1. Introduction
Arrhythmia is a disorder characterized by irregular heart rhythms [
1]. Under normal conditions, the heart’s rhythm is regulated by electrical impulses generated by specialized cells within the heart [
2], and abnormalities in these electrical impulses can lead to irregular heart rates and rhythms. Many arrhythmias are benign and do not cause notable health issues; however, some can disrupt the blood flow, which can result in serious and potentially life-threatening conditions, e.g., stroke, myocardial infarction, heart failure, or sudden cardiac death [
2].
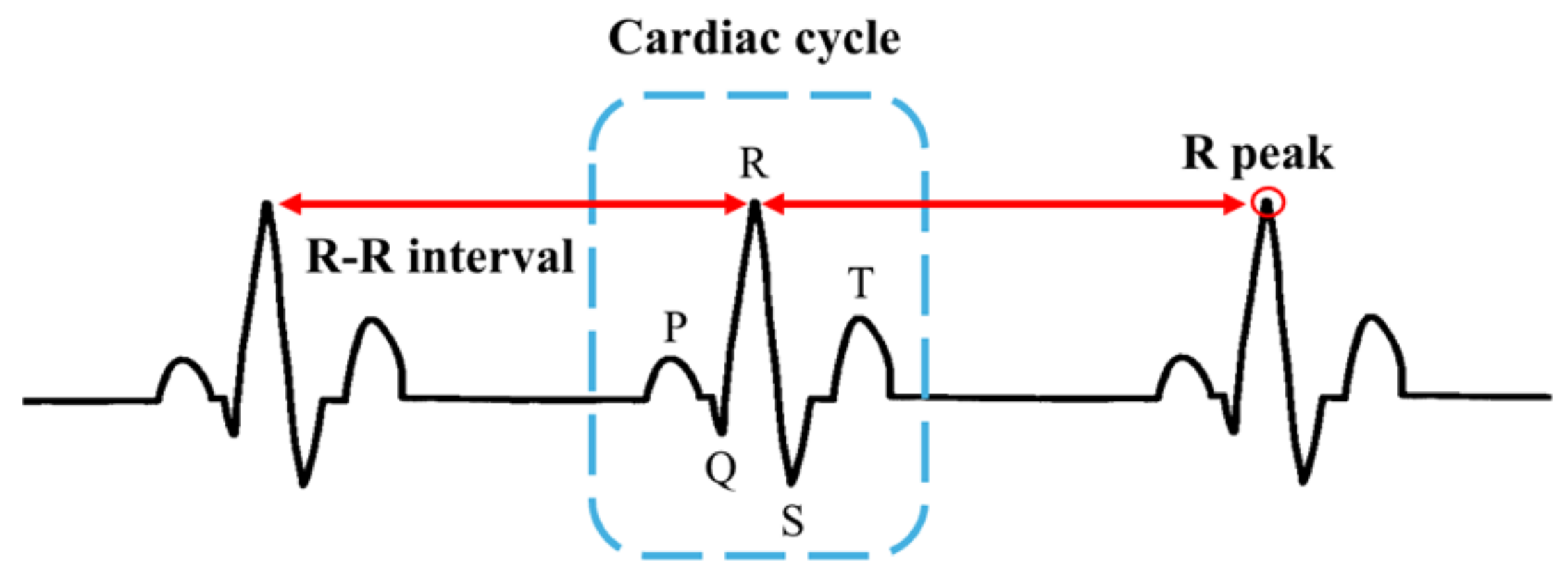
The primary tool employed to detect and diagnose arrhythmia is an electrocardiogram (ECG), which records the electrical activity of the heart [
3], and clinicians can identify various cardiovascular disorders by analyzing the electrical activity of the cardiac cycles captured in ECG data. Despite their utility, interpreting ECGs can be time consuming and complex owing to the notable variability in normal and pathological patterns [
4].
