Proteomic Applications in Aquatic Environment Studies
Abstract
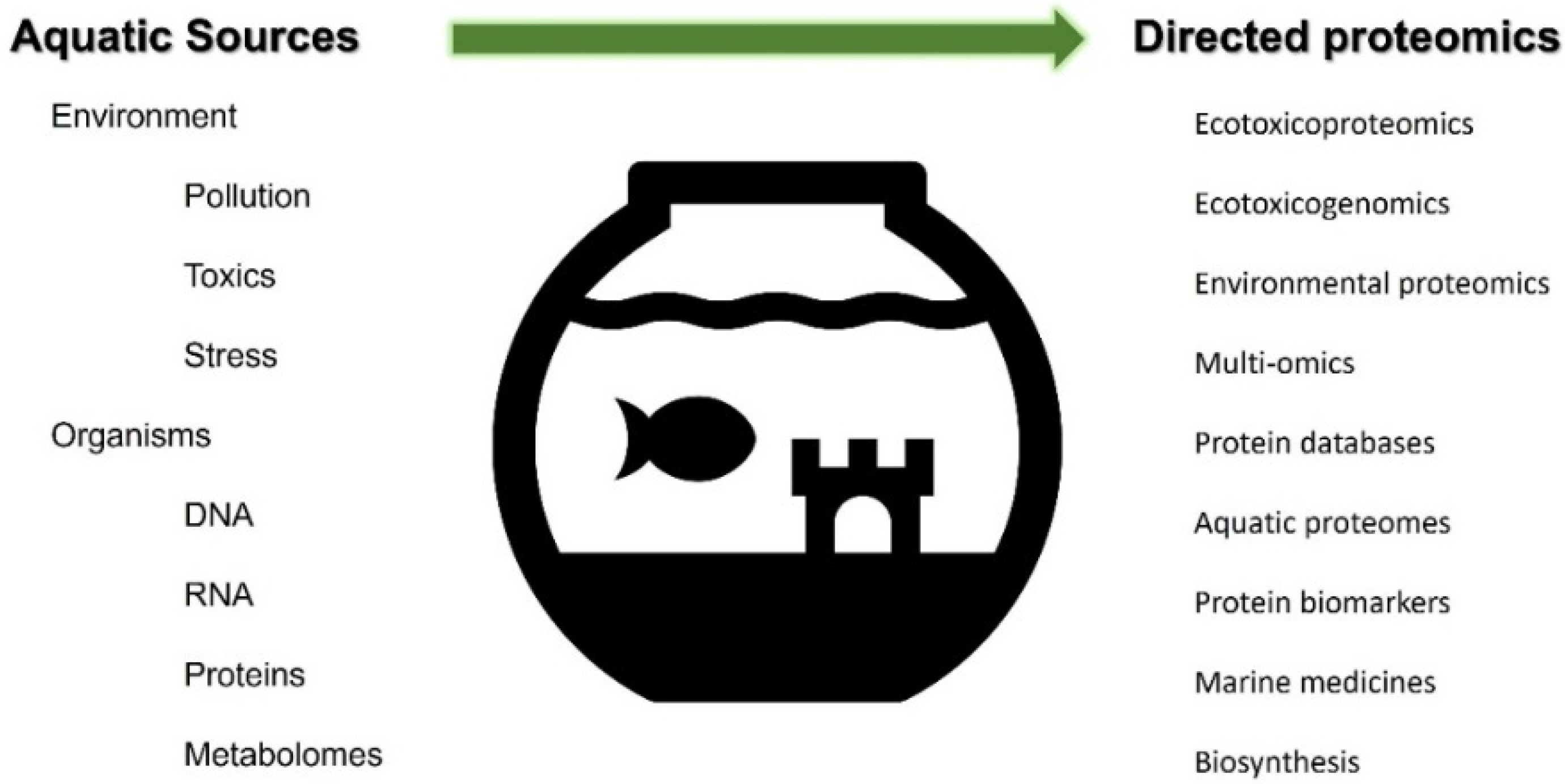
1. Introduction
2. Methods: Search and Selection of Literature
3. Proteomics as a Biomonitoring Tool
4. Applications of Proteomics in Aquatic Studies
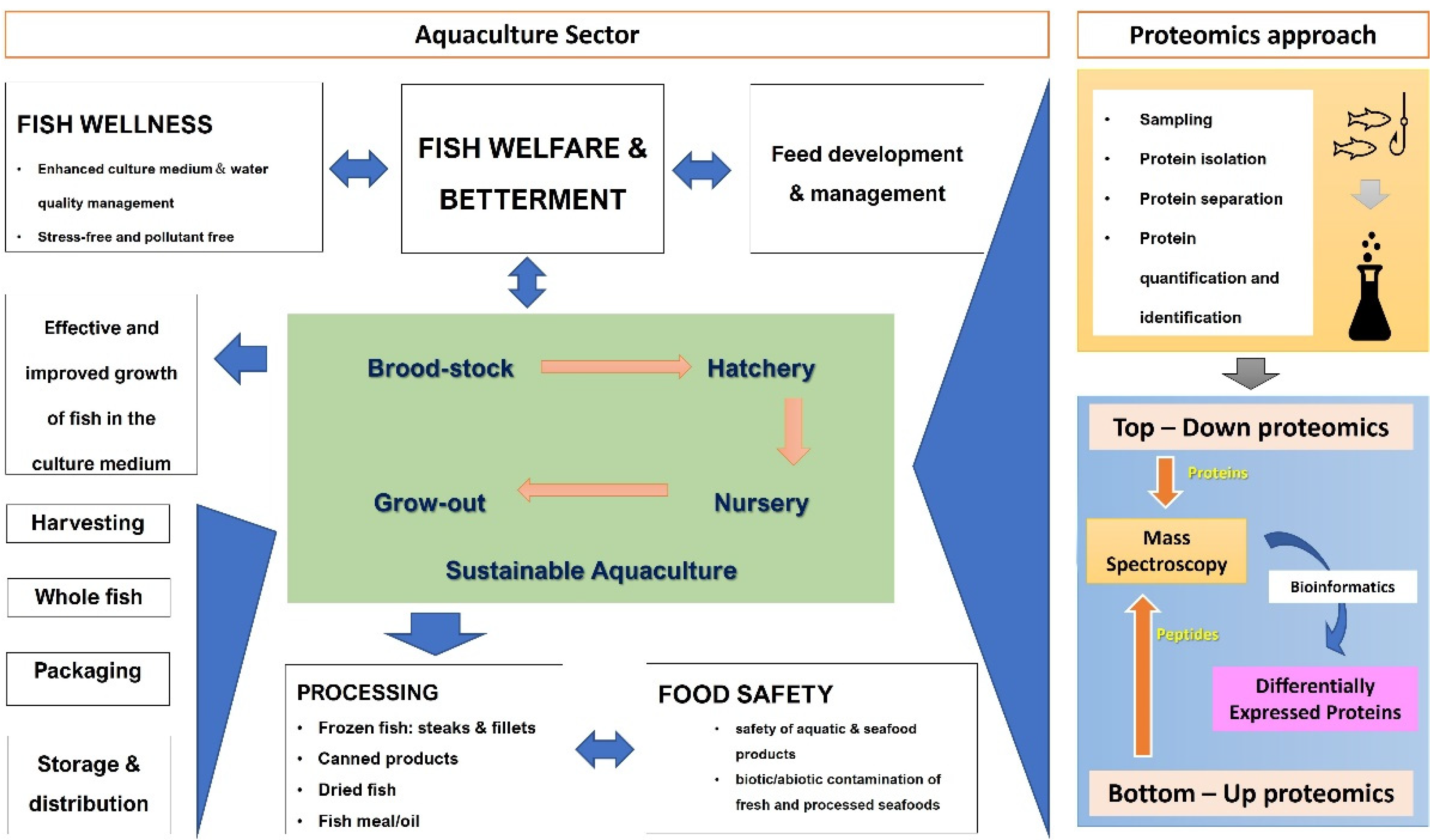
4.1. Proteomics in the Aquatic Food Industry
4.2. Proteomics in Aquatic Environmental Pollution and Monitoring
4.3. Natural Aquatic Proteins and Marine-Derived Medicine
5. Challenges and Recommendations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Swaminathan, S. GFP: The green revolution. Nat. Cell Biol. 2009, 11, S20. [Google Scholar] [CrossRef]
- Yang, Y.J.; Jung, D.; Yang, B.; Hwang, B.H.; Cha, H.J. Aquatic proteins with repetitive motifs provide insights to bioengineering of novel biomaterials. Biotechnol. J. 2014, 9, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Swinbanks, D. Australia backs innovation, shuns telescope. Nature 1995, 378, 653. [Google Scholar] [CrossRef] [PubMed]
- Phimister, B. Four companies announce discovery of β-secretase gene. Nat. Biotechnol. 2000, 18, 16. [Google Scholar] [CrossRef]
- Smalla, K.; Sobecky, P.A. The prevalence and diversity of mobile genetic elements in bacterial communities of different environmental habitats: Insights gained from different methodological approaches. FEMS Microbiol. Ecol. 2002, 42, 165–175. [Google Scholar] [CrossRef]
- Rotchell, J.M.; Ostrander, G.K. Molecular markers of endocrine disruption in aquatic organisms. J. Toxicol. Environ. Health—Part B Crit. Rev. 2003, 6, 453–495. [Google Scholar] [CrossRef]
- Gouveia, D.; Almunia, C.; Cogne, Y.; Pible, O.; Degli-Esposti, D.; Salvador, A.; Cristobal, S.; Sheehan, D.; Chaumot, A.; Geffard, O.; et al. Ecotoxicoproteomics: A decade of progress in our understanding of anthropogenic impact on the environment. J. Proteom. 2019, 198, 66–77. [Google Scholar] [CrossRef]
- Nesatyy, V.J.; Suter, M.J.F. Proteomics for the analysis of environmental stress responses in organisms. Environ. Sci. Technol. 2007, 41, 6891–6900. [Google Scholar] [CrossRef]
- Mizukami, H.; Hathway, B.; Procopio, N. Aquatic Decomposition of Mammalian Corpses: A Forensic Proteomic Approach. J. Proteome Res. 2020, 19, 2122–2135. [Google Scholar] [CrossRef]
- Humbert, J.-F.; Dorigo, U. Biodiversity and aquatic ecosystem functioning: A mini-review. Aquat. Ecosyst. Health Manag. 2005, 8, 367–374. [Google Scholar] [CrossRef]
- Korsgaard, L.; Schou, J.S. Economic valuation of aquatic ecosystem services in developing countries. Water Policy 2009, 12, 20–31. [Google Scholar] [CrossRef]
- Sandrin, T.R.; Dowd, S.E.; Herman, D.C.; Maier, R.M. Aquatic Environments. In Environmental Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 103–122. [Google Scholar] [CrossRef]
- Mooij, W.M.; van Wijk, D.; Beusen, A.H.; Brederveld, R.J.; Chang, M.; Cobben, M.M.; DeAngelis, D.L.; Downing, A.S.; Green, P.; Gsell, A.S.; et al. Modeling water quality in the Anthropocene: Directions for the next-generation aquatic ecosystem models. Curr. Opin. Environ. Sustain. 2019, 36, 85–95. [Google Scholar] [CrossRef]
- Wu, Y. Indicators for Monitoring Aquatic Ecosystem. In Periphyton; Elsevier: Amsterdam, The Netherlands, 2017; pp. 71–106. [Google Scholar] [CrossRef]
