Evaluating Protein Extraction Techniques for Elucidating Proteomic Changes in Yeast Deletion Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Yeast Strains
2.2. Growth Conditions
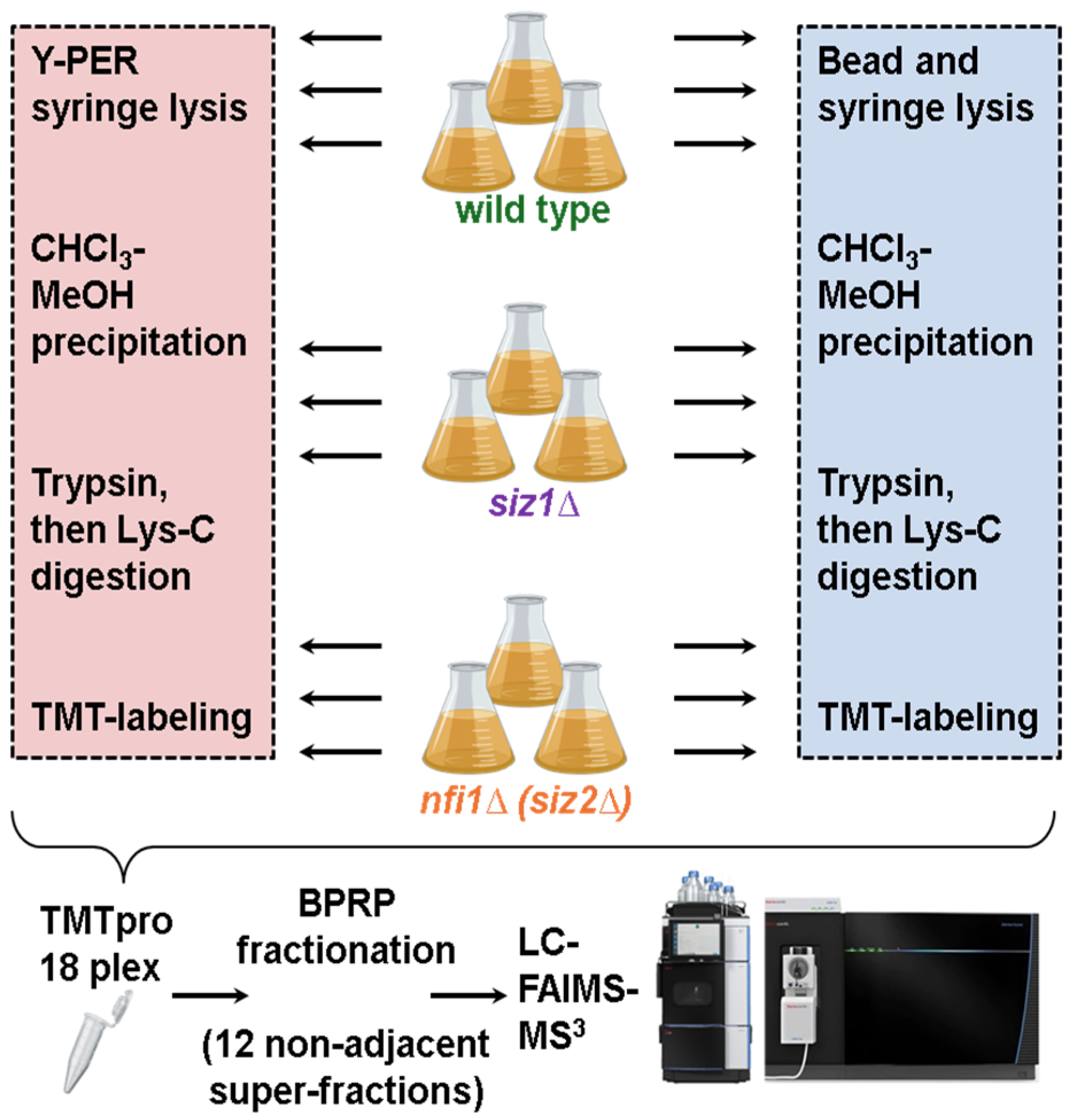
2.3. Cell Lysis and Protein Extraction
2.4. Protein Digestion, TMT Labeling, and Sample Processing
2.5. Basic pH Reversed-Phase (BPRP) Fractionation—Peptide Pre-Fractionation Was Carried out Using Basic pH Reversed-Phase (BPRP) High-Performance Liquid Chromatography (HPLC)
2.6. Liquid Chromatography and Mass Spectrometry Analysis
2.7. Mass Spectrometry Data Analysis
3. Results and Discussion
3.1. Sample Preparation Was Performed in Parallel with Two Lysis Methods
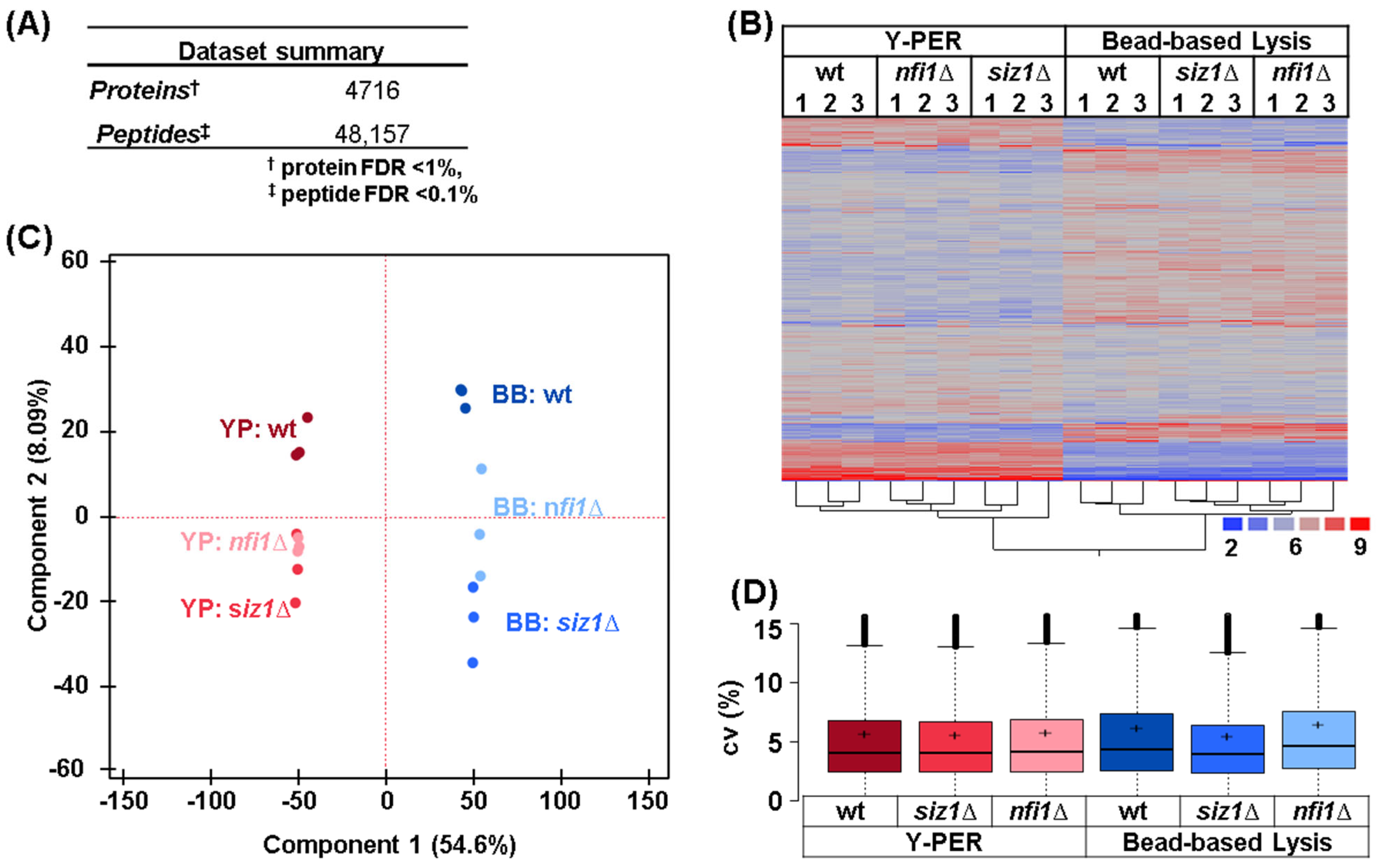
3.2. Hierarchical Clustering and Principal Component Analysis Revealed Distinct Impacts of Lysis Methods on Protein Profiles
3.3. Detergent-Based Lysis Demonstrated a Slight Enhancement in Protein Extraction Efficiency
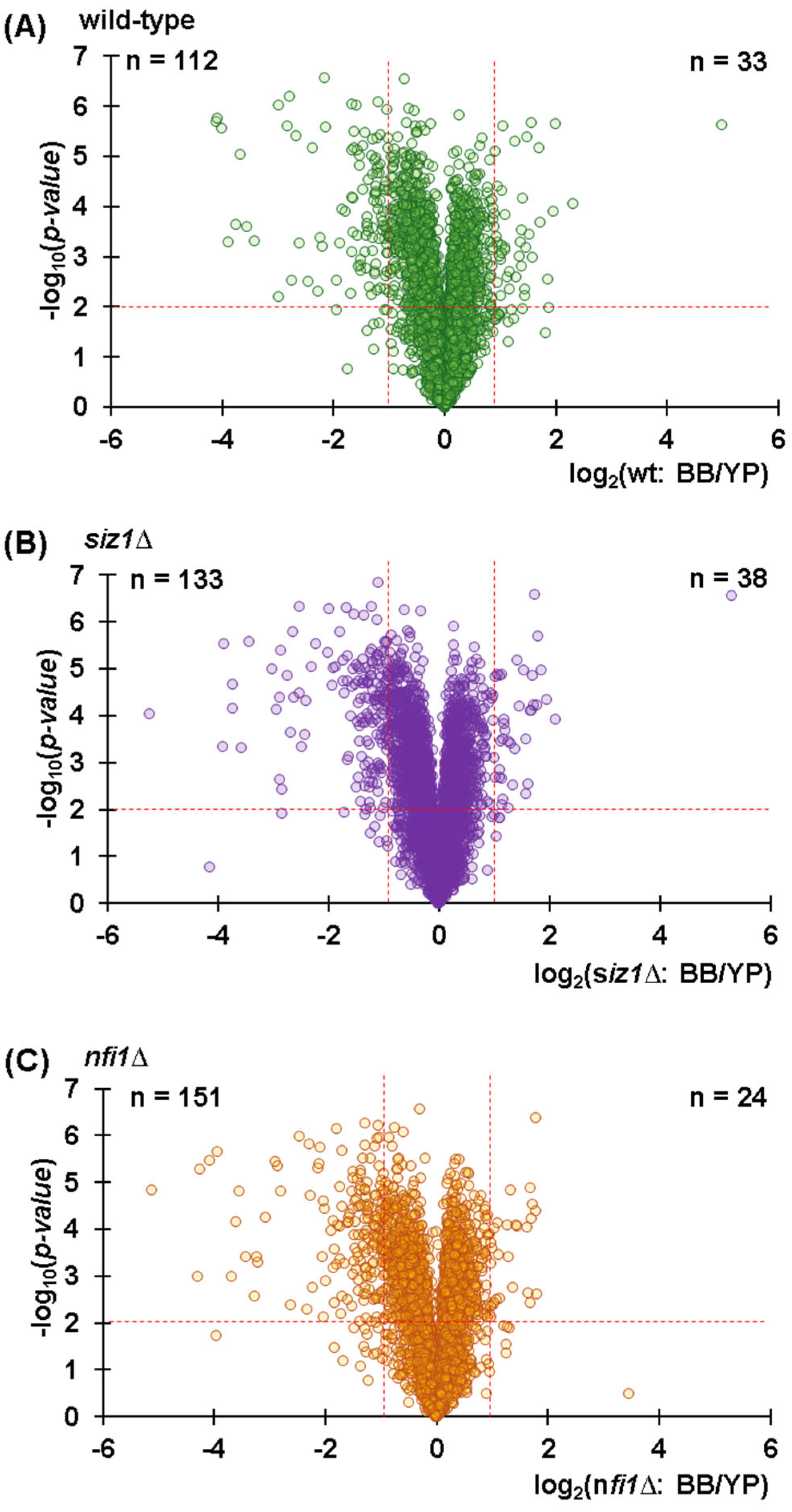
3.4. Deleting SUMO E3 Ligases SIZ1 and NFI1 Resulted in Minimal Proteomic Alterations, Indicating Potential Redundant Function
3.5. Several Proteins Show Significant Abundance Differences Across Strains
3.6. Functional Enrichment Analysis Underscored the Effects of Gene Deletions on Cellular Processes
3.7. Conclusions and Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement:
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Botstein, D.; Fink, G.R. Yeast: An experimental organism for 21st Century biology. Genetics 2011, 189, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Karboune, S. A comparative study for the isolation and characterization of mannoproteins from Saccharomyces cerevisiae yeast cell wall. Int. J. Biol. Macromol. 2018, 119, 654–661. [Google Scholar] [CrossRef]
