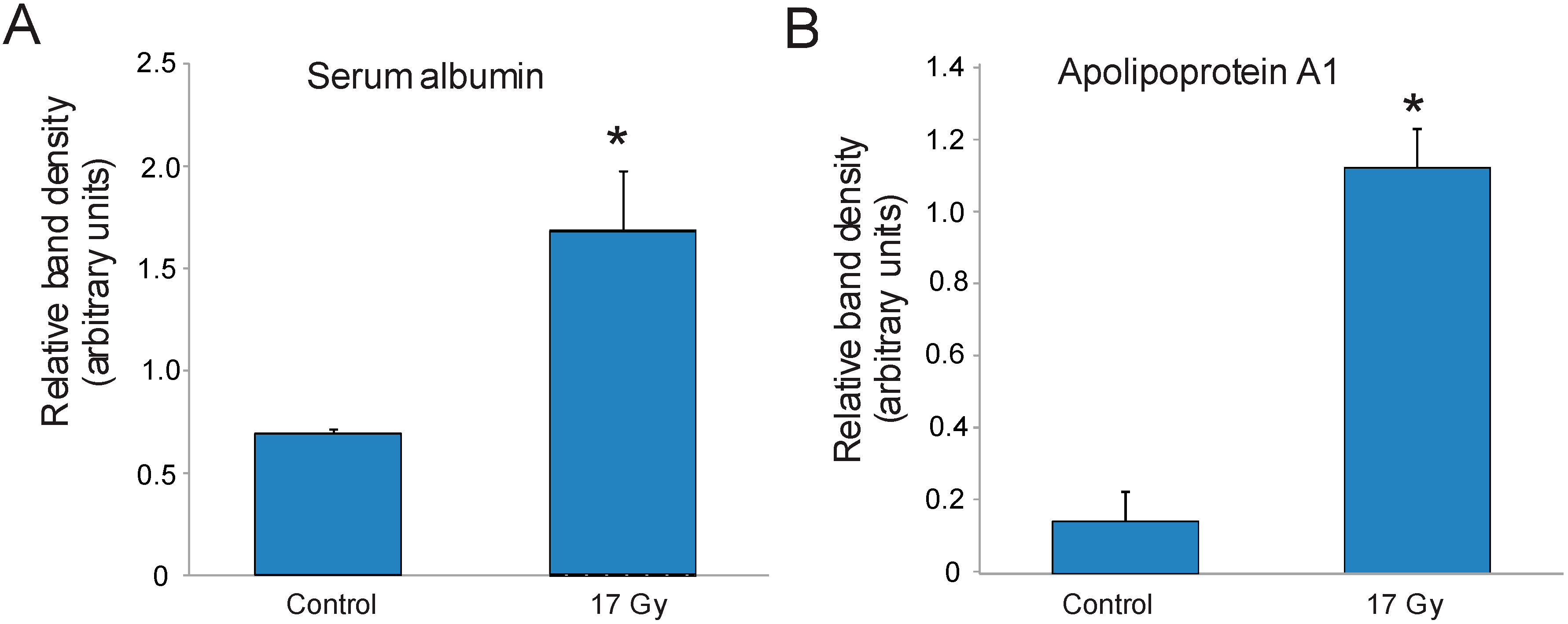
3.3. Identification of Carbonylated Proteins from Control and Irradiated Lung Tissue
| Spot No. | Protein ID | Number of Peptides | % Coverage |
|---|---|---|---|
| 1 | Serum albumin | 20 | 40 |
| 2 | Selenium binding protein-1 (SBP1) | 12 | 36 |
| 6 | Alpha-1-antitrypsin | 9 | 26 |
| 7 | Actin, cytoplasmic | 9 | 36 |
| 9 | Carbonic anhydrase 2 (CAII) | 9 | 50 |
| 11 | Peroxiredoxin-6 | 16 | 75 |
| 12 | Apolipoprotein A1 | 15 | 43 |
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Movsas, B.; Raffin, T.A.; Epstein, A.H.; Link, C.J., Jr. Pulmonary radiation injury. Chest 1997, 111, 1061–1076. [Google Scholar] [CrossRef] [PubMed]
- Bentzen, S.M.; Skoczylas, J.Z.; Bernier, J. Quantitative clinical radiobiology of early and late lung reactions. Int. J. Radiat. Biol. 2000, 76, 453–462. [Google Scholar] [PubMed]
- Marks, L.B.; Yu, X.; Vujaskovic, Z.; Small, W., Jr.; Folz, R.; Anscher, M.S. Radiation-induced lung injury. Semin. Radiat. Oncol. 2003, 13, 333–345. [Google Scholar] [CrossRef]
- Claude, L.; Perol, D.; Ginestet, C.; Falchero, L.; Arpin, D.; Vincent, M.; Martel, I.; Hominal, S.; Cordier, J.F.; Carrie, C. A prospective study on radiation pneumonitis following conformal radiation therapy in non-small-cell lung cancer: Clinical and dosimetric factors analysis. Radiother. Oncol. 2004, 71, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.N.; Yan, P.; Yuan, G.H.; Lv, X.Y.; Gong, H.; Zhao, H.; Wang, Y.M. Advantages of CyberKnife for inoperable stage I peripheral non-small-cell lung cancer compared to three-dimensional conformal radiotherapy. Mol. Clin. Oncol. 2015, 3, 442–448. [Google Scholar] [PubMed]
- Abdulla, S.; Salavati, A.; Saboury, B.; Basu, S.; Torigian, D.A.; Alavi, A. Quantitative assessment of global lung inflammation following radiation therapy using FDG PET/CT: A pilot study. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Robnett, T.J.; Machtay, M.; Vines, E.F.; McKenna, M.G.; Algazy, K.M.; McKenna, W.G. Factors predicting severe radiation pneumonitis in patients receiving definitive chemoradiation for lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2000, 48, 89–94. [Google Scholar] [CrossRef]
- Hughes-Davies, L.; Tarbell, N.J.; Coleman, C.N.; Silver, B.; Shulman, L.N.; Linggood, R.; Canellos, G.P.; Mauch, P.M. Stage IA–IIB Hodgkin’s disease: Management and outcome of extensive thoracic involvement. Int. J. Radiat. Oncol. Biol. Phys. 1997, 39, 361–369. [Google Scholar] [CrossRef]
- Ghafoori, P.; Marks, L.B.; Vujaskovic, Z.; Kelsey, C.R. Radiation-induced lung injury. Assessment, management, and prevention. Oncology 2008, 22, 37–47. [Google Scholar] [PubMed]