- Valavanidis, P.; Vlachogianni, T. Integrated Biomarkers in Aquatic Organisms as a Tool for Biomonitoring Environmental Pollution and Improved Ecological Risk Assessment. Sci. Adv. Environ. Toxicol. Ecotoxicol. 2010, 10, 325–333. [Google Scholar]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant abiotic stress proteomics: The major factors determining alterations in cellular proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef]
- Hillebrand, H.; Jacob, U.; Leslie, H.M. Integrative research perspectives on marine conservation: Marine Conservation. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190444. [Google Scholar] [CrossRef]
- Gebremedhin, S.; Bruneel, S.; Getahun, A.; Anteneh, W.; Goethals, P. Scientific methods to understand fish population dynamics and support sustainable fisheries management. Water 2021, 13, 574. [Google Scholar] [CrossRef]
- Rudovica, V.; Rotter, A.; Gaudêncio, S.P.; Novoveská, L.; Akgül, F.; Akslen-Hoel, L.K.; Alexandrino, D.A.M.; Anne, O.; Arbidans, L.; Atanassova, M.; et al. Valorization of Marine Waste: Use of Industrial By-Products and Beach Wrack Towards the Production of High Added-Value Products. Front. Mar. Sci. 2021, 8, 723333. [Google Scholar] [CrossRef]
- Nyman, T.A. The role of mass spectrometry in proteome studies. Biomol. Eng. 2001, 18, 221–227. [Google Scholar] [CrossRef]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef]
- Avissar, S.; Schreiber, G. G protein measurements: An empirical tool for the diagnosis of affective disorders. Drug Dev. Res. 2000, 50, 316–323. [Google Scholar] [CrossRef]
- Legutki, J.B.; Zhao, Z.G.; Greving, M.; Woodbury, N.; Johnston, S.A.; Stafford, P. Scalable high-density peptide arrays for comprehensive health monitoring. Nat. Commun. 2014, 5, 4785. [Google Scholar] [CrossRef] [PubMed]
- Oonk, S.; Schuurmans, T.; Pabst, M.; de Smet, L.C.P.M.; de Puit, M. Proteomics as a new tool to study fingermark ageing in forensics. Sci. Rep. 2018, 8, 16425. [Google Scholar] [CrossRef] [PubMed]
- Moreira, M.; Schrama, D.; Farinha, A.P.; Cerqueira, M.; de Magalhães, C.R.; Carrilho, R.; Rodrigues, P. Fish pathology research and diagnosis in aquaculture of farmed fish; a proteomics perspective. Animals 2021, 11, 125. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Silva, T.S.; Dias, J.; Jessen, F. PROTEOMICS in aquaculture: Applications and trends. J. Proteom. 2012, 75, 4325–4345. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, D.; Chaumot, A.; Charnot, A.; Almunia, C.; François, A.; Navarro, L.; Armengaud, J.; Salvador, A.; Geffard, O. Ecotoxico-Proteomics for Aquatic Environmental Monitoring: First in Situ Application of a New Proteomics-Based Multibiomarker Assay Using Caged Amphipods. Environ. Sci. Technol. 2017, 51, 13417–13426. [Google Scholar] [CrossRef]
- Bestel-Corre, G.; Dumas-Gaudot, E.; Gianinazzi, S. Proteomics as a tool to monitor plant-microbe endosymbioses in the rhizosphere. Mycorrhiza 2004, 14, 1–10. [Google Scholar] [CrossRef]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and their applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef]
- Yadav, S.; Srivastava, A.; Biswas, S.; Chaurasia, N.; Singh, S.K.; Kumar, S.; Srivastava, V.; Mishra, Y. Comparison and optimization of protein extraction and two-dimensional gel electrophoresis protocols for liverworts. BMC Res. Notes 2020, 13, 60. [Google Scholar] [CrossRef]
- An, Y.; Scherer, P. Mouse Adipose Tissue Protein Extraction. Bio-Protocol 2020, 10, e3631. [Google Scholar] [CrossRef]