- Mukherjee, M.; Nandi, A.; Chandra, K.; Saikia, S.K.; Jana, C.K.; Das, N. Protein extraction from Saccharomyces cerevisiae at different growth phases. J. Microbiol. Methods 2020, 172, 105906. [Google Scholar] [CrossRef]
- Rossio, V.; Paulo, J.A. Comparison of the Proteomes and Phosphoproteomes of S. cerevisiae Cells Harvested with Different Strategies. Proteomes 2023, 11, 28. [Google Scholar] [CrossRef]
- Zhao, Y.; Han, Z.; Zhu, X.; Chen, B.; Zhou, L.; Liu, X.; Liu, H. Yeast Proteins: Proteomics, Extraction, Modification, Functional Characterization, and Structure: A Review. J. Agric. Food Chem. 2024, 72, 18774–18793. [Google Scholar] [CrossRef]
- Papanayotou, I.; Sun, B.; Roth, A.F.; Davis, N.G. Protein aggregation induced during glass bead lysis of yeast. Yeast 2010, 27, 801–816. [Google Scholar] [CrossRef]
- Arachea, B.T.; Sun, Z.; Potente, N.; Malik, R.; Isailovic, D.; Viola, R.E. Detergent selection for enhanced extraction of membrane proteins. Protein Expr. Purif. 2012, 86, 12–20. [Google Scholar] [CrossRef]
- Schuster, M.; Tielker, D.; Schaffer, S. Extraction of yeast mitochondrial membrane proteins by nondenaturing detergent/polymer two-phase system partitioning. Anal. Biochem. 2009, 385, 67–74. [Google Scholar] [CrossRef]
- Boukli, N.M.; Delgado, Y.; Vázquez, A.; Ortiz, J.G.; Rodríguez, L.V. Reliable Approach for Pure Yeast Cell Wall Protein Isolation from Saccharomyces cerevisiae. Int. J. Mol. Sci. 2022, 23, 9435. [Google Scholar] [CrossRef]