To address these challenges, machine learning models have been developed and employed for classifying various cardiovascular disorders, including arrhythmia. Numerous studies have demonstrated that machine learning models, particularly deep learning approaches, achieve high arrhythmia classification accuracy [
5,
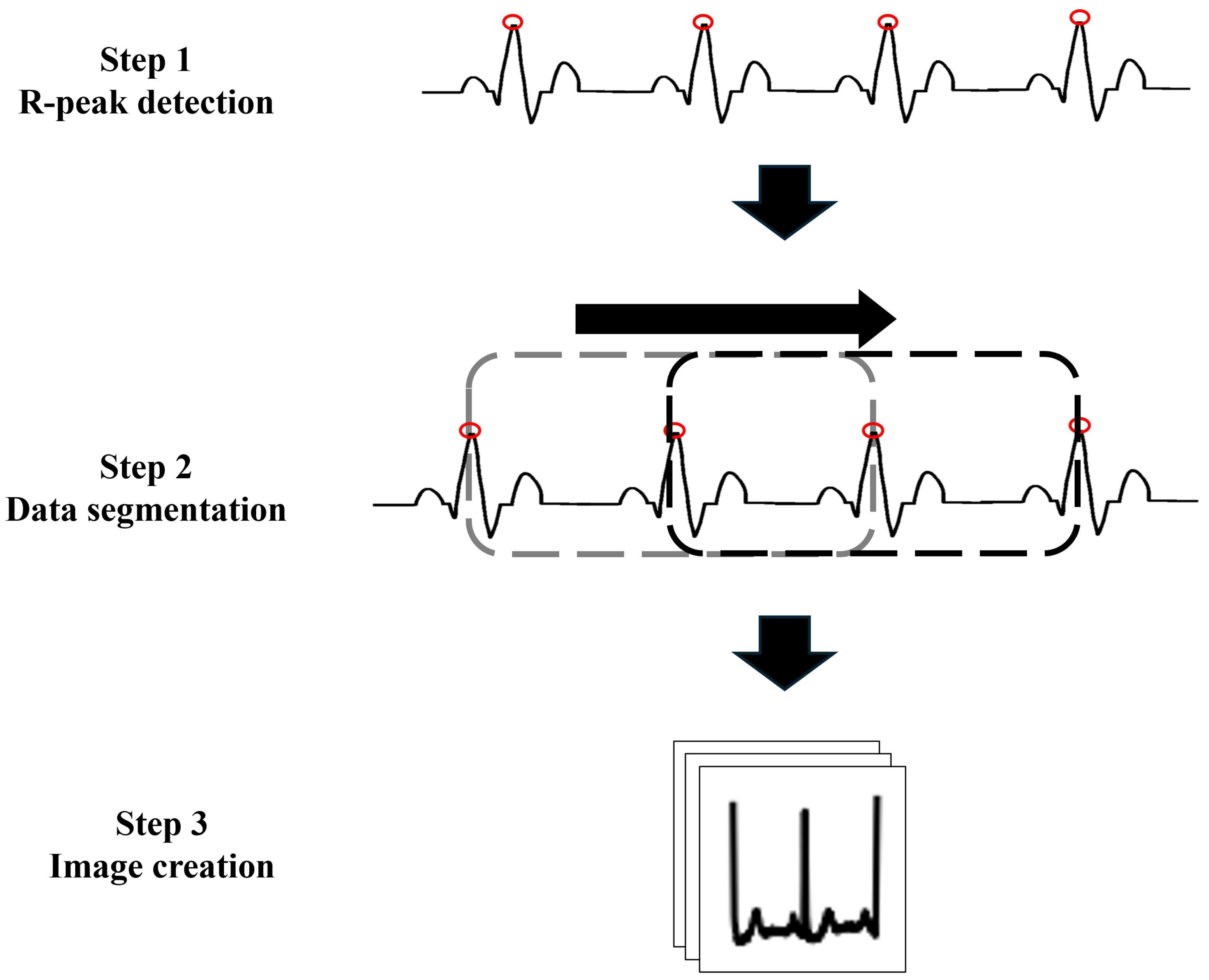
6]. Typically, ECG data are segmented into individual waveforms representing each cardiac cycle, which are used as the input for training models. However, abnormal activity may also occur in the intervals between cardiac cycles; thus, using only an individual cardiac cycle as an input feature may not capture all relevant features that appear between the cardiac cycles [
7].