- Bose, U.; Broadbent, J.A.; Juhász, A.; Karnaneedi, S.; Johnston, E.B.; Stockwell, S.; Byrne, K.; Limviphuvadh, V.; Maurer-Stroh, S.; Lopata, A.L.; et al. Protein extraction protocols for optimal proteome measurement and arginine kinase quantitation from cricket Acheta domesticus for food safety assessment. Food Chem. 2021, 348, 129110. [Google Scholar] [CrossRef]
- Rodrigues-Ribeiro, L.; Melo-Braga, M.N.; Kjeldsen, F.; Gómez-Mendoza, D.P.; Verano-Braga, T. Assessment of protein extraction and digestion efficiency of well-established shotgun protocols for heart proteomics. Anal. Biochem. 2019, 578, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ji, C.; Wei, L.; Zhao, J. Evaluation of protein extraction protocols for 2DE in marine ecotoxicoproteomics. Proteomics 2013, 13, 3205–3210. [Google Scholar] [CrossRef] [PubMed]
- Betti, M.; Ciacci, C.; Abramovich, S.; Frontalini, F. Protein extractions from Amphistegina lessonii: Protocol development and optimization. Life 2021, 11, 418. [Google Scholar] [CrossRef]
- Sedelnikova, S. Protocols and Tips in Protein Purification or How to Purify Protein in One Day; Department of Molecular Biology and Biotechnology of The University of Sheffield: Sheffield, UK, 2018. [Google Scholar]
- Harnedy, P.A.; FitzGerald, R.J. Extraction of protein from the macroalga Palmaria palmata. LWT—Food Sci. Technol. 2013, 51, 375–382. [Google Scholar] [CrossRef]
- Sanchez, B.C.; Ralston-Hooper, K.; Sepúlveda, M.S. Review of recent proteomic applications in aquatic toxicology. Environ. Toxicol. Chem. 2011, 30, 274–282. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, L.; Xia, X.; Hu, W.; Zhou, P. Effect of geographic variation on the proteome of sea cucumber (Stichopus japonicus). Food Res. Int. 2020, 136, 109498. [Google Scholar] [CrossRef]
- Popović, N.T.; Kazazić, S.P.; Strunjak-Perović, I.; Čož-Rakovac, R. Differentiation of environmental aquatic bacterial isolates by MALDI-TOF MS. Environ. Res. 2017, 152, 7–16. [Google Scholar] [CrossRef]
- Rodriguez-Temporal, D.; Perez-Risco, D.; Struzka, E.A.; Mas, M.; Alcaide, F. Evaluation of two protein extraction protocols based on freezing and mechanical disruption for identifying nontuberculous mycobacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry from liquid and solid cultures. J. Clin. Microbiol. 2018, 56, 4. [Google Scholar] [CrossRef]
- Dieme, C.; Yssouf, A.; Vega-Rúa, A.; Berenger, J.M.; Failloux, A.B.; Raoult, D.; Parola, P.; Almeras, L. Accurate identification of Culicidae at aquatic developmental stages by MALDI-TOF MS profiling. Parasites Vectors 2014, 7, 544. [Google Scholar] [CrossRef]
- Colatriano, D.; Walsh, D.A. An aquatic microbial metaproteomics workflow: From cells to tryptic peptides suitable for tandem mass spectrometry-based analysis. J. Vis. Exp. 2015, 2015, e52827. [Google Scholar] [CrossRef]
- Leduc, A.; Fournier, V.; Henry, J. A standardized, innovative method to characterize the structure of aquatic protein hydrolysates. Heliyon 2020, 6, e04170. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, S.C.; He, L.; Li, M. Fingerprinting protein structures effectively and efficiently. Bioinformatics 2014, 30, 949–955. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Niessen, K.A.; Xu, M.; George, D.K.; Chen, M.C.; Ferré-D’Amaré, A.R.; Snell, E.H.; Cody, V.; Pace, J.; Schmidt, M.; Markelz, A.G. Protein and RNA dynamical fingerprinting. Nat. Commun. 2019, 10, 1026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zong, R.; He, H.; Huang, T. Effects of hydrogen peroxide on Scenedesmus obliquus: Cell growth, antioxidant enzyme activity and intracellular protein fingerprinting. Chemosphere 2022, 287, 132185. [Google Scholar] [CrossRef]