- Packeiser, H.; Lim, C.; Balagurunathan, B.; Wu, J.; Zhao, H. An extremely simple and effective colony PCR procedure for bacteria, yeasts, and microalgae. Appl. Biochem. Biotechnol. 2013, 169, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Silver, H.R.; Xiong, L.; Belichenko, I.; Adegite, C.; Johnson, E.S. Topoisomerase I-dependent viability loss in Saccharomyces cerevisiae mutants defective in both SUMO conjugation and DNA repair. Genetics 2007, 177, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Reindle, A.; Belichenko, I.; Bylebyl, G.R.; Chen, X.L.; Gandhi, N.; Johnson, E.S. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 2006, 119, 4749–4757. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Toh, E.A.; Kikuchi, Y. Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 2003, 133, 415–422. [Google Scholar] [CrossRef]
- Rossio, V.; Liu, X.; Paulo, J.A. Comparative Proteomic Analysis of Two Commonly Used Laboratory Yeast Strains: W303 and BY4742. Proteomes 2023, 11, 30. [Google Scholar] [CrossRef]
- McAlister, G.C.; Huttlin, E.L.; Haas, W.; Ting, L.; Jedrychowski, M.P.; Rogers, J.C.; Kuhn, K.; Pike, I.; Grothe, R.A.; Blethrow, J.D.; et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 2012, 84, 7469–7478. [Google Scholar] [CrossRef]
- Edwards, A.; Haas, W. Multiplexed Quantitative Proteomics for High-Throughput Comprehensive Proteome Comparisons of Human Cell Lines. Methods Mol. Biol. 2016, 1394, 1–13. [Google Scholar]
- Zhang, T.; Liu, X.; Rossio, V.; Dawson, S.L.; Gygi, S.P.; Paulo, J.A. Enhancing Proteome Coverage by Using Strong Anion-Exchange in Tandem with Basic-pH Reversed-Phase Chromatography for Sample Multiplexing-Based Proteomics. J. Proteome Res. 2024, 23, 2870–2881. [Google Scholar] [CrossRef]