Additionally, data characteristics are another factor that can affect the learning ability of the involved model. Data imbalance due to the rarity of certain types of disorder can negatively impact classification performance [
4]. Moreover, the ECG pattern of each patient is influenced by their demographic background [
8]. In cases of insufficient generalizability, the model’s classification performance is typically poor when evaluating unseen data from patients because each patient exhibits unique patterns.
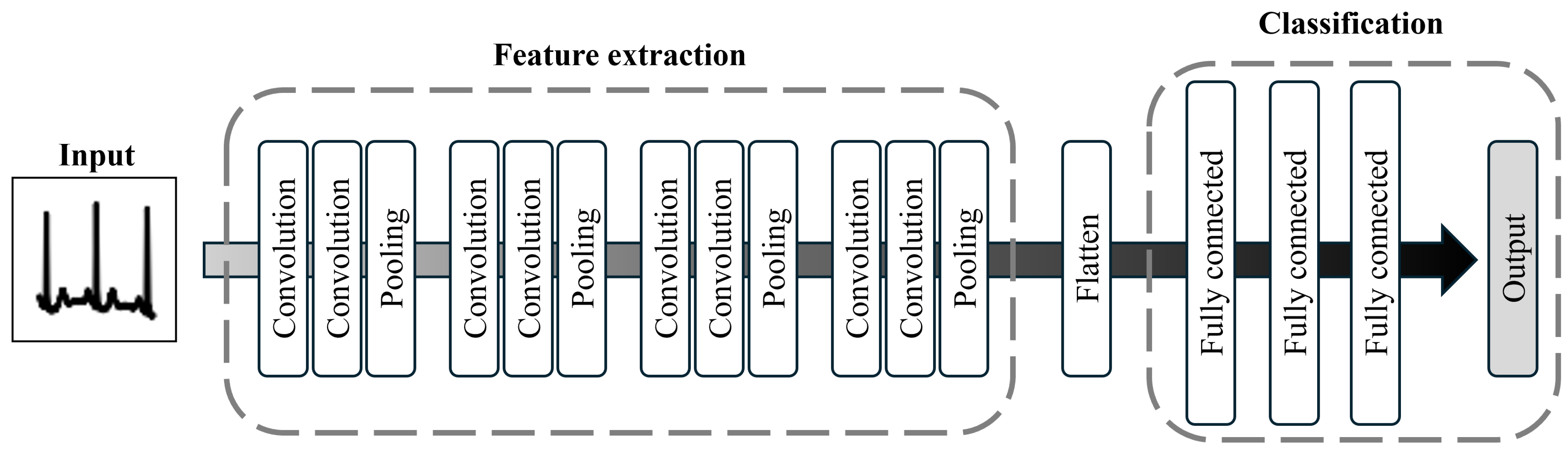
To address these limitations and enhance the performance of classification models, we develop a machine learning approach that addresses the challenges of input representation and data imbalance. Herein, we propose a convolutional neural network (CNN)-based arrhythmia classification model that incorporates two specialized modules to address these issues. The first module focuses on data preparation. Here, rather than using individual cardiac cycles, images of three consecutive cardiac cycles are utilized as the input to expand the learning frame. The second module addresses model training by introducing a focal loss (FL) function to assign notable importance to the minority class [
9]. These two innovations allow the model to capture abnormal patterns both within and between cardiac cycles, thereby improving the classification performance for the minority class.
To evaluate the proposed model, we assess its classification performance across eight arrhythmia classes using three metrics—accuracy, specificity, and sensitivity. In addition, we compare the performance of the proposed model with that of another CNN-based model that employs a different loss function, i.e., the categorical cross-entropy error (CCE).
The primary contributions of this study are summarized as follows. First, extending the learning frame with images that include multiple cardiac cycles enables the model to extract more features from the ECG data, which improves classification performance. Second, as discussed in a previous study [
5], utilizing images as input helps mitigate noise and trend effects in the ECG data. Transforming numerical time-series data into images reduces the model’s dependence on complex and time-consuming data preprocessing steps, which is advantageous in terms of realizing real-time classification. Third, the implementation of the FL function improves classification efficiency by up-weighting the minority class, enhancing the overall performance of the model in imbalanced-data scenarios.