- Piñeiro, C.; Barros-Velázquez, J.; Vázquez, J.; Figueras, A.; Gallardo, J.M. Proteomics as a tool for the investigation of seafood and other marine products. J. Proteome Res. 2003, 2, 127–135. [Google Scholar] [CrossRef]
- Golden, C.D.; Koehn, J.Z.; Shepon, A.; Passarelli, S.; Free, C.M.; Viana, D.F.; Matthey, H.; Eurich, J.G.; Gephart, J.A.; Fluet-Chouinard, E.; et al. Aquatic foods to nourish nations. Nature 2021, 598, 315–320. [Google Scholar] [CrossRef]
- Fiorella, K.J.; Okronipa, H.; Baker, K.; Heilpern, S. Contemporary aquaculture: Implications for human nutrition. Curr. Opin. Biotechnol. 2021, 70, 83–90. [Google Scholar] [CrossRef]
- Abdelrahman, H.; ElHady, M.; Alcivar-Warren, A.; Allen, S.; Al-Tobasei, R.; Bao, L.; Beck, B.; Blackburn, H.; Bosworth, B.; Buchanan, J.; et al. Aquaculture genomics, genetics and breeding in the United States: Current status, challenges, and priorities for future research. BMC Genom. 2017, 18, 191. [Google Scholar] [CrossRef]
- Carrera, M.; Pazos, M.; Gasset, M. Proteomics-based methodologies for the detection and quantification of seafood allergens. Foods 2020, 9, 1134. [Google Scholar] [CrossRef]
- Dalmo, R.A.; Bøgwald, J. ß-Glucans As Conductors of Immune Symphonies. Fish Shellfish Immunol. 2008, 25, 384–396. [Google Scholar] [CrossRef]
- Ghaedi, G.; Keyvanshokooh, S.; Mohammadi Azarm, H.; Akhlaghi, M. Proteomic analysis of muscle tissue from rainbow trout (Oncorhynchus mykiss) fed dietary β-glucan. Iran. J. Vet. Res. 2016, 17, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Morais, S.; Silva, T.; Cordeiro, O.; Rodrigues, P.; Guy, D.R.; Bron, J.E.; Taggart, J.B.; Bell, J.G.; Tocher, D.R. Effects of genotype and dietary fish oil replacement with vegetable oil on the intestinal transcriptome and proteome of Atlantic salmon (Salmo salar). BMC Genom. 2012, 13, 448. [Google Scholar] [CrossRef] [PubMed]
- Yesiltas, B.; Gregersen, S.; Lægsgaard, L.; Brinch, M.L.; Olsen, T.H.; Marcatili, P.; Overgaard, M.T.; Hansen, E.B.; Jacobsen, C.; García-Moreno, P.J. Emulsifier peptides derived from seaweed, methanotrophic bacteria, and potato proteins identified by quantitative proteomics and bioinformatics. Food Chem. 2021, 362, 130217. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; You, W.; Chen, N.; Cao, M.; Tang, X.; Guan, X.; Qu, W.; Chen, R.; Mao, Y.; Poetsch, A. Comparative Quantitative Proteomics Reveals the Desiccation Stress Responses of the Intertidal Seaweed NEOPORPHYRA haitanensis. J. Phycol. 2020, 56, 1664–1675. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, C.Y.; Sun, L.; He, Z.; Yang, P.L.; Liao, H.J.; Feng, Y. Edible aquatic insects: Diversities, nutrition, and safety. Foods 2021, 10, 3033. [Google Scholar] [CrossRef]
- Liu, X.; Afonso, L.; Altman, E.; Johnson, S.; Brown, L.; Li, J. O-acetylation of sialic acids in N-glycans of Atlantic salmon (Salmo salai) serum is altered by handling stress. Proteomics 2008, 8, 2849–2857. [Google Scholar] [CrossRef]
- Wulff, T.; Jessen, F.; Roepstorff, P.; Hoffmann, E.K. Long term anoxia in rainbow trout investigated by 2-DE and MS/MS. Proteomics 2008, 8, 1009–1018. [Google Scholar] [CrossRef]
- Silva, T.S.; Cordeiro, O.; Richard, N.; Conceição, L.E.C.; Rodrigues, P.M. Changes in the soluble bone proteome of reared white seabream (Diplodus sargus) with skeletal deformities. Comp. Biochem. Physiol.—Part D Genom. Proteom. 2011, 6, 82–91. [Google Scholar] [CrossRef]