- Abid, S.H.; Malhotra, V.; Perry, M.C. Radiation-induced and chemotherapy-induced pulmonary injury. Curr. Opin. Oncol. 2001, 13, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Carlson, K.; Backlund, L.; Smedmyr, B.; Oberg, G.; Simonsson, B. Pulmonary function and complications subsequent to autologous bone marrow transplantation. Bone Marrow Transpl. 1994, 14, 805–811. [Google Scholar]
- Sampath, S.; Schultheiss, T.E.; Wong, J. Dose response and factors related to interstitial pneumonitis after bone marrow transplant. Int. J. Radiat. Oncol. Biol. Phys. 2005, 63, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Baranov, A.E.; Selidovkin, G.D.; Butturini, A.; Gale, R.P. Hematopoietic recovery after 10-Gy acute total body radiation. Blood 1994, 83, 596–599. [Google Scholar] [PubMed]
- Uozaki, H.; Fukayama, M.; Nakagawa, K.; Ishikawa, T.; Misawa, S.; Doi, M.; Maekawa, K. The pathology of multi-organ involvement: Two autopsy cases from the Tokai-mura criticality accident. BJR Suppl. 2005, 27, 13–16. [Google Scholar] [CrossRef]
- Crawford, S.W. Diagnosis and management of pulmonary problems associated with radiation injury. In The Medical Basis for Radiation-Accident Preparedness. The Clinical Care of Victims; Ricks, R.C., Berger, M.E., O’Hara, F.M., Eds.; The Parthenon Publishing Group: Boca Raton, FL, USA, 2002; pp. 131–138. [Google Scholar]
- Selman, M.; Thannickal, V.J.; Pardo, A.; Zisman, D.A.; Martinez, F.J.; Lynch, J.P., 3rd. Idiopathic pulmonary fibrosis: Pathogenesis and therapeutic approaches. Drugs 2004, 64, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, T.; Baumann, M.; Voigtmann, L.; Knorr, A. Effect of irradiated volume on lung damage in pigs. Radiother. Oncol. 1997, 44, 35–40. [Google Scholar] [CrossRef]
- Augustine, A.D.; Gondre-Lewis, T.; McBride, W.; Miller, L.; Pellmar, T.C.; Rockwell, S. Animal models for radiation injury, protection and therapy. Radiat. Res. 2005, 164, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Calveley, V.L.; Khan, M.A.; Yeung, I.W.; Vandyk, J.; Hill, R.P. Partial volume rat lung irradiation: Temporal fluctuations of in-field and out-of-field DNA damage and inflammatory cytokines following irradiation. Int. J. Radiat. Biol. 2005, 81, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 2002, 33, 1–14. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Leapman, R.D.; Lai, B.; Ravel, B.; Li, S.M.; Kemner, K.M.; et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007, 5, e92. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Gebicki, J.M. Proteins are major initial cell targets of hydroxyl free radicals. Int. J. Biochem. Cell Biol. 2004, 36, 2334–2343. [Google Scholar] [CrossRef] [PubMed]