The remainder of this paper is structured as follows.
Section 2 provides background information on relevant topics.
Section 3 describes the methodology of the proposed CNN model. The experimental results are presented in
Section 4, and the results are analyzed and discussed in detail in
Section 5, including a performance comparison with an existing method. Finally, the paper is concluded in
Section 6.
5. Discussion
5.1. Performance Comparison Between the CNN-CCE and CNN-FL Models
Herein, the CCE and FL loss functions were implemented in the construction of two CNN models for the classification of multicycle ECG images. The results obtained for the two CV settings revealed that these models demonstrated considerably different performances.
The first setting corresponded to 10-fold CV, which allowed data from the same patient to be included in both the training and test sets. Here, both the CNN-CCE and CNN-FL models demonstrated high performance, with scores over 96% for all evaluation metrics; however, the proposed CNN-FL model exhibited slightly better performance. As shown in
Table 4, the accuracy, specificity, and sensitivity values obtained by the proposed CNN-FL were 0.12%, 0.09%, and 0.16% higher than those of the CNN-CCE model, respectively. The confusion matrix showed similar results. The individual accuracy scores for most classes, especially for the extreme minority classes such as VFW and VEB, demonstrated that the proposed CNN-FL model outperformed the CNN-CCE model.
Then, leave-one-person-out CV was performed in the second experiment. Owing to the unique ECG patterns of each patient, it was difficult for the models to identify unseen data. Compared with setting 1 (10-fold CV), the classification performance of the models in this setting noticeably decreased. Nonetheless, the proposed CNN-FL model still outperformed the CNN-CCE model. According to the results, the specificity score of the proposed CNN-FL model was 2.83% less than that of the CNN-CCE model; however, the accuracy, sensitivity, and F1-score values obtained by the proposed CNN-FL model were 2.19%, 8.49%, and 5.33% higher than those of the CNN-CCE model, respectively. With the higher sensitivity values, the proposed CNN-FL model demonstrated better performance in terms of avoiding false negative predictions. Furthermore, the better F1-score indicated improved ability to handle imbalanced data. In addition, the proposed CNN-FL model identified the minority class, i.e., LBBB, more accurately than the CNN-CCE model (
Table 9).
According to the evaluation results, both loss functions performed effectively in terms of identifying different types of arrhythmia; however, after carefully considering the handling of the imbalance problem, the results indicate that the proposed CNN-FL model demonstrated better performance. As mentioned previously, the FL function prioritizes classes based on the difficulty of classifying them. In other words, less difficult classes are assigned lower weight, and this property may have contributed to the more accurate classification of the minority classes.
In addition, the results obtained for setting 2 proved that the proposed CNN-FL model was useful in terms of recognizing and identifying the unseen data correctly. Thus, it can be concluded that using the FL loss function improved the correctness and robustness of the CNN model for the arrhythmia classification task.
5.2. Performance Comparison Between the Proposed CNN and Existing Methods
As discussed in the previous section, the proposed CNN-FL model outperformed the CNN-CCE model. Herein, the proposed CNN-FL model is compared with other existing methods.
For setting 1,
Table 10 presents the 10-fold CV results from seven existing methods. As can be seen, the performance of the proposed CNN-FL model is comparable to that of the compared methods. In addition, when compared with existing methods that included the same number of classes, the sensitivity and specificity values obtained by the proposed CNN-FL model were the highest, and the proposed model performed better when classifying the minority class. As shown in
Table 11, the accuracy value obtained for the APC class using the proposed model was higher than those obtained in previous research [
5,
41,
42].
Table 12 compares the results of the approach involving leave-one-person-out CV with those of two existing studies that conducted similar experiments. As shown, the accuracy and specificity values obtained by the proposed CNN-FL model did not exceed those of the compared methods. However, the sensitivity score obtained by the proposed CNN-FL model was substantially higher than those of the compared methods. It can be concluded that the proposed CNN-FL model was more effective in terms of preventing false negatives than the existing methods. Nevertheless, at the same time, CNN-FL showed a higher rate of false positive predictions.