- Erickson, B.K.; Mintseris, J.; Schweppe, D.K.; Navarrete-Perea, J.; Erickson, A.R.; Nusinow, D.P.; Paulo, J.A.; Gygi, S.P. Active Instrument Engagement Combined with a Real-Time Database Search for Improved Performance of Sample Multiplexing Workflows. J. Proteome Res. 2019, 18, 1299–1306. [Google Scholar] [CrossRef]
- Navarrete-Perea, J.; Gygi, S.P.; Paulo, J.A. Temporal proteomic changes induced by nicotine in human cells: A quantitative proteomics approach. J. Proteom. 2021, 241, 104244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Gygi, S.P.; Paulo, J.A. Temporal Proteomic Profiling of SH-SY5Y Differentiation with Retinoic Acid Using FAIMS and Real-Time Searching. J. Proteome Res. 2021, 20, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Navarrete-Perea, J.; Yu, Q.; Gygi, S.P.; Paulo, J.A. Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J. Proteome Res. 2018, 17, 2226–2236. [Google Scholar] [CrossRef]
- Paulo, J.A.; O’Connell, J.D.; Everley, R.A.; O’Brien, J.; Gygi, M.A.; Gygi, S.P. Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J. Proteom. 2016, 148, 85–93. [Google Scholar] [CrossRef]
- Paulo, J.A.; O’Connell, J.D.; Gygi, S.P. A Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments. J. Am. Soc. Mass Spectrom. 2016, 27, 1620–1625. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010, 428, 133–145. [Google Scholar] [CrossRef]
- Zhou, W.; Ryan, J.J.; Zhou, H. Global Analyses of Sumoylated Proteins in Saccharomyces cerevisiae: Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 2004, 279, 32262–32268. [Google Scholar] [CrossRef]
- Pinder, J.B.; McQuaid, M.E.; Dobson, M.J. Deficient sumoylation of yeast 2-micron plasmid proteins Rep1 and Rep2 associated with their loss from the plasmid-partitioning locus and impaired plasmid inheritance. PLoS ONE 2013, 8, e60384. [Google Scholar] [CrossRef]