- Panganiban, R.A.; Mungunsukh, O.; Day, R.M. X-irradiation induces ER stress, apoptosis, and senescence in pulmonary artery endothelial cells. Int. J. Radiat. Biol. 2013, 89, 656–667. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lorimore, S.A.; Evans, C.A.; Whetton, A.D.; Wright, E.G. A proteomic analysis of murine bone marrow and its response to ionizing radiation. Proteomics 2005, 5, 4254–4263. [Google Scholar] [CrossRef] [PubMed]
- Barshishat-Kupper, M.; Tipton, A.J.; McCart, E.A.; McCue, J.; Mueller, G.P.; Day, R.M. Effect of ionizing radiation on liver protein oxidation and metabolic function in C57BL/6J mice. Int. J. Radiat. Biol. 2014, 90, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Barshishat-Kupper, M.; McCart, E.A.; Mueller, G.P.; Day, R.M. Bone marrow protein oxidation in response to ionizing radiation in C57BL/6J mice. Proteomes 2014, 2, 291–302. [Google Scholar] [CrossRef]
- McPherson, C. Reduction of Psuedomonas aeruginosa and coliform bacteria in mouse drinking water following treatment with hyrdochloric acid and chlorine. Lab. Anim. Care 1963, 13, 737–744. [Google Scholar] [PubMed]
- Ma, C.M.; Coffey, C.W.; DeWerd, L.A.; Liu, C.; Nath, R.; Seltzer, S.M.; Seuntjens, J.P. AAPM protocol for 40–300 kV X-ray beam dosimetry in radiotherapy and radiobiology. Med. Phys. 2001, 28, 868–893. [Google Scholar] [CrossRef] [PubMed]
- Standard practice for use of an alanine-EPR dosimetry system. In ISO/ASTM International Standard 51608–2013(E); ISO International: Geneva, Switzerland; ASTM: West Conshohocken, PA, USA, 2013; p. 7.
- Muth, E.; Driscoll, W.J.; Smalstig, A.; Goping, G.; Mueller, G.P. Proteomic analysis of rat atrial secretory granules: A platform for testable hypotheses. Biochim. Biophys. Acta 2004, 1699, 263–275. [Google Scholar] [CrossRef]
- ProteinProspector MS-Fit Search Engine. Available online: http://prospector.ucsf.edu/prospector/mshome.htm (accessed on 26 August 2012).
- Clauser, K.R.; Baker, P.; Burlingame, A.L. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 1999, 71, 2871–2882. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Cheema, A.K.; Zhang, L.; Suzuki, Y.J. Protein carbonylation as a novel mechanism in redox signaling. Circ. Res. 2008, 102, 310–318. [Google Scholar] [CrossRef] [PubMed]
- ImageJ softward. Available online: http://www.uhnresearch.ca/facilities/wcif/index.htm (accessed on 12 December 2013).