- Addis, M.F.; Cappuccinelli, R.; Tedde, V.; Pagnozzi, D.; Porcu, M.C.; Bonaglini, E.; Roggio, T.; Uzzau, S. Proteomic analysis of muscle tissue from gilthead sea bream (Sparus aurata, L.) farmed in offshore floating cages. Aquaculture 2010, 309, 245–252. [Google Scholar] [CrossRef]
- Xu, Z.N.; Zheng, G.D.; Wu, C.B.; Jiang, X.Y.; Zou, S.M. Identification of proteins differentially expressed in the gills of grass carp (Ctenopharyngodon idella) after hypoxic stress by two-dimensional gel electrophoresis analysis. Fish Physiol. Biochem. 2019, 45, 743–752. [Google Scholar] [CrossRef]
- Jiang, H.; Li, F.; Xie, Y.; Huang, B.; Zhang, J.; Zhang, J.; Zhang, C.; Li, S.; Xiang, J. Comparative proteomic profiles of the hepatopancreas in Fenneropenaeus chinensis response to hypoxic stress. Proteomics 2009, 9, 3353–3367. [Google Scholar] [CrossRef] [PubMed]
- Timmins-Schiffman, E.B.; Crandall, G.A.; Vadopalas, B.; Riffle, M.E.; Nunn, B.L.; Roberts, S.B. Integrating Discovery-driven Proteomics and Selected Reaction Monitoring to Develop a Noninvasive Assay for Geoduck Reproductive Maturation. J. Proteome Res. 2017, 16, 3298–3309. [Google Scholar] [CrossRef] [PubMed]
- Spencer, L.H.; Horwith, M.; Lowe, A.T.; Venkataraman, Y.R.; Timmins-Schiffman, E.; Nunn, B.L.; Roberts, S.B. Pacific geoduck (Panopea generosa) resilience to natural pH variation. Comp. Biochem. Physiol.—Part D Genom. Proteom. 2019, 30, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Forné, I.; Abián, J.; Cerdà, J. Fish proteome analysis: Model organisms and non-sequenced species. Proteomics 2010, 10, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Hicks, D.T. Seafood safety and quality: The consumer’s role. Foods 2016, 5, 71. [Google Scholar] [CrossRef]
- Gao, P.; Mohd Noor, N.Q.I.; Shaarani, S.M. Current status of food safety hazards and health risks connected with aquatic food products from Southeast Asian region. Crit. Rev. Food Sci. Nutr. 2022, 62, 3471–3489. [Google Scholar] [CrossRef]
- Keyvanshokooh, S.; Kalbassi, M.R.; Hosseinkhani, S.; Vaziri, B. Comparative proteomics analysis of male and female Persian sturgeon (Acipenser persicus) gonads. Anim. Reprod. Sci. 2009, 111, 361–368. [Google Scholar] [CrossRef]
- Martinez, I.; Friis, T.J. Application of proteome analysis to seafood authentication. Proteomics 2004, 4, 347–354. [Google Scholar] [CrossRef]
- Lei, Q.Y.; Lü, S.H. Molecular ecological responses of dinoflagellate, Karenia mikimotoi to environmental nitrate stress. Mar. Pollut. Bull. 2011, 62, 2692–2699. [Google Scholar] [CrossRef]
- Duan, Y.; Xiong, D.; Wang, Y.; Zhang, Z.; Li, H.; Dong, H.; Zhang, J. Toxicological effects of microplastics in Litopenaeus vannamei as indicated by an integrated microbiome, proteomic and metabolomic approach. Sci. Total Environ. 2021, 761, 143311. [Google Scholar] [CrossRef]
- Barros, D.; Pradhan, A.; Mendes, V.M.; Manadas, B.; Santos, P.M.; Pascoal, C.; Cássio, F. Proteomics and antioxidant enzymes reveal different mechanisms of toxicity induced by ionic and nanoparticulate silver in bacteria. Environ. Sci. Nano 2019, 6, 1207–1218. [Google Scholar] [CrossRef]
- Saleh, S.; Staes, A.; Deborggraeve, S.; Gevaert, K. Targeted Proteomics for Studying Pathogenic Bacteria. Proteomics 2019, 19, 1800435. [Google Scholar] [CrossRef]
