Biomarkers in Ovarian Cancer: Towards Personalized Medicine
Abstract
1. Ovarian Cancer: A Rare but Fatal Malignancy
1.1. Classification of Ovarian Carcinomas
1.2. Strategies for the Treatment of Ovarian Cancer
- Cytoreductive surgery. At the initial stages of tumor development, the most common procedure is the resection of the tumoral mass by laparotomy [31].
- Chemotherapy. For more advanced stages, the tumor resection procedure is often combined with chemotherapeutic approaches [31]. Among them, platinum-based treatments (cisplatin and carboplatin) in combination with paclitaxel are the first line of treatment for OC. This therapy has been applied during the last 20 years with no other treatment outperforming it [32]. Only bevacizumab, an anti-angiogenic drug, was introduced in 2011 to complement the platinum/paclitaxel combination [33,34]. Despite the high-rate effectiveness of this first-line treatment, any therapeutic improvements are still welcome to improve drug outcomes, being drug transport efficiencies the most important limiting factors in existing treatments. In this regard, hyperthermic intraperitoneal chemotherapy (HIPEC) allows the single administration of high doses of the cytostatic while also exploiting the effect of hyperthermia (41–43 °C for 30–120 min), improving the drug cytotoxicity. The clinical trial OVHIPEC-1, performed in The Netherlands and Belgium, showed higher disease-free survival and overall survival rates in the patients undergoing surgery resection and HIPEC vs. those who underwent surgery alone [35,36,37]. Likewise, the use of drug nanocarriers appears as a promising alternative to ensure the successful delivery of drug-based treatments [38,39,40]. This option will be further discussed in Section 4.
- Immunotherapy. New immune-based therapies are also under investigation for OC. Thus, the inhibition of specific proteins (like PD-1) by drugs (e.g., nivolumab) that results in the promotion of anti-tumor immunity is showing promising results in the field, with ~15% of OC cases positively responding to the treatment [41]. Other therapies using immune modulators or immune checkpoint inhibitors are also applied to patients.
- Targeted therapies. In 2014, poly(ADP-ribose) polymerase (PARP) inhibitors were approved as maintenance therapy for patients with recurrent disease after platinum treatments. PARP inhibition leads to the accumulation of double-strand breaks that cannot be repaired in cells that are homologous recombination repair deficient (HRD), finally leading to cell death. Considering around 50% of HGSC tumors are HRD, this therapy has been reported as an important alternative [42]. Three clinical trials in phase III showed promising results leading to the approval of niraparib [43], olaparib [44], and rucaparib [45] drugs. Recently, PARP inhibitors are also under evaluation in the front-line setting (rather than maintenance therapy) via four phase-III clinical trials [46,47,48,49].Other targeted therapies include the inhibition of proteins of the tropomyosin receptor kinase (TRK) family. The binding of neurotrophins to TRK receptors activates Ras, PI3K and phospholipase C-γ1 signaling cascades in a normal state. However, any rearrangement of these receptors may lead to cell malfunctioning and tumorigenesis due to overactivation of signal transduction [50].
- Hormonal therapies. Since OC progression depends on hormones released from the hypothalamic-pituitary-ovarian axis and considering the demonstrated efficacy of hormone therapies in breast and endometrial cancers, these therapeutic strategies have been stated for the treatment of patients showing platinum resistance and tumor recurrences. While gonadotropins, estrogens, and androgens promote OC advancement, gonadotropin-releasing hormone (GnRH) and progesterone might have a protective role [51]. Thus, analogues of GnRH (e.g., triptorelin), or inhibitors of estrogen (e.g., tamoxifen) and androgen (e.g., flutamide), are used in the clinics [52].
2. Biomarkers in Ovarian Cancer: From Diagnosis to Prognosis
| Biomarker | Full Name | Features | Specificity/ Sensitivity | Diagnostic/ Prognostic Marker? | References | |
|---|---|---|---|---|---|---|
| Serum markers | CA-125 | Carbohydrate antigen 125 | - Highly present in 80% of late-stage epithelial OC - Present in other non-tumoral conditions (e.g., endometriosis, normal menstruation, pregnancy) → no longer recommended for screening and diagnosis | 90%/60% | yes/yes | [7,62,63,64,65,66] |
| HE4 | Human epididymis protein | - Expressed in endometrioid and serous OC - Present in some postmenopausal conditions | 95%/73% | yes/no | [62,63,67,68,69] | |
| KLK | Kallikrein | Upregulated in OC (serum and ascites) with poor prognosis and chemoresistance to paclitaxel | 75%/77% | yes/no | [62,70] | |
| PSN | Prostasin | Expression levels > 100x in epithelial and stromal OC vs. normal condition | 94%/51% | yes/no | [62,71] | |
| TTR | Transthyretin | Low levels in OC | 69%/79% | yes/no | [62,72] | |
| Transferrin | Transferrin | Low levels in OC | 74%/73% | yes/no | [62] | |
| VEGF | Vascular endothelial growth factor | Direct correlation with OC | 74%/79% | yes/yes | [7,62] | |
| Bikunin | - | High levels related to favorable prognosis | 70%/75% | no/yes | [62,73] | |
| CKB | Creatine kinase B | Highly expressed in early tumoral phases | 94%/92% | es/yes | [62,74] | |
| Plasma markers | apoA-I | Apolipoprotein A-I | Low levels in OC | 98%/94% | yes/no | [62,75] |
| OPN | Osteopontin | Highly expressed in OC | 34%/81% | yes/yes | [62,76] |
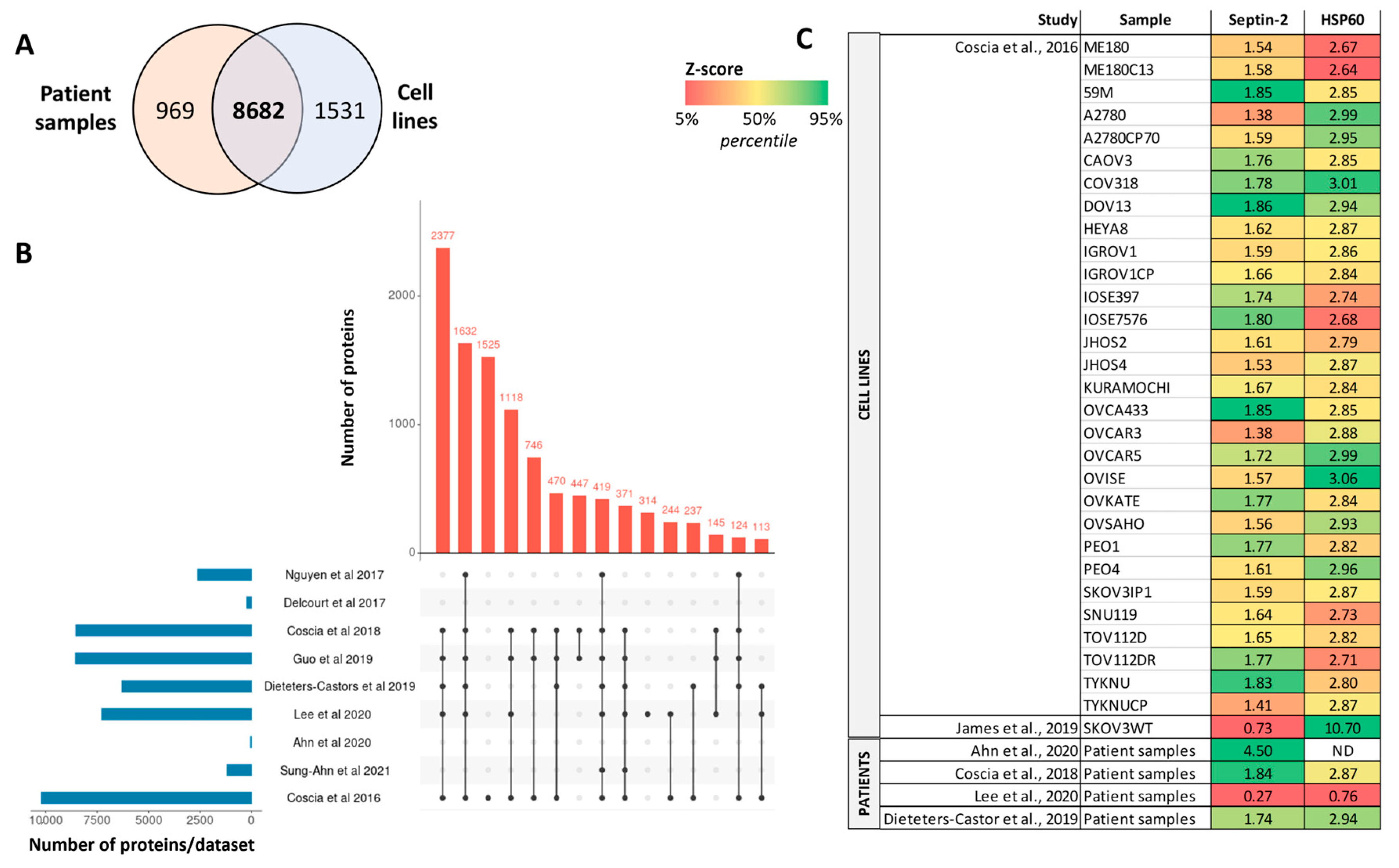
3. Mass Spectrometry-Based Proteomics Studies in Ovarian Cancer
| Sample Type | Sample Origin | OC Subtype | Studied Analytes | MS Technology | Outcome Summary | Reference |
|---|---|---|---|---|---|---|
| Tumor tissue | Patients (25 cases) and cell lines | HGSC | Proteins and phosphoproteins | LC-MS/MS | 8190 quantified proteins | [88] |
| Patients (103 cases) | Mesenchymal HGSC | Proteins | SWATH/DIA-MS and iTRAQ-DDA | 4363 by iTRAQ-DDA and 1659 by SWATH/DIA-MS (1599 in common) | [95] | |
| Patients (30 cases) | HGSC | Proteins and phosphoproteins | TMT-based LC-MS/MS | 7290 proteins and 12,914 phosphosites | [88,89] | |
| Patients (11-paired normal and tumoral cases) | Serous, clear cell, endometrioid carcinomas | Proteins | TMT-based LC-MS/MS | 7719 proteins | [92] | |
| Patients (20 cases) | HGSC and endometrioid carcinoma | Proteins | LC-MS/MS | 8-marker panel for discrimination between HGSOC and endometrioid carcinoma | [96] | |
| Patients (31 cases) | Serous OC | Proteins | MALDI imaging MS | 3844 proteins | [97] | |
| Blood | Patients (20 cases) | HGSC | Plasma metabolites and proteins | Nano-LC-ESI–MS/MS and MRM-MS | 34 metabolites (L-carnitine and PC-O) and 197 proteins (PPCS, PMP2, and TUBB) | [87] |
| Ascites | Patients (70 cases) | HGSC | Macrophage secretome | LC-MS/MS | Focus on TGFB1, TNC and FN1 (low levels relate to better survival rates) | [98] |
| Cell culture | Patient (2 patient-derived primary cell lines) | HGSC | Proteins and phosphoproteins | LC-MS/MS | 4151 quantified proteins, and 2905 phosphorylation sites | [99] |
| Patients (8 cases) and cell lines (30) | HGSC | Proteins | LC-MS/MS | >10,000 proteins (67-protein signature) | [100] | |
| Cell line (SKOV3WT) | Serous and clear-cell OC | Proteins | LC-MS/MS | Septin-2 as protein target to reduce tumorigenesis | [94] | |