- Paun, A.; Lemay, A.M.; Haston, C.K. Gene expression profiling distinguishes radiation-induced fibrosing alveolitis from alveolitis in mice. Radiat. Res. 2010, 173, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Madian, A.G.; Myracle, A.D.; Diaz-Maldonado, N.; Rochelle, N.S.; Janle, E.M.; Regnier, F.E. Differential carbonylation of proteins as a function of in vivo oxidative stress. J. Proteome Res. 2011, 10, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- England, K.; Cotter, T. Identification of carbonylated proteins by MALDI-TOF mass spectroscopy reveals susceptibility of ER. Biochem. Biophys. Res. Commun. 2004, 320, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Oikawa, M.; Tamagami, T.; Kumaki, H.; Nakaune, R.; Amano, J.; Akinaga, Y.; Fukui, K.; Abe, K.; Urano, S. Oxidized proteins in astrocytes generated in a hyperbaric atmosphere induce neuronal apoptosis. J. Alzheimers Dis. 2007, 11, 165–174. [Google Scholar] [PubMed]
- Linares, M.; Marin-Garciia, P.; Mendez, D.; Puyet, A.; Diez, A.; Bautista, J.M. Proteomic approaches to identifying carbonylated proteins in brain tissue. J. Proteome Res. 2011, 10, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.S.; Kunwar, A.; Ahmad, A.; Kumbhare, L.B.; Jain, V.K.; Priyadarsini, K.I. In vitro radioprotection studies of organoselenium compounds: Differences between mono- and diselenides. Radiat. Environ. Biophys. 2009, 48, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Clerici, M.; Colombo, G.; Secundo, F.; Gagliano, N.; Colombo, R.; Portinaro, N.; Giustarini, D.; Milzani, A.; Rossi, R.; Dalle-Donne, I. Cigarette smoke induces alterations in the drug-binding properties of human serum albumin. Blood Cells Mol. Dis. 2014, 52, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Colombo, G.; Aldini, G.; Orioli, M.; Giustarini, D.; Gornati, R.; Rossi, R.; Colombo, R.; Carini, M.; Milzani, A.; Dalle-Donne, I. Water-Soluble α,β-unsaturated aldehydes of cigarette smoke induce carbonylation of human serum albumin. Antioxid. Redox Signal. 2010, 12, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Albarello, K.; dos Santos, G.A.; Bochi, G.V.; Sangoi, M.B.; Almeida, T.C.; Paz da Silva, J.E.; Garcia, S.C.; Moresco, R.N. Ischemia modified albumin and carbonyl protein as potential biomarkers of protein oxidation in hemodialysis. Clin. Biochem. 2012, 45, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Pavone, B.; Sirolli, V.; Giardinelli, A.; Bucci, S.; Forli, F.; Di Cesare, M.; Sacchetta, P.; Di Pietro, N.; Pandolfi, A.; Urbani, A.; Bonomini, M. Plasma protein carbonylation in chronic uremia. J. Nephrol. 2011, 24, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, Y.; Terawaki, H.; Terada, T.; Era, S. Albumin thiol oxidation and serum protein carbonyl formation are progressively enhanced with advancing stages of chronic kidney disease. Clin. Exp. Nephrol. 2009, 13, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Mendez, L.; Pazos, M.; Giralt, M.; Nogues, M.R.; Perez-Jimenez, J.; Torres, J.L.; Gallardo, J.M.; Medina, I. Targets of protein carbonylation in spontaneously hypertensive obese Koletsky rats and healthy Wistar counterparts: A potential role on metabolic disorders. J. Proteom. 2014, 106, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Mendez, L.; Pazos, M.; Molinar-Toribio, E.; Sanchez-Martos, V.; Gallardo, J.M.; Rosa Nogues, M.; Torres, J.L.; Medina, I. Protein carbonylation associated to high-fat, high-sucrose diet and its metabolic effects. J. Nutr. Biochem. 