- Sangsuriya, P.; Huang, J.Y.; Chu, Y.F.; Phiwsaiya, K.; Leekitcharoenphon, P.; Meemetta, W.; Senapin, S.; Huang, W.P.; Withyachumnarnkul, B.; Flegel, T.W.; et al. Construction and application of a protein interaction map for white spot syndrome virus (WSSV). Mol. Cell. Proteomics 2014, 13, 269–282. [Google Scholar] [CrossRef]
- Chan, L.L.; Sit, W.H.; Lam, P.K.S.; Hsieh, D.P.H.; Hodgkiss, I.J.; Wan, J.M.F.; Ho, A.Y.T.; Choi, N.M.C.; Wang, D.Z.; Dudgeon, D. Identification and characterization of a “biomarker of toxicity” from the proteome of the paralytic shellfish toxin-producing dinoflagellate Alexandrium tamarense (Dinophyceae). Proteomics 2006, 6, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Elbehiry, A.; Marzouk, E.; Abdeen, E.; Al-Dubaib, M.; Alsayeqh, A.; Ibrahem, M.; Hamada, M.; Alenzi, A.; Moussa, I.; Hemeg, H.A. Proteomic characterization and discrimination of Aeromonas species recovered from meat and water samples with a spotlight on the antimicrobial resistance of Aeromonas hydrophila. Microbiologyopen 2019, 8, e782. [Google Scholar] [CrossRef]
- Zhu, W.; Zhou, S.; Chu, W. Comparative proteomic analysis of sensitive and multi-drug resistant Aeromonas hydrophila isolated from diseased fish. Microb. Pathog. 2020, 139, 103930. [Google Scholar] [CrossRef]
- Sun, L.; Chen, H.; Lin, W.; Lin, X. Quantitative proteomic analysis of Edwardsiella tarda in response to oxytetracycline stress in biofilm. J. Proteom. 2017, 150, 141–148. [Google Scholar] [CrossRef]
- Peng, B.; Li, H.; Peng, X. Proteomics approach to understand bacterial antibiotic resistance strategies. Expert Rev. Proteom. 2019, 16, 829–839. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Gallardo, J.M. Rapid direct detection of the major fish allergen, parvalbumin, by selected MS/MS ion monitoring mass spectrometry. J. Proteom. 2012, 75, 3211–3220. [Google Scholar] [CrossRef]
- Rahman, A.M.A.; Kamath, S.; Lopata, A.L.; Helleur, R.J. Analysis of the allergenic proteins in black tiger prawn (Penaeus monodon) and characterization of the major allergen tropomyosin using mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 1457–1466. [Google Scholar] [CrossRef]
- Tomanek, L. Proteomics to study adaptations in marine organisms to environmental stress. J. Proteom. 2014, 105, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Chia, M.A.; Lorenzi, A.S.; Ameh, I.; Dauda, S.; Cordeiro-Araújo, M.K.; Agee, J.T.; Okpanachi, I.Y.; Adesalu, A.T. Susceptibility of phytoplankton to the increasing presence of active pharmaceutical ingredients (APIs) in the aquatic environment: A review. Aquat. Toxicol. 2021, 234, 105809. [Google Scholar] [CrossRef] [PubMed]
- Picotti, P.; Aebersold, R. Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 2012, 9, 555–566. [Google Scholar] [CrossRef] [PubMed]
- De Marchi, L.; Coppola, F.; Soares, A.M.V.M.; Pretti, C.; Monserrat, J.M.; della Torre, C.; Freitas, R. Engineered nanomaterials: From their properties and applications, to their toxicity towards marine bivalves in a changing environment. Environ. Res. 2019, 178, 108683. [Google Scholar] [CrossRef]
- Trapp, J.; Armengaud, J.; Salvador, A.; Chaumot, A.; Geffard, O. Next-generation proteomics: Toward customized biomarkers for environmental biomonitoring. Environ. Sci. Technol. 2014, 48, 13560–13572. [Google Scholar] [CrossRef]
- López-Pedrouso, M.; Varela, Z.; Franco, D.; Fernández, J.A.; Aboal, J.R. Can proteomics contribute to biomonitoring of aquatic pollution? A critical review. Environ. Pollut. 2020, 267, 115473. [Google Scholar] [CrossRef]