| Cell line (OVCAR-3, SKOV-3) | HGSC and non-serous OC | Proteins and phosphoproteins | LC-MS/MS | 3324 proteins, 2978 phosphopeptides | [101] | |
| Cell lines (8) | Epithelial OC | ECM1-interacting proteins | LC-MS/MS | ECM1a, integrin aXb2, hnRNPLL, and ABCG1 as potential targets | [102] |
4. New Therapeutic Strategies in OC Based on Drug-Nanodelivery Systems
- Liposomes. These vesicles composed of lipid bilayers present the ability to encapsulate both hydrophobic and hydrophilic substances, making them particularly adaptable for the targeted delivery of therapeutic agents. Liposomes provide a protective environment, shielding the drugs from degradation and ensuring their stability [108]. This property is especially advantageous in OC treatment, where the delivery of chemotherapeutic drugs is critical for effective tumor regress [109]. Likewise, liposomal formulations of drugs such as doxorubicin and paclitaxel have been developed to address challenges related to drug solubility, bioavailability, and toxicity [110,111]. Some examples have gained clinical approval for OC, including Doxil and Lipo-PTX, liposomal formulations of doxorubicin and paclitaxel, respectively [112,113,114]. These formulations have shown promising outcomes in clinical settings, offering prolonged circulation times, reduced toxicities, and improved therapeutic indices compared to their conventional counterparts. Another clinical study evaluated the combined use of paclitaxel liposomes and carboplatin with the administration of the free drugs, reporting a significantly enhanced response in the encapsulated condition with reduced side effects [115]. Furthermore, one notable benefit of using liposomes is the reduction of systemic side effects associated with chemotherapy. By facilitating targeted drug delivery, liposomes minimize the exposure of healthy tissues to potent chemotherapeutic agents, leading to a more favorable safety profile. Likewise, by targeting overexpressed receptors (e.g., luteinizing hormone-releasing hormone receptor, LHRHR), liposomes increase their uptake rate, enhancing cell apoptosis, as shown in in vitro studies in A2780 cells [116]. However, the use of liposomes in OC therapy presents some stability issues and the transition from laboratory-scale to large-scale production presents obstacles that require ongoing research and technological advancements [117].
| Nanocarrier | Features | Advantages | Disadvantages | Examples | References |
|---|---|---|---|---|---|
| Liposome | Encapsulation of hydrophobic and hydrophilic substances | - biocompatibility - surface modification - reduction of side effects | - stability issues - hard transition to large-scale production | Doxil, Lipo-PTX | [112,113,114] |
| Nanoparticle | Delivery of drugs attached to its surface or by encapsulation | - easy synthesis - size control - surface modification | - heterogeneous synthesis processes - concentration-dependent toxicity for patients | Cis-platin coated iron nanoparticles, gold nanoparticles, albumin-based nanoparticles | [38,118,119,120,121,122] |
| Micelle | Encapsulation of hydrophobic drugs | - biocompatibility - drug stabilization - surface modification | - difficult delivery of hydrophilic substances | Genexol-PM, PEG-based micelles, poly(propylene oxide) (PPO)-based micelles | [123,124] |
| Dendrimer | Functionalization of its dendritic architecture with ligands for targeted drug delivery | - co-delivery of substances - surface modification | - heterogeneous - difficult synthesis processes | Phosphorus (P-dendrimers), polyamidoamines (PAMAM), polypeptides, polyesters | [125,126,127,128] |
- Dendrimers. These nanocarriers are highly branched macromolecules with well-defined structures that offer the potential for targeted drug delivery, and that can be functionalized with ligands for specific interactions with cancer cells [126,127]. Their dendritic architecture allows for precise control over drug loading, enabling the encapsulation of therapeutic agents within their well-defined branches. Like liposomes, dendrimers can also be modified by attaching ligands or antibodies to their surface, improving their specific targeting [129]. This dendritic structure also allows the co-delivery of multiple drugs with different functionalities, enabling synergistic effects and overcoming drug resistance mechanisms often encountered in conventional OC treatment. This aspect makes dendrimers valuable tools for designing personalized therapeutic approaches tailored to the specific characteristics of each cancer case [125]. Several in vitro studies have reported the benefits of using drug-coupled dendrimers. For instance, its combination with cisplatin to treat OVCAR3, SKOV, A2780, and CP70 cells reported a 7-x increase in the expression of apoptotic genes and a 2-x increase in the activity of caspases, ultimately leading to tumoral death [130].
- Nanoparticles. Most nanoparticles used in OC belong to either polymeric, metallic, or albumin-based nanoparticles. Polymeric nanoparticles are constructed from biodegradable polymers like poly(lactic-co-glycolic acid) (PLGA) or polyethylene glycol (PEG) [118,119], while metallic nanoparticles are made of metallic elements such as gold, silver, or iron [120,121,131], and albumin-based nanoparticles, like Abraxane, use albumin aggregates as a carrier. Their versatility allows the controlled release of drugs like paclitaxel, olaparib, or cisplatin, and they can also get their surface modified to allocate specific targeting molecules [38,46,107,132,133]. Moreover, nanoparticles can encapsulate therapeutic agents, preventing premature drug degradation and ensuring their sustained release [134,135]. Also, the small size of nanoparticles contributes to their ability to passively target tumors through the Enhanced Permeability and Retention (EPR) effect. This phenomenon leverages the leaky vasculature surrounding tumors, allowing nanoparticles to accumulate selectively in cancerous tissues. The passive targeting mechanism enhances drug delivery efficiency and ensures a higher concentration of therapeutic agents at the tumor site [136]. As for albumin-based nanoparticles, they benefit from the natural affinity for the albumin receptor on cancer cells, facilitating targeted drug delivery. This approach improves drug solubility, reduces the need for toxic solvents, and enhances the therapeutic effects of drugs like paclitaxel in OC. Additionally, their biocompatibility is a critical factor in minimizing systemic toxicities associated with chemotherapy, as it presents a benefit from the natural origin of this protein, which normally is well-tolerated by the body, reducing the risk of adverse reactions [122,137]. Nanoparticles have been evaluated in multiple in vitro studies showing promising results. For instance, PLGA-based nanoparticles carrying molecules to specifically bind the LHRH receptor (i.e., LHRH-a) and delivering CPT-11, an inhibitor of DNA topoisomerase I, significantly inhibited the cellular proliferation of A2780 cisplatin-resistant cells [138]. Also, degradable mesoporous silica nanoparticles encapsulating paclitaxel showed enhanced toxicity in OVACAR-3 and PA-1 cells [139]. Another investigation in vitro studied the role of the nanoparticle surface charge, reporting that nonionic polymeric nanostructures reduce cancer cell viability at greater levels compared to positively charged formulations [140]. Despite the promising results of these nanostructures, they are not routinely applied for the treatment of OC patients.