Even without data preprocessing, the proposed CNN-FL model was able to identify arrhythmia comparably to existing methods, which can be attributed to two main reasons.
The first reason pertains to data preparation. In the current study, three R-peak ECG images were utilized as the input to capture the shape of the waveform and the intervals between the R-peaks. Thus, the model could capture and learn more characteristics of each arrhythmia. More detail in the ECG image could lead to more accurate disease classification.
The second reason involves the handling of imbalanced data. Herein, two loss functions, i.e., CCE and FL, were compared to investigate which loss function handled this problem more effectively. As discussed previously, with the FL function, more weight is assigned to the minority class than the majority class. This allowed the proposed CNN-FL model to prioritize the class with fewer data, thereby enabling more accurate predictions.
5.3. Challenges and Future Study
Based on the evaluation and comparison results, the proposed CNN-FL model achieved accuracy comparable to or exceeding that of existing methods in the arrhythmia classification task, despite utilizing only ECG images without complex data preprocessing. However, as demonstrated by the results, the values of some metrics decreased, and after investigation of these issues, the causes were identified.
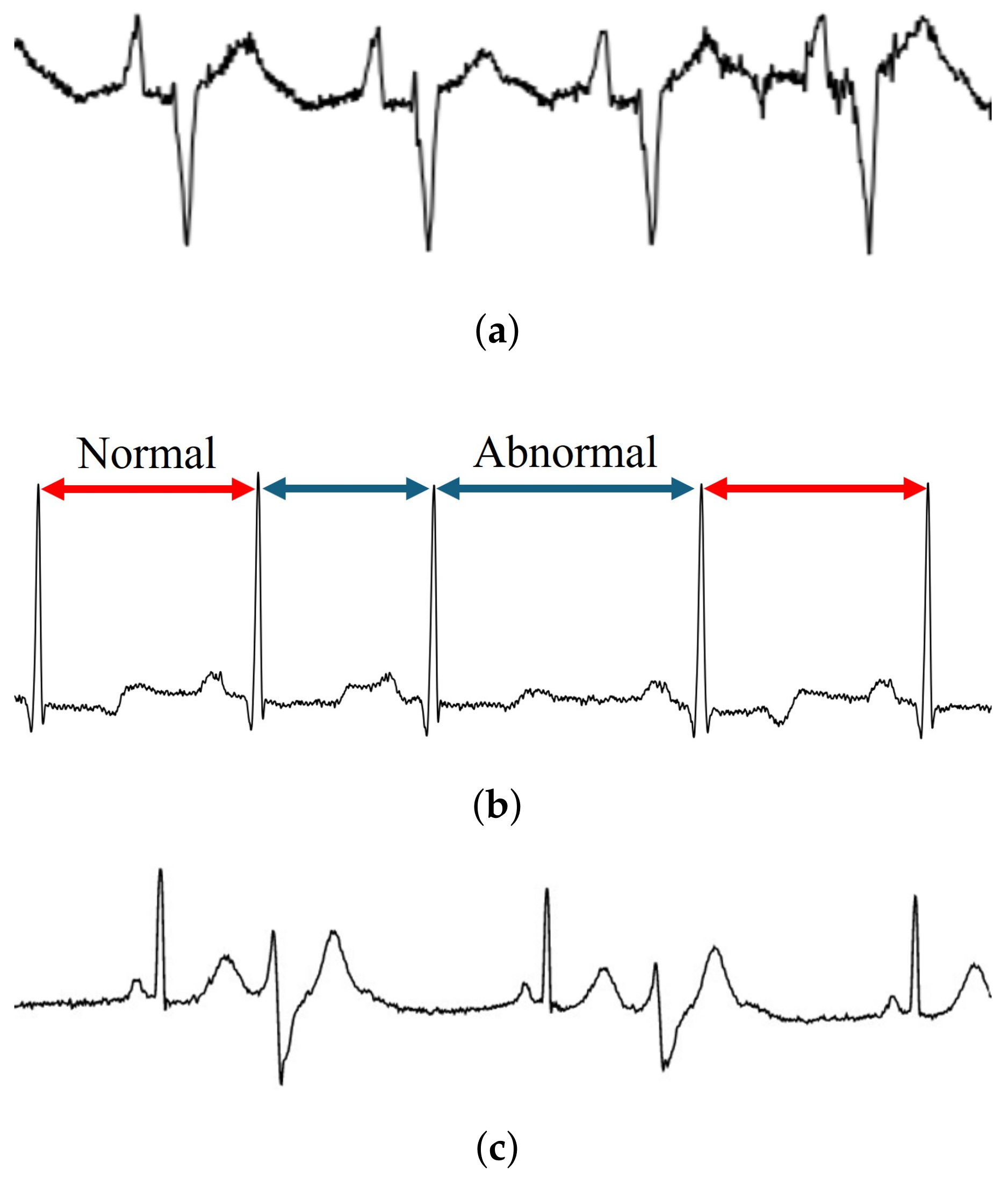
As mentioned previously, ECG images were employed as the model input herein. According to a previous study [
5], transforming signal data to images can reduce the effects of noise and trends; however, during the evaluation, several images were misidentified. Upon further investigation, these misidentified images could be categorized into two types, i.e., strong noise and strong trend (
Figure 6). This suggests that image transformation may not eliminate these effects completely. Thus, the CNN must be improved to prevent the effects from noise and trends. As suggested in previous research [
48], enhancing the denoising and detrending abilities of CNNs can be realized by incorporating residual blocks and attention mechanisms. To solve this problem, implementing these components should be considered in future study.
In addition, to implement the model for practical application, various factors must be considered. As previously mentioned, ECG patterns exhibit high variance: thus, the model should be generalized to handle unseen data from new patients. From the leave-one-person-out results, it can be seen that the proposed CNN-FL model outperformed the compared models; however, the scores were less than 90%. With a small dataset like the MIT-BIH Arrhythmia dataset, the model may not handle the variance in the patterns effectively, thereby leading to poor performance and generalization issues. To address this, future studies could incorporate larger and more diverse datasets, along with data augmentation techniques. These steps could enhance the model’s ability to generalize to new patients and reduce the impact of dataset limitations.