- Culbertson, M.R.; Henry, S.A. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics 1975, 80, 23–40. [Google Scholar] [CrossRef]
- Overkamp, K.M.; Kotter, P.; van der Hoek, R.; Schoondermark-Stolk, S.; Luttik, M.A.; van Dijken, J.P.; Pronk, J.T. Functional analysis of structural genes for NAD(+)-dependent formate dehydrogenase in Saccharomyces cerevisiae. Yeast 2002, 19, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.L.; Fuge, E.K.; Padilla, P.A.; Werner-Washburne, M. A stationary-phase gene in Saccharomyces cerevisiae is a member of a novel, highly conserved gene family. J. Bacteriol. 1996, 178, 6865–6872. [Google Scholar] [CrossRef] [PubMed]
- Wertman, K.F.; Drubin, D.G.; Botstein, D. Systematic mutational analysis of the yeast ACT1 gene. Genetics 1992, 132, 337–350. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossio, V.; Paulo, J.A. Evaluating Protein Extraction Techniques for Elucidating Proteomic Changes in Yeast Deletion Strains. Proteomes 2025, 13, 28. https://doi.org/10.3390/proteomes13030028
Rossio V, Paulo JA. Evaluating Protein Extraction Techniques for Elucidating Proteomic Changes in Yeast Deletion Strains. Proteomes. 2025; 13(3):28. https://doi.org/10.3390/proteomes13030028
Chicago/Turabian StyleRossio, Valentina, and Joao A. Paulo. 2025. "Evaluating Protein Extraction Techniques for Elucidating Proteomic Changes in Yeast Deletion Strains" Proteomes 13, no. 3: 28. https://doi.org/10.3390/proteomes13030028
APA StyleRossio, V., & Paulo, J. A. (2025). Evaluating Protein Extraction Techniques for Elucidating Proteomic Changes in Yeast Deletion Strains. Proteomes, 13(3), 28. https://doi.org/10.3390/proteomes13030028