- Micelles. These nanostructures are formed by the self-assembly of amphiphilic molecules in aqueous solutions and offer a multifaceted approach to addressing key challenges associated with traditional drug delivery [124]. One of their distinctive features is their biocompatibility which contributes to their potential for minimizing systemic toxicities associated with chemotherapies [123]. They also present the ability to encapsulate hydrophobic drugs within their core [141] and to include modifications in their surface [106,142]. Micelles, together with liposomes, are the only nanostructures approved by a national drug administration to be used in patients. Specifically, in 2007, a paclitaxel-carrying PEG-PLA polymeric micelle (Genexol-PM) was approved in South Korea for breast, lung, and OC treatment [143,144]. Other investigations have also shown promising results in vivo and in vitro, such as the encapsulation of paclitaxel in micelles with epidermal growth factor (EGF) as targeting molecule that showed an improved uptake by SKOV3 cells subsequently inhibiting their proliferation [145].
Surface Modification of NP to Promote Targeted Active Drug Uptake
- TFR2. It is a transmembrane glycoprotein that plays a pivotal role in the regulation of iron homeostasis within the human body thanks to its interaction with transferrin, a protein responsible for transporting iron in the bloodstream, which facilitates the sensing of iron levels. In this sense, OC cells often exhibit alterations in iron homeostasis to support their rapid proliferation and growth [146,147,148]. Possibly due to this effect, this receptor is overexpressed in some OC cell lines, making it a suitable molecule for targeted therapies [149].
- AXL receptor. It is a member of the family of receptor tyrosine kinases alongside Tyro3 and Mer [150]. AXL is frequently overexpressed in OC cells, and its upregulation has been associated with aggressive tumor behavior, metastasis, and resistance to conventional therapies. The activation of AXL signaling pathways contributes to processes such as epithelial-to-mesenchymal transition (EMT), which enhances the invasive potential of cancer cells. Moreover, AXL has been involved in immune evasion, dampening the antitumor immune response. This receptor’s role in promoting cell survival and inhibiting apoptosis further underscores its significance in OC progression. Thus, the AXL receptor can be employed dually: (i) exploring AXL inhibitors as potential therapeutic agents to counteract the aggressive features of OC [151,152], and (ii) targeting AXL receptor within nanodelivery systems to improve drug incorporation and release efficacies.
- VEGFR. Related to the metastasis field, the receptor for the angiogenic VEGF factor, a biomarker described in Section 2, has become a key strategy in the development of targeted therapies for OC [153,154]. Anti-angiogenic drugs, such as bevacizumab, an anti-VEGF monoclonal antibody, specifically target VEGF or its receptor to block the formation of new blood vessels, thus restricting the access of oxygen and nutrients to the tumor, thus making bevacizumab an excellent candidate for developing new targeted therapies [47,155,156,157,158]. Moreover, the receptor could be used as a potential target for drug delivery.
- Folate receptor. The overexpression of this receptor on OC is often associated with increased tumor aggressiveness, and poor prognosis and can be leveraged by the specific binding affinity to some drugs like mirvetuximab soravtansine-gynx, an antibody-drug conjugate designed to selectively deliver a chemotherapy agent to cancer cells that overexpress this receptor. This targeted approach aims to enhance the efficacy of chemotherapy while minimizing damage to healthy cells [159,160].
- Others. Multiple other receptors can be targeted to enhance treatments. For instance, the follicle-stimulating hormone receptor (FSHR) that is overexpressed in OC cells can be targeted by including the binding peptide domain of FSH (FSH33) onto the nanostructure surface, like dendrimers, exhibiting an increased tumoral selectivity [161]. Likewise, biotin functionalization of the surface might enhance the biotin receptor-mediated endocytosis uptake of nanosystems, as demonstrated in an in vitro study with OVCAR3 cells by Yellepeddi et al. [162].
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schoutrop, E. Molecular, Cellular and Systemic Aspects of Epithelial Ovarian Cancer and Its Tumor Microenvironment. Semin. Cancer Biol. 2022, 86, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Shimada, C.; Xu, R.; Al-Alem, L.; Stasenko, M.; Spriggs, D.R.; Rueda, B.R. Galectins and Ovarian Cancer. Cancers 2020, 12, 1421. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A. Ovarian cancer statistics. Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Ovarian Cancer Statistics. World Cancer Research Fund International. Available online: https://www.wcrf.org/cancer-trends/ovarian-cancer-statistics/ (accessed on 10 February 2024).
- Webb, P.M.; Jordan, S.J. Epidemiology of Epithelial Ovarian Cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 3–14. [Google Scholar] [CrossRef]
- Zhang, Y. Global Patterns and Trends in Ovarian Cancer Incidence: Age, Period and Birth Cohort Analysis. BMC Cancer 2019, 19, 984. [Google Scholar] [CrossRef]
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef]
- Fan, C.A.; Reader, J.; Roque, D.M. Review of Immune Therapies Targeting Ovarian Cancer. Curr. Treat. Options Oncol. 2018, 19, 74. [Google Scholar] [CrossRef]
- Saorin, A.; Di Gregorio, E.; Miolo, G.; Steffan, A.; Corona, G. Emerging Role of Metabolomics in Ovarian Cancer Diagnosis. Metabolites 2020, 10, 419. [Google Scholar] [CrossRef]
- Kim, J.; Park, E.Y.; Kim, O.; Schilder, J.M.; Coffey, D.M.; Cho, C.H.; Bast, R.C., Jr. Cell Origins of High-Grade Serous Ovarian Cancer. Cancers 2018, 10, 433. [Google Scholar] [CrossRef]
- Guo, L.; Wang, J.; Li, N.; Cui, J.; Su, Y. Peptides for diagnosis and treatment of ovarian cancer. Front. Oncol. 2023, 13, 1135523. [Google Scholar] [CrossRef]
- Meinhold-Heerlein, I.; Fotopoulou, C.; Harter, P.; Kurzeder, C.; Mustea, A.; Wimberger, P.; Hauptmann, S.; Sehouli, J. The New WHO Classification of Ovarian, Fallopian Tube, and Primary Peritoneal Cancer and Its Clinical Implications. Arch. Gynecol. Obstet. 2016, 293, 695–700. [Google Scholar] [CrossRef]
- Köbel, M.; Kang, E.Y. The Evolution of Ovarian Carcinoma Subclassification. Cancers 2022, 14, 416. [Google Scholar] [CrossRef]
- Ravindran, F.; Choudhary, B. Ovarian Cancer: Molecular Classification and Targeted Therapy; IntechOpen Limited: London, UK, 2021; pp. 8–11. [Google Scholar]
- Koshiyama, M.; Matsumura, N.; Konishi, I. Recent Concepts of Ovarian Carcinogenesis: Type I and Type II. BioMed Res. Int. 2014, 2014, 934261. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.M. The Dualistic Model of Ovarian Carcinogenesis Revisited, Revised, and Expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef]
- Cho, K.R. Ovarian Cancer Update: Lessons from Morphology, Molecules, and Mice. Proc. Arch. Pathol. Lab. Med. 2009, 133, 1775–1781. [Google Scholar] [CrossRef]
- Gadducci, A.; Multinu, F.; Cosio, S.; Carinelli, S.; Ghioni, M.; Aletti, G.D. Clear Cell Carcinoma of the Ovary: Epidemiology, Pathological and Biological Features, Treatment Options and Clinical Outcomes. Gynecol. Oncol. 2021, 162, 741–750. [Google Scholar] [CrossRef]
- Berns, K.; Caumanns, J.J.; Hijmans, E.M.; Gennissen, A.M.C.; Severson, T.M.; Evers, B.; Wisman, G.B.A.; Jan Meersma, G.; Lieftink, C.; Beijersbergen, R.L.; et al. ARID1A Mutation Sensitizes Most Ovarian Clear Cell Carcinomas to BET Inhibitors. Oncogene 2018, 37, 4611–4625. [Google Scholar] [CrossRef]