Another point to consider is resource consumption. Deep learning models receive considerable attention owing to their high performance; however, processing such models requires huge computational resources. In this study, a high-performance computer was used to develop and evaluate the proposed CNN-FL model. Thus, it may be challenging to implement the proposed CNN-FL model on small devices for real-time classification tasks. To address this limitation, the size and complexity of the model should be reduced using model compression techniques.
Regarding investigation and model performance evaluation, future work should address several points to improve the effectiveness of the classification model for arrhythmia. Herein, the proposed model was evaluated using a single dataset, MIT-BIH Arrhythmia; however, future work should consider testing the model on additional datasets to improve generalizability. Exploring the possibility of using ECG data in its native format as input could provide deeper insights and improve the model’s performance. Moreover, an ablation study is needed to confirm the contributions of image usage and the FL function as well as to evaluate how these factors influence the model’s performance. Additionally, using techniques such as Grad-CAM [
49] could offer more interpretability in understanding the model’s decision-making process.
Finally, to improve model performance and reduce the computational burden, the use of transfer learning could be explored by leveraging pretrained models, particularly in scenarios with limited data. This approach may help improve the model’s accuracy while maintaining efficiency.
6. Conclusions
Herein, a CNN-based model was proposed to classify different types of arrhythmia. Based on the evaluation experiments, the primary conclusions are summarized as follows.
Using ECG images containing three R-peaks, the proposed CNN-FL model can capture the waveform shape and R-peak intervals, thereby effectively learning ECG abnormalities associated with arrhythmia.
The FL function enables effective learning of arrhythmia ECGs from minority classes.
Compared with those corresponding to existing methods, the sensitivity of the proposed CNN-FL model, i.e., the model’s arrhythmia detection capability, is substantially higher.
Thus, we expect that the proposed CNN-FL model will be an effective tool for discriminating different types of arrhythmia in practical application. In the future, the architecture of the model will be modified to enhance its ability to prevent noise and trend effects. In addition, effective model compression techniques should be considered to implement the proposed model for practical application in real-world situations.