- Brandão-Dias, P.F.P.; Rosi, E.J.; Shogren, A.J.; Tank, J.L.; Fischer, D.T.; Egan, S.P. Fate of Environmental Proteins (eProteins) from Genetically Engineered Crops in Streams is Controlled by Water pH and Ecosystem Metabolism. Environ. Sci. Technol. 2021, 55, 4688–4697. [Google Scholar] [CrossRef]
- Casabianca, S.; Bellingeri, A.; Capellacci, S.; Sbrana, A.; Russo, T.; Corsi, I.; Penna, A. Ecological implications beyond the ecotoxicity of plastic debris on marine phytoplankton assemblage structure and functioning. Environ. Pollut. 2021, 290, 118101. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, Y.; Shi, Z.; Li, Z.; Liang, X. Ecotoxicoproteomic assessment of microplastics and plastic additives in aquatic organisms: A review. Comp. Biochem. Physiol.—Part D Genom. Proteom. 2020, 36, 100713. [Google Scholar] [CrossRef]
- Khrunyk, Y.; Lach, S.; Petrenko, I.; Ehrlich, H. Progress in Modern Marine Biomaterials Research. Mar. Drugs 2020, 18, 589. [Google Scholar] [CrossRef]
- Ren, X.B.; Dang, Y.R.; Liu, S.S.; Huang, K.X.; Qin, Q.L.; Chen, X.L. Identification and Characterization of Three Chitinases with Potential in Direct Conversion of Crystalline Chitin into N, N′-Diacetylchitobiose. Mar. Drugs 2022, 20, 165. [Google Scholar] [CrossRef] [PubMed]
- Baghban, N.; Khoradmehr, A.; Nabipour, I.; Tamadon, A.; Ullah, M. The potential of marine-based gold nanomaterials in cancer therapy: A mini-review. Gold Bull. 2022, 55, 53–63. [Google Scholar] [CrossRef]
- Liu, S.; Lau, C.S.; Liang, K.; Wen, F.; Teoh, S.H. Marine collagen scaffolds in tissue engineering. Curr. Opin. Biotechnol. 2022, 74, 92–103. [Google Scholar] [CrossRef]
- Papon, N.; Copp, B.R.; Courdavault, V. Marine drugs: Biology, pipelines, current and future prospects for production. Biotechnol. Adv. 2022, 54, 107871. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Mandhare, A.; Bagalkote, V. Marine natural products as source of new drugs: An updated patent review (July 2018–July 2021). Expert Opin. Ther. Pat. 2022, 32, 317–363. [Google Scholar] [CrossRef]
- Barbarino, E.; Lourenço, S.O. An evaluation of methods for extraction and quantification of protein from marine macro- and microalgae. J. Appl. Phycol. 2005, 17, 447–460. [Google Scholar] [CrossRef]
- Knight, M.I.; Chambers, P.J. Problems associated with determining protein concentration: A comparison of techniques for protein estimations. Appl. Biochem. Biotechnol.—Part B Mol. Biotechnol. 2003, 23, 19–28. [Google Scholar] [CrossRef]
- Martyniuk, C.J.; Alvarez, S.; Denslow, N.D. DIGE and iTRAQ as biomarker discovery tools in aquatic toxicology. Ecotoxicol. Environ. Saf. 2012, 76, 3–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fröhlich, T.; Arnold, G.J.; Fritsch, R.; Mayr, T.; Laforsch, C. LC-MS/MS-based proteome profiling in Daphnia pulex and Daphnia longicephala: The Daphnia pulex genome database as a key for high throughput proteomics in Daphnia. BMC Genom. 2009, 10, 171. [Google Scholar] [CrossRef]
- Kwon, D.; Park, J.M.; Duong, V.A.; Hong, S.J.; Cho, B.K.; Lee, C.G.; Choi, H.K.; Kim, D.M.; Lee, H. Comparative proteomic profiling of marine and freshwater Synechocystis strains using liquid chromatography-tandem mass spectrometry. J. Mar. Sci. Eng. 2020, 8, 790. [Google Scholar] [CrossRef]