- Merritt, M.A.; Cramer, D.W. Molecular Pathogenesis of Endometrial and Ovarian Cancer. Cancer Biomark. 2011, 9, 287–305. [Google Scholar] [CrossRef]
- Ioffe, Y.J.; Chiappinelli, K.B.; Mutch, D.G.; Zighelboim, I.; Goodfellow, P.J. Phosphatase and Tensin Homolog (PTEN) Pseudogene Expression in Endometrial Cancer: A Conserved Regulatory Mechanism Important in Tumorigenesis? Gynecol. Oncol. 2012, 124, 340–346. [Google Scholar] [CrossRef]
- Liao, X.; Siu, M.K.Y.; Au, C.W.H.; Chan, Q.K.Y.; Chan, H.Y.; Wong, E.S.Y.; Ip, P.P.C.; Ngan, H.Y.S.; Cheung, A.N.Y. Aberrant Activation of Hedgehog Signaling Pathway Contributes to Endometrial Carcinogenesis through Β-Catenin. Mod. Pathol. 2009, 22, E72–E78. [Google Scholar] [CrossRef] [PubMed]
- Babaier, A.; Ghatage, P. Mucinous Cancer of the Ovary: Overview and Current Status. Diagnostics 2020, 10, 52. [Google Scholar] [CrossRef]
- Hammel, P.; Voitot, H.; Vilgrain, V.; Lévy, P.; Ruszniewski, P.; Bernades, P. Diagnostic Value of CA 72-4 and Carcinoembryonic Antigen Determination in the Fluid of Pancreatic Cystic Lesions. Eur. J. Gastroenterol. Hepatol. 1998, 10, 1230–1235. [Google Scholar] [CrossRef]
- Pandey, D.; Sharma, R.; Sharma, S.; Salhan, S. Unusually High Serum Levels of CA 19-9 in an Ovarian Tumour: Malignant or Benign? J. Clin. Diagn. Res. 2017, 11, QD8–QD10. [Google Scholar] [CrossRef]
- Huang, Q.; Chen, X.; Lu, W.; Lai, M.; Lu, B. Expression of REG4 in Ovarian Mucinous Tumors. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 295–301. [Google Scholar] [CrossRef]
- Amante, S.; Santos, F.; Cunha, T.M. Low-Grade Serous Epithelial Ovarian Cancer: A Comprehensive Review and Update for Radiologists. Insights Imaging 2021, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Lisio, M.A.; Fu, L.; Goyeneche, A.; Gao, Z.H.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed]
- Beirne, J.P.; McArt, D.G.; Roddy, A.; McDermott, C.; Ferris, J.; Buckley, N.E.; Coulter, P.; McCabe, N.; Eddie, S.L.; Dunne, P.D.; et al. Defining the Molecular Evolution of Extrauterine High Grade Serous Carcinoma. Gynecol. Oncol. 2019, 155, 305–317. [Google Scholar] [CrossRef]
- Ersoy, E.; Cao, Q.J.; Otis, C.N. HER2 Protein Overexpression and Gene Amplification in Tubo-Ovarian High-Grade Serous Carcinomas. Int. J. Gynecol. Pathol. 2022, 41, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Orr, B.; Edwards, R.P. Diagnosis and Treatment of Ovarian Cancer. Hematol. Oncol. Clin. N. Am. 2018, 32, 943–964. [Google Scholar] [CrossRef]
- Ledermann, J.A. First-Line Treatment of Ovarian Cancer: Questions and Controversies to Address. Ther. Adv. Med. Oncol. 2018, 10, 558–565. [Google Scholar] [CrossRef]
- Burger, R.A. Incorporation of Bevacizumab in the Primary Treatment of Ovarian Cancer. N. Engl. J. Med. 2011, 365, 2473–2483. [Google Scholar] [CrossRef]
- Perren, T.J. A Phase 3 Trial of Bevacizumab in Ovarian Cancer. N. Engl. J. Med. 2011, 365, 2484–2496. [Google Scholar] [CrossRef]
- Aronson, S.L.; Lopez-Yurda, M.; Koole, S.N.; Schagen van Leeuwen, J.H.; Schreuder, H.W.R.; Hermans, R.H.M.; de Hingh, I.H.J.T.; van Gent, M.D.J.M.; Arts, H.J.G.; van Ham, M.A.P.C.; et al. Cytoreductive Surgery with or without Hyperthermic Intraperitoneal Chemotherapy in Patients with Advanced Ovarian Cancer (OVHIPEC-1): Final Survival Analysis of a Randomised, Controlled, Phase 3 Trial. Lancet Oncol. 2023, 378, 230–240. [Google Scholar] [CrossRef]
- Ghirardi, V.; Fagotti, A.; Scambia, G. Hyperthermic Intraperitoneal Chemotherapy for Ovarian Cancer: Long-Term Findings from the OVHIPEC-1 Trial. Lancet Oncol. 2023, 24, 5256–5262. [Google Scholar] [CrossRef]
- Aronson, L.; Lopez-Yurda, M.I.; Koole, S.N.; Schagen van Leeuwen, J.H.; Schreuder, H.; Hermans, R.; de Hingh, I.H.J.T.; Van Gent, M.D.J.M.; Arts, H.J.G.; van Ham, M.; et al. Final Survival Analysis of the Phase III OVHIPEC-1 Trial of Hyperthermic Intraperitoneal Chemotherapy in Ovarian Cancer after Ten Year Follow-Up. J. Clin. Oncol. 2023, 41, 5509. [Google Scholar] [CrossRef]
- Turiel-Fernández, D.; Gutiérrez-Romero, L.; Corte-Rodriguez, M.; Bettmer, J.; Montes-Bayón, M. Ultrasmall Iron Oxide Nanoparticles Cisplatin (IV) Prodrug Nanoconjugate: ICP-MS Based Strate-Gies to Evaluate the Formation and Drug Delivery Capabilities in Single Cells. Anal. Chim. Acta 2021, 1159, 338356. [Google Scholar] [CrossRef]
- Wu, Y. Nanoparticle-Based Combination Therapy for Ovarian Cancer. Int. J. Nanomed. 2023, 18, 1965–1987. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Romero, L.; Rivas-García, L.; Sánchez-González, C.; Llopis, J.; Blanco, E.; Montes-Bayón, M. Cellular Toxicity Mechanisms and the Role of Autophagy in Pt(IV) Prodrug-Loaded Ultrasmall Iron Oxide Nanoparticles Used for Enhanced Drug Delivery. Pharmaceutics 2021, 13, 1730. [Google Scholar] [CrossRef]
- Herrera, F.G.; Irving, M.; Kandalaft, L.E.; Coukos, G. Rational Combinations of Immunotherapy with Radiotherapy in Ovarian Cancer. Lancet Oncol. 2019, 20, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Hockings, H.; Miller, R.E. The Role of PARP Inhibitor Combination Therapy in Ovarian Cancer. Ther. Adv. Med. Oncol. 2023, 15, 1758–8340. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.R.; Monk, B.J.; Herrstedt, J.; Oza, A.M.; Mahner, S.; Redondo, A.; Fabbro, M.; Ledermann, J.A.; Lorusso, D.; Vergote, I.; et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016, 375, 2154–2164. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib Tablets as Maintenance Therapy in Patients with Platinum-Sensitive, Relapsed Ovarian Cancer and a BRCA1/2 Mutation (SOLO2/ENGOT-Ov21): A Double-Blind, Randomised, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib Maintenance Treatment for Recurrent Ovarian Carcinoma after Response to Platinum Therapy (ARIEL3): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef]
- Moore, K. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef]
- Ray-Coquard, I. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Coleman, R.L.; Fleming, G.F.; Brady, M.F.; Swisher, E.M.; Steffensen, K.D.; Friedlander, M.; Okamoto, A.; Moore, K.N.; Efrat Ben-Baruch, N.; Werner, T.L.; et al. Veliparib with First-Line Chemotherapy and as Maintenance Therapy in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2403–2415. [Google Scholar] [CrossRef]
- Lange, A.M.; Lo, H.W. Inhibiting TRK Proteins in Clinical Cancer Therapy. Cancers 2018, 10, 105. [Google Scholar] [CrossRef]
- Saeaib, N.; Peeyananjarassri, K.; Liabsuetrakul, T.; Buhachat, R.; Myriokefalitaki, E. Hormone Replacement Therapy after Surgery for Epithelial Ovarian Cancer. Cochrane Database Syst. Rev. 2020, 1, CD012559. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.; Wang, Y.; Zhao, X.; Qi, X. Hormone Therapy for Ovarian Cancer: Emphasis on Mechanisms and Applications. Oncol. Rep. 2021, 46, 223. [Google Scholar] [CrossRef] [PubMed]
- Durno, K.; Powell, M.E. The Role of Radiotherapy in Ovarian Cancer. Int. J. Gynecol. Cancer 2022, 32, 366–371. [Google Scholar] [CrossRef]
- Ryu, J.; Thomas, S.N. Quantitative Mass Spectrometry-Based Proteomics for Biomarker Development in Ovarian Cancer. Molecules 2021, 26, 2674. [Google Scholar] [CrossRef]
- Zhang, Y. Germline Variants Profiling of BRCA1 and BRCA2 in Chinese Hakka Breast and Ovarian Cancer Patients. BMC Cancer 2022, 22, 842. [Google Scholar] [CrossRef]
- Petrucelli, N.; Daly, M.B.; Pal, T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. Gene Rev. 1998. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1247/ (accessed on 10 February 2024).