2014, 25, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Betsuyaku, T.; Konno, S.; Ito, Y.; Nasuhara, Y.; Hizawa, N.; Kondo, T.; Nishimura, M. Diversity of protein carbonylation in allergic airway inflammation. Free Radic. Res. 2008, 42, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Leber, B.; Stadlbauer, V.; Stiegler, P.; Stanzer, S.; Mayrhauser, U.; Koestenbauer, S.; Leopold, B.; Sereinigg, M.; Puntschart, A.; Stojakovic, T.; et al. Effect of oxidative stress and endotoxin on human serum albumin in brain-dead organ donors. Transl. Res. 2012, 159, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Bansal, M.P.; Mukhopadhyay, T.; Scott, J.; Cook, R.G.; Mukhopadhyay, R.; Medina, D. DNA sequencing of a mouse liver protein that binds selenium: Implications for selenium’s mechanism of action in cancer prevention. Carcinogenesis 1990, 11, 2071–2073. [Google Scholar] [CrossRef] [PubMed]
- Papp, L.V.; Holmgren, A.; Khanna, K.K. Selenium and selenoproteins in health and disease. Antioxid. Redox Signal. 2010, 12, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fang, W.; Huang, Y.; Hu, F.; Ying, Q.; Yang, W.; Xiong, B. Reduction of selenium-binding protein 1 sensitizes cancer cells to selenite via elevating extracellular glutathione: A novel mechanism of cancer-specific cytotoxicity of selenite. Free Radic. Biol. Med. 2015, 79, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Ghouse, R.; Chu, A.; Wang, Y.; Perlmutter, D.H. Mysteries of alpha1-antitrypsin deficiency: Emerging therapeutic strategies for a challenging disease. Dis. Model. Mech. 2014, 7, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Banfi, C.; Brioschi, M.; Barcella, S.; Veglia, F.; Biglioli, P.; Tremoli, E.; Agostoni, P. Oxidized proteins in plasma of patients with heart failure: Role in endothelial damage. Eur. J. Heart Fail. 2008, 10, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.L.; Chertkow, H.M.; Bergman, H.; Schipper, H.M. Aberrant profiles of native and oxidized glycoproteins in Alzheimer plasma. Proteomics 2003, 3, 2240–2248. [Google Scholar] [CrossRef] [PubMed]
- Korolainen, M.A.; Nyman, T.A.; Nyyssonen, P.; Hartikainen, E.S.; Pirttila, T. Multiplexed proteomic analysis of oxidation and concentrations of cerebrospinal fluid proteins in Alzheimer disease. Clin. Chem. 2007, 53, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Steiner, S.R.; Philbert, M.A. Proteomic identification of carbonylated proteins in 1,3-dinitrobenzene neurotoxicity. Neurotoxicology 2011, 32, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, E.; Hussain, S.N. Protein carbonylation in skeletal muscles: Impact on function. Antioxid. Redox Signal. 2010, 12, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.G.; Chae, H.Z.; Kim, K. Peroxiredoxins: A historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005, 38, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Marcocci, L.; Das, D.; Wang, X.; Luo, H.; Zungu-Edmondson, M.; Suzuki, Y.J. Mechanism of protein decarbonylation. Free Radic. Biol. Med. 2013, 65, 1126–1133. [Google Scholar] [CrossRef] [PubMed]
- De Waal, E.M.; Liang, H.; Pierce, A.; Hamilton, R.T.; Buffenstein, R.; Chaudhuri, A.R. Elevated protein carbonylation and oxidative stress do not affect protein structure and function in the long-living naked-mole rat: A proteomic approach. Biochem. Biophys. Res. Commun. 2013, 434, 815–819. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, X.; He, S.; Liu, Y.; An, M.; Ji, J. Differential proteomics analysis of specific carbonylated proteins in the temporal cortex of aged rats: The deterioration of antioxidant system. Neurochem. Res. 2010, 35, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Tran-Dinh, A.; Diallo, D.; Delbosc, S.; Varela-Perez, L.M.; Dang, Q.B.; Lapergue, B.; Burillo, E.; Michel, J.B.; Levoye, A.; Martin-Ventura, J.L.; et al. HDL and endothelial protection. Br. J. Pharmacol. 2013, 169, 493–511. [Google Scholar] [CrossRef] [PubMed]