- Pappireddi, N.; Martin, L.; Wühr, M. A Review on Quantitative Multiplexed Proteomics. ChemBioChem 2019, 20, 1210–1224. [Google Scholar] [CrossRef] [PubMed]
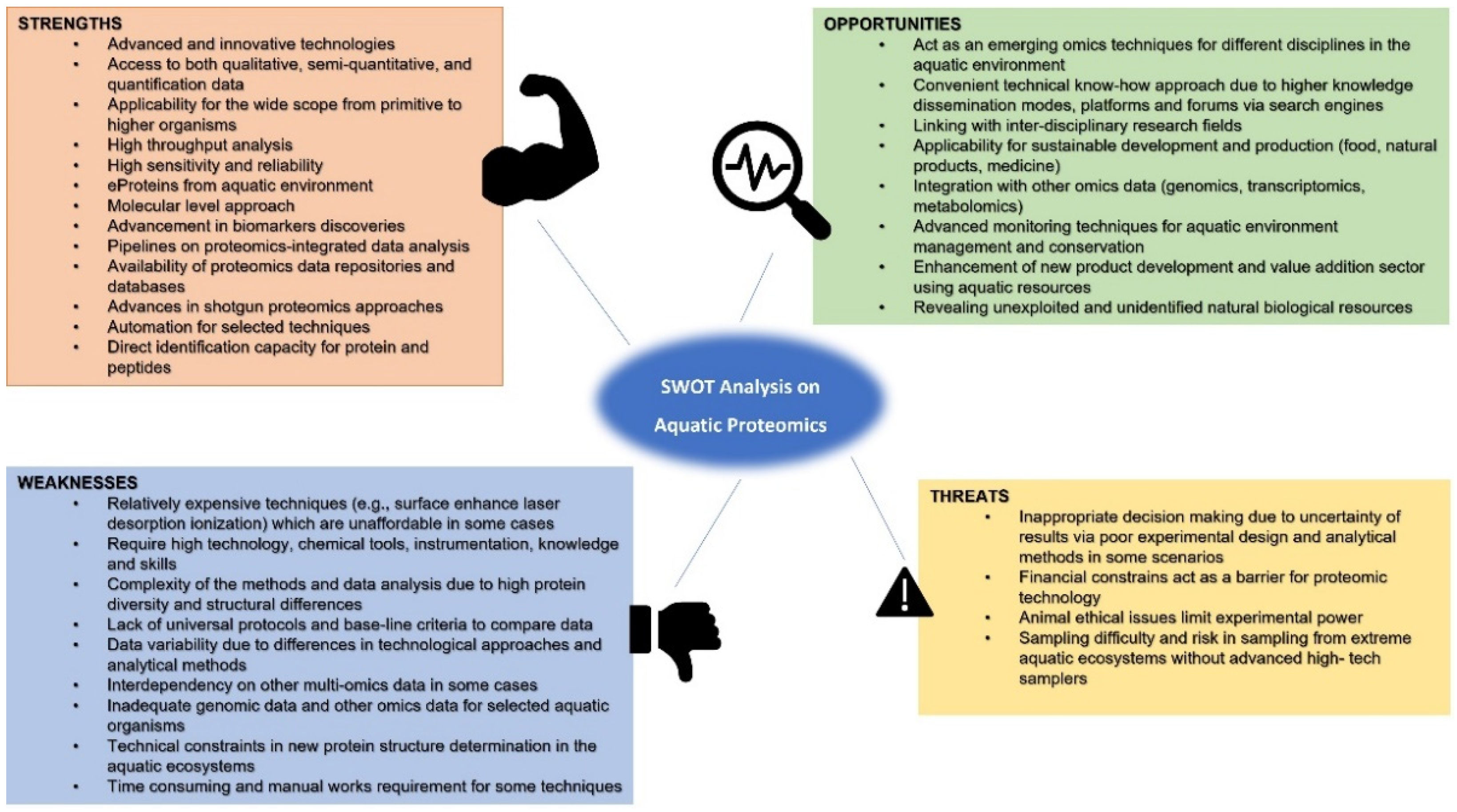
| Sector | Challenges | Recommendations |
|---|---|---|
| Food industry | Seafood safety and human health risks. Sustainable aquaculture development through modified feed, disease management, and pollution control. Quality of the aquatic product. Pathogens and biotoxins in aquatic food. Animal welfare and ethics. | Seafood allergies testing and ELISA assay development to check the quality of seafood with the application of proteomics. Enhancing aquatic feed quality and efficiency using alternative protein sources. Screening production with appropriate biomarkers as a diagnostic tool for infectious fish diseases in aquaculture farms. Use of macroalgae-based seaweed proteomics to identify novel aquaculture species. Sustainable aquatic, genetically modified food product development and conservation of edible aquatic animals using proteomic-based approaches. Application of advanced, high-throughput proteomic techniques for disease management and welfare monitoring. |
| Environmental monitoring | Monitoring pollutants and impact assessment. Conservation and management of aquatic life. Antibiotic resistance in the aquatic environment. Aquatic biodiversity assessment. Environmental toxicity. | Application of multiple biomarkers for aquatic toxicology studies. Species-specific proteomic database development for biomarker species. Developing global protein baseline data for the impacts of biological processes resulting from changes in the proteomes of aquatic organisms in response to environmental stresses. Explore unidentified novel species using proteomic data. Application of eProteins for biomonitoring and impact assessment. |
| Natural products | Novel natural product isolation and identification. Evaluation of biological activities. Drug development from natural aquatic products. Predicting protein domain architecture. Impact on biodiversity by harnessing. Nonanimal protein identification. | Estimation of biodiversity and interactions among organisms using proteomics in the study area prior to sampling. Extraction of minimum quantities from sensitive aquatic ecosystems as a conservation strategy. Implement bio or chemical synthesis processes. Develop and use high-tech equipment for underwater sampling. Use NMR spectroscopy to determine molecular structure. Seaweed proteomics for novel protein and peptide isolation. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajahin Gamage, N.T.; Miyashita, R.; Takahashi, K.; Asakawa, S.; Senevirathna, J.D.M. Proteomic Applications in Aquatic Environment Studies. Proteomes 2022, 10, 32. https://doi.org/10.3390/proteomes10030032
Gajahin Gamage NT, Miyashita R, Takahashi K, Asakawa S, Senevirathna JDM. Proteomic Applications in Aquatic Environment Studies. Proteomes. 2022; 10(3):32. https://doi.org/10.3390/proteomes10030032
Chicago/Turabian StyleGajahin Gamage, Nadeeka Thushari, Rina Miyashita, Kazutaka Takahashi, Shuichi Asakawa, and Jayan Duminda Mahesh Senevirathna. 2022. "Proteomic Applications in Aquatic Environment Studies" Proteomes 10, no. 3: 32. https://doi.org/10.3390/proteomes10030032
APA StyleGajahin Gamage, N. T., Miyashita, R., Takahashi, K., Asakawa, S., & Senevirathna, J. D. M. (2022). Proteomic Applications in Aquatic Environment Studies. Proteomes, 10(3), 32. https://doi.org/10.3390/proteomes10030032