- Dann, R.B.; Deloia, J.A.; Timms, K.M.; Zorn, K.K.; Potter, J.; Flake, D.D.; Lanchbury, J.S.; Krivak, T.C. BRCA1/2 Mutations and Expression: Response to Platinum Chemotherapy in Patients with Advanced Stage Epithelial Ovarian Cancer. Proc. Gynecol. Oncol. 2012, 125, 677–682. [Google Scholar] [CrossRef]
- Andrikopoulou, A.; Zografos, E.; Apostolidou, K.; Kyriazoglou, A.; Papatheodoridi, A.M.; Kaparelou, M.; Koutsoukos, K.; Liontos, M.; Dimopoulos, M.A.; Zagouri, F. Germline and Somatic Variants in Ovarian Carcinoma: A next-Generation Sequencing (NGS) Analysis. Front Oncol. 2022, 12, 1030786. [Google Scholar] [CrossRef]
- McLaughlin, J.R.; Rosen, B.; Moody, J.; Pal, T.; Fan, I.; Shaw, P.A.; Risch, H.A.; Sellers, T.A.; Sun, P.; Narod, S.A. Long-Term Ovarian Cancer Survival Associated with Mutation in BRCA1 or BRCA2. J. Natl. Cancer Inst. 2013, 105, 341–345. [Google Scholar] [CrossRef]
- Kotsopoulos, J.; Rosen, B.; Fan, I.; Moody, J.; McLaughlin, J.R.; Risch, H.; May, T.; Sun, P.; Narod, S.A. Ten-Year Survival after Epithelial Ovarian Cancer Is Not Associated with BRCA Mutation Status. Gynecol. Oncol. 2016, 140, 964–971. [Google Scholar] [CrossRef]
- Wang, Y.; Li, N.; Ren, Y.; Zhao, J. Association of BRCA1/2 Mutations with Prognosis and Surgical Cytoreduction Outcomes in Ovarian Cancer Patients: An Updated Meta-Analysis. J. Obstet. Gynaecol. Res. 2022, 48, 2270–2284. [Google Scholar] [CrossRef]
- Atallah, G.A.; Aziz, N.H.A.; Teik, C.K.; Shafiee, M.N.; Kampan, N.C. New Predictive Biomarkers for Ovarian Cancer. Diagnostics 2021, 11, 465. [Google Scholar] [CrossRef]
- Radu, M.R. Ovarian Cancer: Biomarkers and Targeted Therapy. Biomedicines 2021, 9, 693. [Google Scholar] [CrossRef]
- Bai, H. The Prognostic Value of Pretreatment CA-125 Levels and CA-125 Normalization in Ovarian Clear Cell Carcinoma: A Two-Academic-Institute Study. Oncotarget 2016, 7, 15566–15576. [Google Scholar] [CrossRef]
- Piatek, S. Rising Serum CA-125 Levels within the Normal Range Is Strongly Associated Recurrence Risk and Survival of Ovarian Cancer. J. Ovarian Res. 2020, 13, 102. [Google Scholar] [CrossRef]
- Dochez, V.; Caillon, H.; Vaucel, E.; Dimet, J.; Winer, N.; Ducarme, G. Biomarkers and Algorithms for Diagnosis of Ovarian Cancer: CA125, HE4, RMI and ROMA, a Review. J. Ovarian Res. 2019, 12, 28. [Google Scholar] [CrossRef]
- Hellström, I. The HE4 (WFDC2) Protein Is a Biomarker for Ovarian Carcinoma. Cancer Res. 2003, 63, 3695–3700. [Google Scholar]
- Behrouzi, R.; Barr, C.E.; Crosbie, E.J. HE4 as a Biomarker for Endometrial Cancer. Cancers 2021, 13, 4764. [Google Scholar] [CrossRef]
- Granato, T.; Porpora, M.G.; Longo, F.; Angeloni, A.; Manganaro, L.; Anastasi, E. HE4 in the Differential Diagnosis of Ovarian Masses. Clin. Chim. Acta 2015, 6, 44. [Google Scholar] [CrossRef]
- Riedel, M.; Bronger, H.; Magdolen, V.; Dreyer, T. The Prognostic and Diagnostic Potential of Kallikrein-Related Peptidases in Ovarian Cancer. Expert Rev. Mol. Diagn. 2021, 21, 535–545. [Google Scholar] [CrossRef]
- Tamir, A.; Gangadharan, A.; Balwani, S.; Tanaka, T.; Patel, U.; Hassan, A.; Benke, S.; Agas, A.; D’Agostino, J.; Shin, D.; et al. The Serine Protease Prostasin (PRSS8) Is a Potential Biomarker for Early Detection of Ovarian Cancer. J. Ovarian Res. 2016, 25, 781–787. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, S.; Li, L.; Liu, X.; Liu, X.; Dai, S.; Zhang, P.; Lu, H.; Lin, Z.; Yu, Y.; et al. Evaluation of HE4 and TTR for Diagnosis of Ovarian Cancer: Comparison with CA-125. J. Gynecol. Obstet. Hum. Reprod. 2018, 47, e555–e567. [Google Scholar] [CrossRef]
- Press, J.Z.; Wurz, K.; Norquist, B.M.; Lee, M.K.; Pennil, C.; Garcia, R.; Welcsh, P.; Goff, B.A.; Swisher, E.M. Identification of a Preneoplastic Gene Expression Profile in Tubal Epithelium of BRCA1 Mutation Carriers. Neoplasia 2010, 12, 993–1002. [Google Scholar] [CrossRef]
- Li, X.H.; Chen, X.J.; Ou, W.B.; Zhang, Q.; Lv, Z.R.; Zhan, Y.; Ma, L.; Huang, T.; Yan, Y.B.; Zhou, H.M. Knockdown of Creatine Kinase B Inhibits Ovarian Cancer Progression by Decreasing Glycolysis. Int. J. Biochem. Cell Biol. 2013, 45, 291–297. [Google Scholar] [CrossRef]
- Ren, L.; Yi, J.; Li, W.; Zheng, X.; Liu, J.; Wang, J.; Du, G. Apolipoproteins and Cancer. Cancer Med. 2019, 8, 268–272. [Google Scholar] [CrossRef]
- Cerne, K.; Hadzialjevic, B.; Skof, E.; Verdenik, I.; Kobal, B. Potential of Osteopontin in the Management of Epithelial Ovarian Cancer. Radiol. Oncol. 2019, 53, 105–115. [Google Scholar] [CrossRef]
- Russell, M.R.; Graham, C.; D’Amato, A.; Gentry-Maharaj, A.; Ryan, A.; Kalsi, J.K.; Ainley, C.; Whetton, A.D.; Menon, U.; Jacobs, I.; et al. A Combined Biomarker Panel Shows Improved Sensitivity for the Early Detection of Ovarian Cancer Allowing the Identification of the Most Aggressive Type II Tumours. Br. J. Cancer 2017, 117, 666–674. [Google Scholar] [CrossRef]
- Muinao, T.; Boruah, H.P.D.; Pal, M. Multi-Biomarker Panel Signature as the Key to Diagnosis of Ovarian Cancer. Heliyon 2019, 5, 2826. [Google Scholar] [CrossRef]
- Russell, M.R. Diagnosis of Epithelial Ovarian Cancer Using a Combined Protein Biomarker Panel. Br. J. Cancer 2019, 121, 483–489. [Google Scholar] [CrossRef]
- Zhang, Z.; Chan, D.W. The Road from Discovery to Clinical Diagnostics: Lessons Learned from the First FDA-Cleared In Vitro Diagnostic Multivariate Index Assay of Proteomic Biomarkers. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2995–2999. [Google Scholar] [CrossRef]
- Karlsen, M.A. Evaluation of HE4, CA125, Risk of Ovarian Malignancy Algorithm (ROMA) and Risk of Malignancy Index (RMI) as Diagnostic Tools of Epithelial Ovarian Cancer in Patients with a Pelvic Mass. Gynecol. Oncol. 2012, 127, 379–383. [Google Scholar] [CrossRef]
- Moore, R.G. Comparison of a Novel Multiple Marker Assay vs the Risk of Malignancy Index for the Prediction of Epithelial Ovarian Cancer in Patients with a Pelvic Mass. Am. J. Obstet. Gynecol. 2010, 203, 228.e1–228.e6. [Google Scholar] [CrossRef]
- Pinsky, P.F. Potential Effect of the Risk of Ovarian Cancer Algorithm (ROCA) on the Mortality Outcome of the Prostate, Lung, Colorectal and Ovarian (PLCO) Trial. Int. J. Cancer 2013, 132, 2127–2133. [Google Scholar] [CrossRef]
- Bast, R.C., Jr.; Lu, Z.; Han, C.Y.; Lu, K.H.; Anderson, K.S.; Drescher, C.W.; Skates, S.J. Biomarkers and Strategies for Early Detection of Ovarian Cancer. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2504–2512. [Google Scholar] [CrossRef]