- Aluise, C.D.; Miriyala, S.; Noel, T.; Sultana, R.; Jungsuwadee, P.; Taylor, T.J.; Cai, J.; Pierce, W.M.; Vore, M.; Moscow, J.A.; et al. 2-Mercaptoethane sulfonate prevents doxorubicin-induced plasma protein oxidation and TNF-α release: Implications for the reactive oxygen species-mediated mechanisms of chemobrain. Free Radic. Biol. Med. 2011, 50, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Perluigi, M.; di Domenico, F.; Fiorini, A.; Cocciolo, A.; Giorgi, A.; Foppoli, C.; Butterfield, D.A.; Giorlandino, M.; Giorlandino, C.; Schinina, M.E.; et al. Oxidative stress occurs early in Down syndrome pregnancy: A redox proteomics analysis of amniotic fluid. Proteom. Clin. Appl. 2011, 5, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Thomas-Moya, E.; Gomez-Perez, Y.; Fiol, M.; Gianotti, M.; Llado, I.; Proenza, A.M. Gender related differences in paraoxonase 1 response to high-fat diet-induced oxidative stress. Obesity 2008, 16, 2232–2238. [Google Scholar] [CrossRef] [PubMed]
- Yassine, H.N.; Jackson, A.M.; Borges, C.R.; Billheimer, D.; Koh, H.; Smith, D.; Reaven, P.; Lau, S.S.; Borchers, C.H. The application of multiple reaction monitoring and multi-analyte profiling to HDL proteins. Lipids Health Dis. 2014, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Sartore, G.; Seraglia, R.; Burlina, S.; Bolis, A.; Marin, R.; Manzato, E.; Ragazzi, E.; Traldi, P.; Lapolla, A. High-density lipoprotein oxidation in type 2 diabetic patients and young patients with premature myocardial infarction. Nutr. Metab. Cardiovasc. Dis. 2015, 25, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Vivekananda, J.; Awasthi, V.; Awasthi, S.; Smith, D.B.; King, R.J. Hepatocyte growth factor is elevated in chronic lung injury and inhibits surfactant metabolism. Am. J. Physiol. Lung Cell Mol. Physiol. 2000, 278, L382–L392. [Google Scholar] [PubMed]
- Miyazaki, A.; Sagae, N.; Usami, Y.; Sato, M.; Kameda, T.; Yoshimoto, A.; Ishimine, N.; Matsuda, K.; Sugano, M.; Hara, M.; et al. N-homocysteinylation of apolipoprotein A-I impairs the protein’s antioxidant ability but not its cholesterol efflux capacity. Biol. Chem. 2014, 395, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; DiDonato, J.A.; Levison, B.S.; Schmitt, D.; Li, L.; Wu, Y.; Buffa, J.; Kim, T.; Gerstenecker, G.S.; Gu, X.; et al. An abundant dysfunctional apolipoprotein A1 in human atheroma. Nat. Med. 2014, 20, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Vuilleumier, N.; Dayer, J.M.; von Eckardstein, A.; Roux-Lombard, P. Pro- or anti-inflammatory role of apolipoprotein A-1 in high-density lipoproteins? Swiss Med. Wkly. 2013, 143. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barshishat-Kupper, M.; McCart, E.A.; Freedy, J.G.; Tipton, A.J.; Nagy, V.; Kim, S.-Y.; Landauer, M.R.; Mueller, G.P.; Day, R.M. Protein Oxidation in the Lungs of C57BL/6J Mice Following X-Irradiation. Proteomes 2015, 3, 249-265. https://doi.org/10.3390/proteomes3030249
Barshishat-Kupper M, McCart EA, Freedy JG, Tipton AJ, Nagy V, Kim S-Y, Landauer MR, Mueller GP, Day RM. Protein Oxidation in the Lungs of C57BL/6J Mice Following X-Irradiation. Proteomes. 2015; 3(3):249-265. https://doi.org/10.3390/proteomes3030249
Chicago/Turabian StyleBarshishat-Kupper, Michal, Elizabeth A. McCart, James G. Freedy, Ashlee J. Tipton, Vitaly Nagy, Sung-Yop Kim, Michael R. Landauer, Gregory P. Mueller, and Regina M. Day. 2015. "Protein Oxidation in the Lungs of C57BL/6J Mice Following X-Irradiation" Proteomes 3, no. 3: 249-265. https://doi.org/10.3390/proteomes3030249
APA StyleBarshishat-Kupper, M., McCart, E. A., Freedy, J. G., Tipton, A. J., Nagy, V., Kim, S.-Y., Landauer, M. R., Mueller, G. P., & Day, R. M. (2015). Protein Oxidation in the Lungs of C57BL/6J Mice Following X-Irradiation. Proteomes, 3(3), 249-265. https://doi.org/10.3390/proteomes3030249