- Akter, S. Recent Advances in Ovarian Cancer: Therapeutic Strategies, Potential Biomarkers, and Technological Improvements. Cells 2022, 11, 650. [Google Scholar] [CrossRef]
- Sugimoto, S.; Uchiyama, T.; Kawahara, N.; Ohbayashi, C.; Kobayashi, H. Immunohistochemical Expression Status of p53, CD44v9, and Ki-67 in a Series of Fallopian Tube Lesions of High-grade Serous Carcinoma. Int. J. Gynecol. Pathol. 2021, 40, 419–426. [Google Scholar] [CrossRef]
- Ahn, H.-S.; Yeom, J.; Yu, J.; Kwon, Y.-I.; Kim, J.-H.; Kim, K. Convergence of Plasma Metabolomics and Proteomics Analysis to Discover Signatures of High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 3447. [Google Scholar] [CrossRef]
- Coscia, F. Multi-Level Proteomics Identifies CT45 as a Chemosensitivity Mediator and Immunotherapy Target in Ovarian Cancer. Cell 2018, 175, 159–170. [Google Scholar] [CrossRef]
- Lee, S. Molecular Analysis of Clinically Defined Subsets of High-Grade Serous Ovarian Cancer. Cell Rep. 2020, 31, 107502. [Google Scholar] [CrossRef]
- McGee, J.P.; Su, P.; Durbin, K.R.; Hollas, M.A.R.; Bateman, N.W.; Maxwell, G.L.; Conrads, T.P.; Fellers, R.T.; Melani, R.D.; Camarillo, J.M.; et al. Automated Imaging and Identification of Proteoforms Directly from Ovarian Cancer Tissue. Nat. Commun. 2023, 14, 6478. [Google Scholar] [CrossRef]
- Delcourt, V. Combined Mass Spectrometry Imaging and Top-down Microproteomics Reveals Evidence of a Hidden Proteome in Ovarian Cancer. EBioMedicine 2017, 21, 55–64. [Google Scholar] [CrossRef]
- Guo, J. HSP60-Regulated Mitochondrial Proteostasis and Protein Translation Promote Tumor Growth of Ovarian Cancer. Sci. Rep. 2019, 9, 12628. [Google Scholar] [CrossRef]
- Ahn, H.S.; Ho, J.Y.; Yu, J.; Yeom, J.; Lee, S.; Hur, S.Y.; Jung, Y.; Kim, K.; Choi, Y.J. Plasma Protein Biomarkers Associated with Higher Ovarian Cancer Risk in Brca1/2 Carriers. Cancers 2021, 13, 3447. [Google Scholar] [CrossRef]
- James, N.E. Septin-2 Is Overexpressed in Epithelial Ovarian Cancer and Mediates Proliferation via Regulation of Cellular Metabolic Proteins. Oncotarget 2019, 10, 2959–2972. [Google Scholar] [CrossRef][Green Version]
- Thomas, S.N.; Friedrich, B.; Schnaubelt, M.; Chan, D.W.; Zhang, H.; Aebersold, R. Orthogonal Proteomic Platforms and Their Implications for the Stable Classification of High-Grade Serous Ovarian Cancer Subtypes. iScience 2020, 23, 101079. [Google Scholar] [CrossRef]
- Dieters-Castator, D.Z.; Rambau, P.F.; Kelemen, L.E.; Siegers, G.M.; Lajoie, G.A.; Postovit, L.M.; Köbel, M. Proteomics-Derived Biomarker Panel Improves Diagnostic Precision to Classify Endometrioid and High-grade Serous Ovarian Carcinoma. Clin. Cancer Res. 2019, 25, 4309–4319. [Google Scholar] [CrossRef]
- Meding, S. Tryptic Peptide Reference Data Sets for MALDI Imaging Mass Spectrometry on Formalin-Fixed Ovarian Cancer Tissues. J. Proteome Res. 2013, 12, 308–315. [Google Scholar] [CrossRef]
- Steitz, A.M. Tumor-Associated Macrophages Promote Ovarian Cancer Cell Migration by Secreting Transforming Growth Factor Beta Induced (TGFBI) and Tenascin C. Cell Death Dis. 2020, 11, 249. [Google Scholar] [CrossRef]
- Nguyen, E.V. Hyper-Phosphorylation of Sequestosome-1 Distinguishes Resistance to Cis-Platin in Patient Derived High Grade Serous Ovarian Cancer Cells. Mol. Cell. Proteom. 2017, 16, 1377–1392. [Google Scholar] [CrossRef]
- Coscia, F.; Watters, K.; Curtis, M.; Eckert, M.A.; Chiang, C.Y.; Tyanova, S.; Montag, A.; Lastra, R.R.; Lengyel, E.; Mann, M. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat Commun. 2016, 7, 12645. [Google Scholar] [CrossRef]
- Bileck, A. Inward Outward Signaling in Ovarian Cancer: Morpho-Phospho-Proteomic Profiling Upon Application of Hypoxia and Shear Stress Characterizes the Adaptive Plasticity of OVCAR-3 and SKOV-3 Cells. Front. Oncol. 2022, 11, 746411. [Google Scholar] [CrossRef]
- Yin, H. Extracellular Matrix Protein-1 Secretory Isoform Promotes Ovarian Cancer through Increasing Alternative MRNA Splicing and Stemness. Nat. Commun. 2021, 12, 4230. [Google Scholar] [CrossRef]
- Conniot, J. Cancer Immunotherapy: Nanodelivery Approaches for Immune Cell Targeting and Tracking. Front. Chem. 2014, 2, 105. [Google Scholar] [CrossRef]
- Walcher, L. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2014, 11, 1280. [Google Scholar] [CrossRef]
- Rawat, M.; Singh, D.; Saraf, S.; Saraf, S. Nanocarriers: Promising Vehicle for Bioactive Drugs. Biol. Pharm. Bull. 2006, 29, 1790–1798. [Google Scholar] [CrossRef]
- Pérez-Herrero, E.; Fernández-Medarde, A. Advanced Targeted Therapies in Cancer: Drug Nanocarriers, the Future of Chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef]
- Gutierrez-Romero, L.; Díez, P.; Montes-Bayón, M. Bioanalytical Strategies to Evaluate Cisplatin Nanodelivery Systems: From Synthesis to Incorporation in Individual Cells and Biological Response. J. Pharm. Biomed. Anal. 2023, 237, 115760. [Google Scholar] [CrossRef]
- Yingchoncharoen, P.; Kalinowski, D.S.; Richardson, D.R. Lipid-Based Drug Delivery Systems in Cancer Therapy: What Is Available and What Is Yet to Come. Pharmacol. Rev. 2016, 68, 701–787. [Google Scholar] [CrossRef]
- Dasa, S.S.K. Plectin-Targeted Liposomes Enhance the Therapeutic Efficacy of a PARP Inhibitor in the Treatment of Ovarian Cancer. Theranostics 2018, 8, 2782–2798. [Google Scholar] [CrossRef]
- Peter, S.; Alven, S.; Maseko, R.B.; Aderibigbe, B.A. Doxorubicin-Based Hybrid Compounds as Potential Anticancer Agents: A Review. Molecules 2022, 27, 4478. [Google Scholar] [CrossRef]
- Meng, Z. Prodrug Strategies for Paclitaxel. Int. J. Mol. Sci. 2016, 17, 796. [Google Scholar] [CrossRef]
- Green, A.E.; Rose, P.G. Pegylated liposomal doxorubicin in ovarian cancer. Int. J. Nanomed. 2006, 1, 229–239. [Google Scholar]
- Aldughaim, M.S.; Muthana, M.; Alsaffar, F.; Barker, M.D. Specific Targeting of PEGylated Liposomal Doxorubicin (Doxil®) to Tumour Cells Using a Novel TIMP3 Peptide. Molecules 2020, 26, 100. [Google Scholar] [CrossRef]
- Huang, S.-T.; Wang, Y.-P.; Chen, Y.-H.; Lin, C.-T.; Li, W.-S.; Wu, H.-C. Liposomal Paclitaxel Induces Fewer Hematopoietic and Cardiovascular Complications than Bioequivalent Doses of Taxol. Int. J. Oncol. 2018, 53, 1105–1117. [Google Scholar] [CrossRef]
- Tan, X.; Zou, S.; Chen, H.; Chen, D. Comparison of Paclitaxel Liposomes Combined with Carboplatin versus Paclitaxel Combined with Carboplatin in the Treatment of Advanced Ovarian Cancer. J. Int. Med. Res. 2023, 51, 9. [Google Scholar] [CrossRef]
- Ye, H.; Liu, X.; Sun, J.; Zhu, S.; Zhu, Y.; Chang, S. Enhanced Therapeutic Efficacy of LHRHa-Targeted Brucea Javanica Oil Liposomes for Ovarian Cancer. BMC Cancer 2016, 16, 831. [Google Scholar] [CrossRef]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef]
- Ma, C.; Wei, T.; Hua, Y.; Wang, Z.; Zhang, L. Effective Antitumor of Orally Intestinal Targeting Penetrating Peptide-Loaded Tyroserleutide/PLGA Nanoparticles in Hepatocellular Carcinoma. Int. J. Nanomed. 2021, 16, 4495–4513. [Google Scholar] [CrossRef]
- Huang, H.-Y. Collagenase IV and Clusterin-Modified Polycaprolactone-Polyethylene Glycol Nanoparticles for Penetrating Dense Tumor Tissues. Theranostics 2021, 11, 906–924. [Google Scholar] [CrossRef] [PubMed]
- Piktel, E. ROS-Mediated Apoptosis and Autophagy in Ovarian Cancer Cells Treated with Peanut-Shaped Gold Nanoparticles. Int. J. Nanomed. 2021, 16, 1993–2011. [Google Scholar] [CrossRef]
- Choi, Y.-J. Differential Cytotoxic Potential of Silver Nanoparticles in Human Ovarian Cancer Cells and Ovarian Cancer Stem Cells. Int. J. Mol. Sci. 2016, 17, 2077. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, I.; Elzoghby, A. Albumin-Based Nanoparticles: A Promising Strategy to Overcome Cancer Drug Resistance. Cancer Drug Resist. 2020, 3, 930–946. [Google Scholar] [CrossRef] [PubMed]
- Aqeel, R.; Srivastava, N.; Kushwaha, P. Micelles in Cancer Therapy: An Update on Preclinical and Clinical Status. Recent Pat. Nanotechnol. 2022, 16, 283–294. [Google Scholar] [PubMed]
- Oerlemans, C.; Bult, W.; Bos, M.; Storm, G.; Nijsen, J.F.W.; Hennink, W.E. Polymeric Micelles in Anticancer Therapy: Targeting, Imaging and Triggered Release. Pharm. Res. 2010, 27, 2569–2589. [Google Scholar] [CrossRef]
- Madaan, K.; Kumar, S.; Poonia, N.; Lather, V.; Pandita, D. Dendrimers in Drug Delivery and Targeting: Drug-Dendrimer Interactions and Toxicity Issues. J. Pharm. Bioallied Sci. 2014, 6, 139–150. [Google Scholar]
- Mittal, P. Dendrimers: A New Race of Pharmaceutical Nanocarriers. BioMed Res. Int. 2021, 2021, 8844030. [Google Scholar] [CrossRef]
- Chis, A.A. Applications and Limitations of Dendrimers in Biomedicine. Molecules 2020, 25, 3982. [Google Scholar] [CrossRef]
- Bober, Z.; Bartusik-Aebisher, D.; Aebisher, D. Application of Dendrimers in Anticancer Diagnostics and Therapy. Molecules 2022, 27, 3237. [Google Scholar] [CrossRef]
- Mendes, L.P.; Pan, J.; Torchilin, V.P. Dendrimers as Nanocarriers for Nucleic Acid and Drug Delivery in Cancer Therapy. Molecules 2017, 22, 1401. [Google Scholar] [CrossRef]
- Yellepeddi, V.K.; Vangara, K.K.; Palakurthi, S. Poly(Amido)Amine (PAMAM) Dendrimer-Cisplatin Complexes for Chemotherapy of Cisplatin-Resistant Ovarian Cancer Cells. J. Nanoparticle Res. 2013, 15, 1897. [Google Scholar] [CrossRef]
- Evans, E.R.; Bugga, P.; Asthana, V.; Drezek, R. Metallic Nanoparticles for Cancer Immunotherapy. Mater. Today 2019, 21, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, A.; Shah, P.P.; Rai, S.N.; Panguluri, S.K.; Kakar, S.S. MicroRNA Signature of Cis-Platin Resistant vs. Cis-Platin Sensitive Ovarian Cancer Cell Lines. J. Ovarian Res. 2011, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Tossetta, G. Metformin Improves Ovarian Cancer Sensitivity to Paclitaxel and Platinum-Based Drugs: A Review of In Vitro Findings. Int. J. Mol. Sci. 2022, 23, 12893. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, Y.; Zhang, X.; Guo, H.; Gao, H. Nanoparticles in Precision Medicine for Ovarian Cancer: From Chemotherapy to Immunotherapy. Int. J. Pharm. 2020, 591, 119986. [Google Scholar] [CrossRef]
- Chaurasiya, S.; Mishra, V. Biodegradable Nanoparticles as Theranostics of Ovarian Cancer: An Overview. J. Pharm. Pharmacol. 2018, 70, 435–449. [Google Scholar] [CrossRef]
- Shi, Y.; van der Meel, R.; Chen, X.; Lammers, T. The EPR effect and beyond: Strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics 2020, 10, 7921–7924. [Google Scholar] [CrossRef]
- Bessone, F. Albumin Nanoformulations as an Innovative Solution to Overcome Doxorubicin Chemoresistance. Cancer Drug Resist. 2021, 4, 192–207. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yan, S.; Xiao, F.; Xue, M. PLGA Nanoparticles Delivering CPT-11 Combined with Focused Ultrasound Inhibit Platinum Resistant Ovarian Cancer. Transl. Cancer Res. 2021, 10, 1732–1743. [Google Scholar] [CrossRef] [PubMed]
- Salve, R.; Kumar, P.; Chaudhari, B.P.; Gajbhiye, V. Aptamer Tethered Bio-Responsive Mesoporous Silica Nanoparticles for Efficient Targeted Delivery of Paclitaxel to Treat Ovarian Cancer Cells. J. Pharm. Sci. 2023, 112, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Jiang, B.; Li, B.; Li, Z.; Jiang, B.H.; Chen, Y.C. Kaempferol Nanoparticles Achieve Strong and Selective Inhibition of Ovarian Cancer Cell Viability. Int. J. Nanomed. 2012, 7, 3951–3959. [Google Scholar]
- Sawaftah, N.A.; Paul, V.; Awad, N.; Husseini, G.A. Modeling of Anti-Cancer Drug Release Kinetics from Liposomes and Micelles: A Review. IEEE Trans. Nanobioscience 2021, 20, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yang, Q.; Li, J.; Liu, G.; Nikoulin, I.; Jia, S. Clinical Trials of Novel Targeted Therapies in Ovarian Cancer: Moving Beyond Poly ADP Ribose Polymerase (PARP) Inhibitors. Curr. Pharm. Biotechnol. 2018, 19, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.E.; Cummings, N.D.; Sethi, M.; Wang, E.C.; Sukumar, R.; Moore, D.T.; Wang, A.Z. Preclinical Evaluation of Genexol-Pm, a Nanoparticle Formulation of Paclitaxel, as a Novel Radiosensitizer for the Treatment of Non-Small Cell Lung Cancer. Int. J. Radiat. Oncol. Biol Phys. 2013, 86, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Kim, Y.M.; Cho, C.H.; Kim, Y.T.; Kim, S.M.; Hur, S.Y.; Kim, J.H.; Kim, B.G.; Kim, S.C.; Ryu, H.S.; et al. An Open-Label, Randomized, Parallel, Phase Ii Trial to Evaluate the Efficacy and Safety of a Cremophor-Free Polymeric Micelle Formulation of Paclitaxel as First-Line Treatment for Ovarian Cancer: A Korean Gynecologic Oncology Group Study (KGOG-3021). Cancer Res. Treat. 2018, 50, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Liu, X.X.; Yang, X.D.; Tan, S.; Zou, Z.W. A Compound Formulation of EGF-Modified Paclitaxel Micelles and EGF-Modified Emodin Micelles Enhance the Therapeutic Effect of Ovarian Cancer. J. Liposome Res. 2023, 33, 89–101. [Google Scholar] [CrossRef]
- Trinder, D.; Baker, E. Transferrin receptor 2: A new molecule in iron metabolism. Int. J. Biochem. Cell Biol. 2003, 35, 292–296. [Google Scholar] [CrossRef]
- Calzolari, A. Transferrin Receptor 2 Is Frequently Expressed in Human Cancer Cell Lines. Blood Cells Mol. Dis. 2010, 39, 82–91. [Google Scholar] [CrossRef]
- Roetto, A.; Mezzanotte, M.; Pellegrino, R.M. The Functional Versatility of Transferrin Receptor 2 and Its Therapeutic Value. Pharmaceuticals 2018, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Shigeta, S.; Toyoshima, M.; Kitatani, K.; Ishibashi, M.; Usui, T.; Yaegashi, N. Transferrin Facilitates the Formation of DNA Double-Strand Breaks via Transferrin Receptor 1: The Possible Involvement of Transferrin in Carcinogenesis of High-Grade Serous Ovarian Cancer. Oncogene 2016, 35, 3577–3586. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wei, Y.; Wei, X. AXL Receptor Tyrosine Kinase as a Promising Anti-Cancer Approach: Functions, Molecular Mechanisms and Clinical Applications. Mol. Cancer 2019, 18, 153. [Google Scholar] [CrossRef]
- Yeo, X.H. The Effect of Inhibition of Receptor Tyrosine Kinase AXL on DNA Damage Response in Ovarian Cancer. Commun. Biol. 2023, 6, 660. [Google Scholar]
- Byers, L.A. An Epithelial-Mesenchymal Transition Gene Signature Predicts Resistance to EGFR and PI3K Inhibitors and Identifies Axl as a Therapeutic Target for Overcoming EGFR Inhibitor Resistance. Clin. Cancer Res. 2013, 19, 279–290. [Google Scholar] [CrossRef]
- Cheng, K.; Liu, C.-F.; Rao, G.-W. Anti-Angiogenic Agents: A Review on Vascular Endothelial Growth Factor Receptor-2 (VEGFR-2) Inhibitors. Curr. Med. Chem. 2021, 28, 2540–2564. [Google Scholar] [CrossRef]
- Subramanian, J.; Morgensztern, D.; Govindan, R. Vascular Endothelial Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer. Clin. Lung Cancer 2020, 11, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, S.M.; Amini, A.; Morris, D.L.; Pourgholami, M.H. Significance of Vascular Endothelial Growth Factor in Growth and Peritoneal Dissemination of Ovarian Cancer. Cancer Metastasis Rev. 2012, 31, 143–162. [Google Scholar] [CrossRef]
- Kaczmarek, M.M.; Schams, D.; Ziecik, A.J. Role of Vascular Endothelial Growth Factor in Ovarian Physiology—An Overview. Reprod. Biol. 2005, 5, 111–136. [Google Scholar]
- Garcia, J. Bevacizumab (Avastin®) in Cancer Treatment: A Review of 15 Years of Clinical Experience and Future Outlook. Cancer Treat. Rev. 2020, 86, 102017. [Google Scholar] [CrossRef] [PubMed]
- Nakai, H.; Matsumura, N. The Roles and Limitations of Bevacizumab in the Treatment of Ovarian Cancer. Int. J. Clin. Oncol. 2023, 27, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.N. Phase III, Randomized Trial of Mirvetuximab Soravtansine versus Chemothera-Py in Patients with Platinum-Resistant Ovarian Cancer: Primary Analysis of FORWARD I. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, 757–765. [Google Scholar] [CrossRef]
- Heo, Y.-A. Mirvetuximab Soravtansine: First Approval. Drugs 2023, 83, 265–273. [Google Scholar] [CrossRef]
- Modi, D.A.; Sunoqrot, S.; Bugno, J.; Lantvit, D.D.; Hong, S.; Burdette, J.E. Targeting of Follicle Stimulating Hormone Peptide-Conjugated Dendrimers to Ovarian Cancer Cells. Nanoscale 2014, 6, 2812–2820. [Google Scholar] [CrossRef]
- Yellepeddi, V.K.; Kumar, A.; Palakurthi, S. Biotinylated Poly(Amido)Amine (PAMAM) Dendrimers as Carriers for Drug Delivery to Ovarian Cancer Cells in Vitro. Anticancer Res. 2009, 29, 2933–2943. [Google Scholar]
- Hatamikia, S. Ovarian Cancer beyond Imaging: Integration of AI and Multiomics Biomarkers. Eur. Radiol. Exp. 2023, 7, 50. [Google Scholar] [CrossRef]
- Bonilla, D.L.; Reinin, G.; Chua, E. Full Spectrum Flow Cytometry as a Powerful Technology for Cancer Immunotherapy Research. Front. Mol. Biosci. 2021, 7, 612801. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, M.H.; Nolan, G.P. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165, 780–791. [Google Scholar] [CrossRef] [PubMed]
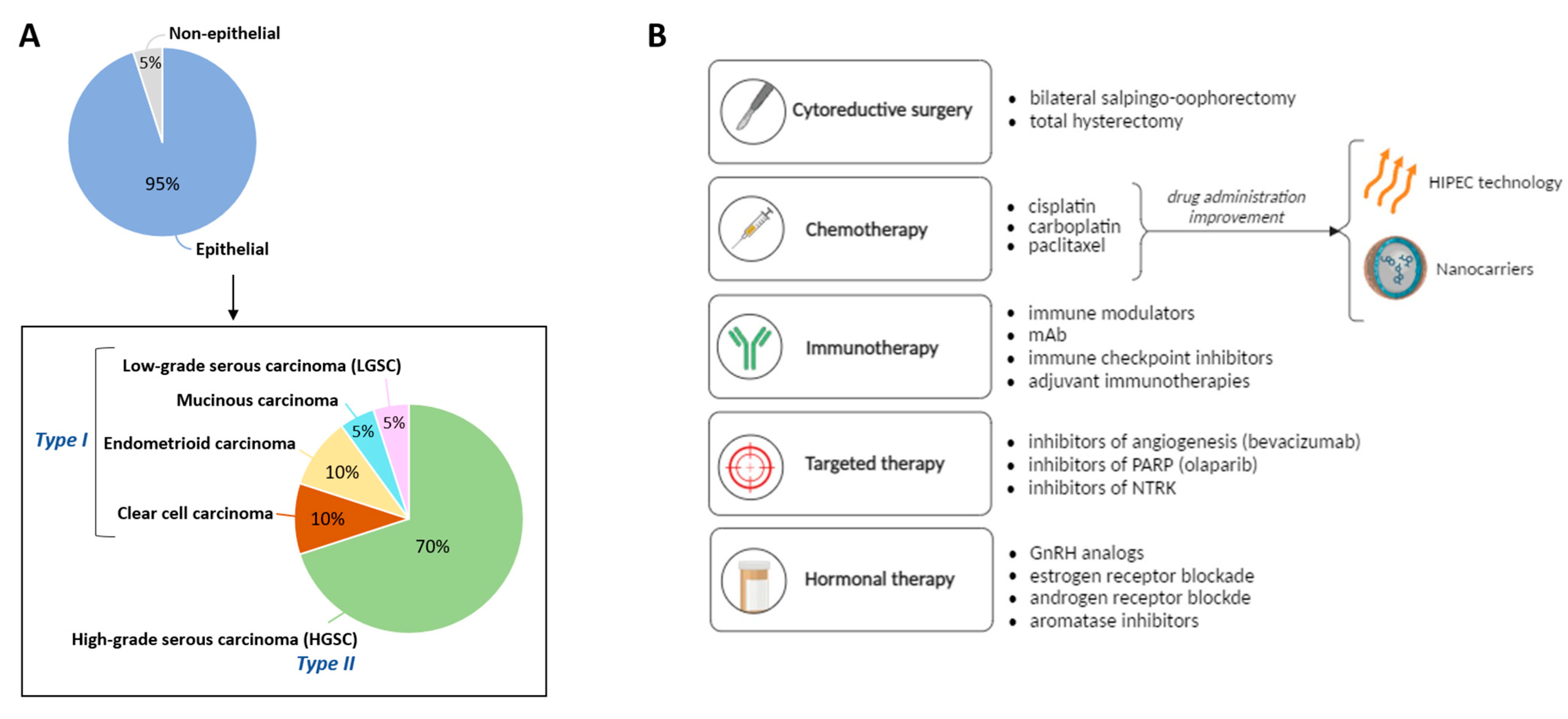
| Molecular and Genetic Classification | Type I Mutations in KRAS, BRAF, PTEN, PIK3CA, CTNNB1, ARID1A Genes, Genetically Stable and Slow Progression | Type II Mutations in TP53, Genetically Unstable and Rapid Progression | |||
|---|---|---|---|---|---|
| Histotype | Clear Cell [17,18,19] | Endometrioid [17,20,21,22] | Mucinous [17,23,24,25,26] | LGSC [27] | HGSC [28,29,30] |
| Tumoral/cellular structure | Clear or hobnail-shaped cells with abundant cytoplasm, often containing glycogen and lipid droplets. | Glandular structures resembling those of the endometrium. | Glandular structures filled with mucin-producing cells. | Bilateral adnexal tumors commonly present as multicystic masses with nodular areas, excrescences, and papillary projections on their inner surface. | Complex papillary architecture characterized by epithelial projections with irregular contours resulting in the formation of multicellular structures. |
| Aggressiveness and proliferation rates | Medium-high aggressiveness behavior and moderate proliferation rates. | Less aggressive compared to HGSC, but more aggressive than LGSC. Proliferation rates vary depending on tumor grade and histological characteristics. | Less aggressive compared to other subtypes. Proliferation rates vary based on tumor grade and histological features. | Low aggressive clinical course and low proliferation rate. | Most common and aggressive subtype. Early dissemination and high rates of recurrence. |
| Genetic aberrations and marker expression | Mutations in ARID1A/B, SMARCA4, ERBB2, PIK3CA, AKT2, PTEN, KRAS, PPP2R1A | Mutations in PTEN, ARID1A, CTNNB1, KRAS/BRAF, PIK3CA; aberrant expression of β-catenin, estrogen and progesterone receptors. | Mutations in KRAS, TP53 PIK3CA/PTEN, ARID1A, BRAF, CTNNB1/APC, elevated levels of CEA, CA-19-9, and REG4. | Mutations in KRAS, BRAF, NRAS, ERBB2, PI3KCA, FFAR1, USP9X, and EIF1AX. | Mutations in TP53 and BRCA1/2; HER2 amplification. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Portugués, C.; Montes-Bayón, M.; Díez, P. Biomarkers in Ovarian Cancer: Towards Personalized Medicine. Proteomes 2024, 12, 8. https://doi.org/10.3390/proteomes12010008
López-Portugués C, Montes-Bayón M, Díez P. Biomarkers in Ovarian Cancer: Towards Personalized Medicine. Proteomes. 2024; 12(1):8. https://doi.org/10.3390/proteomes12010008
Chicago/Turabian StyleLópez-Portugués, Carlos, María Montes-Bayón, and Paula Díez. 2024. "Biomarkers in Ovarian Cancer: Towards Personalized Medicine" Proteomes 12, no. 1: 8. https://doi.org/10.3390/proteomes12010008
APA StyleLópez-Portugués, C., Montes-Bayón, M., & Díez, P. (2024). Biomarkers in Ovarian Cancer: Towards Personalized Medicine. Proteomes, 12(1), 8. https://doi.org/10.3390/proteomes12010008









