A Comparison of Monte Carlo-Based and PINN Parameter Estimation Methods for Malware Identification in IoT Networks
Abstract
:1. Introduction
- Our study provides a detailed and rigorous comparative evaluation of two well known approaches to parameter estimation.
- We identify the benefits of both approaches as well as the time required to perform parameter estimation. This information can help cybersecurity professionals to make informed decisions and develop more efficient strategies to protect IoT networks.
- We hope to inspire other researchers to further explore the intersection of epidemiology, cybersecurity, and data science by highlighting the benefits and limitations of each approach.
2. Materials and Methods
2.1. Synthetic Data
2.2. Data Generation and Mathematical Models
2.3. Propagation Model Identification Methodology
2.3.1. Monte Carlo Method
| Algorithm 1: Monte Carlo parameter estimation with MSE loss function |
 |
| Algorithm 2: Monte Carlo parameter estimation with log square loss function |
 |
2.3.2. Physics-Informed Neural Networks
3. Results and Discussion
3.1. Experimental Setup
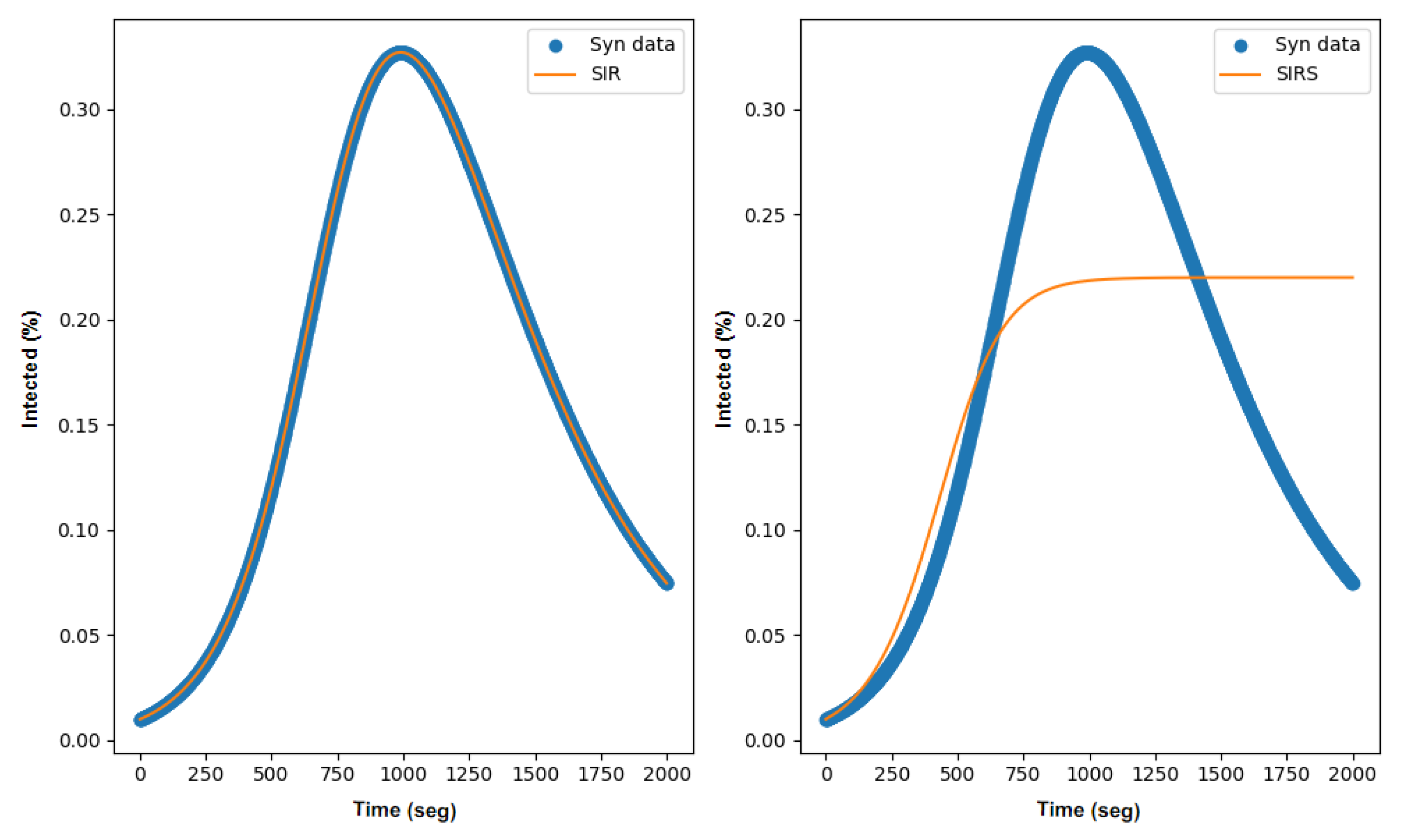
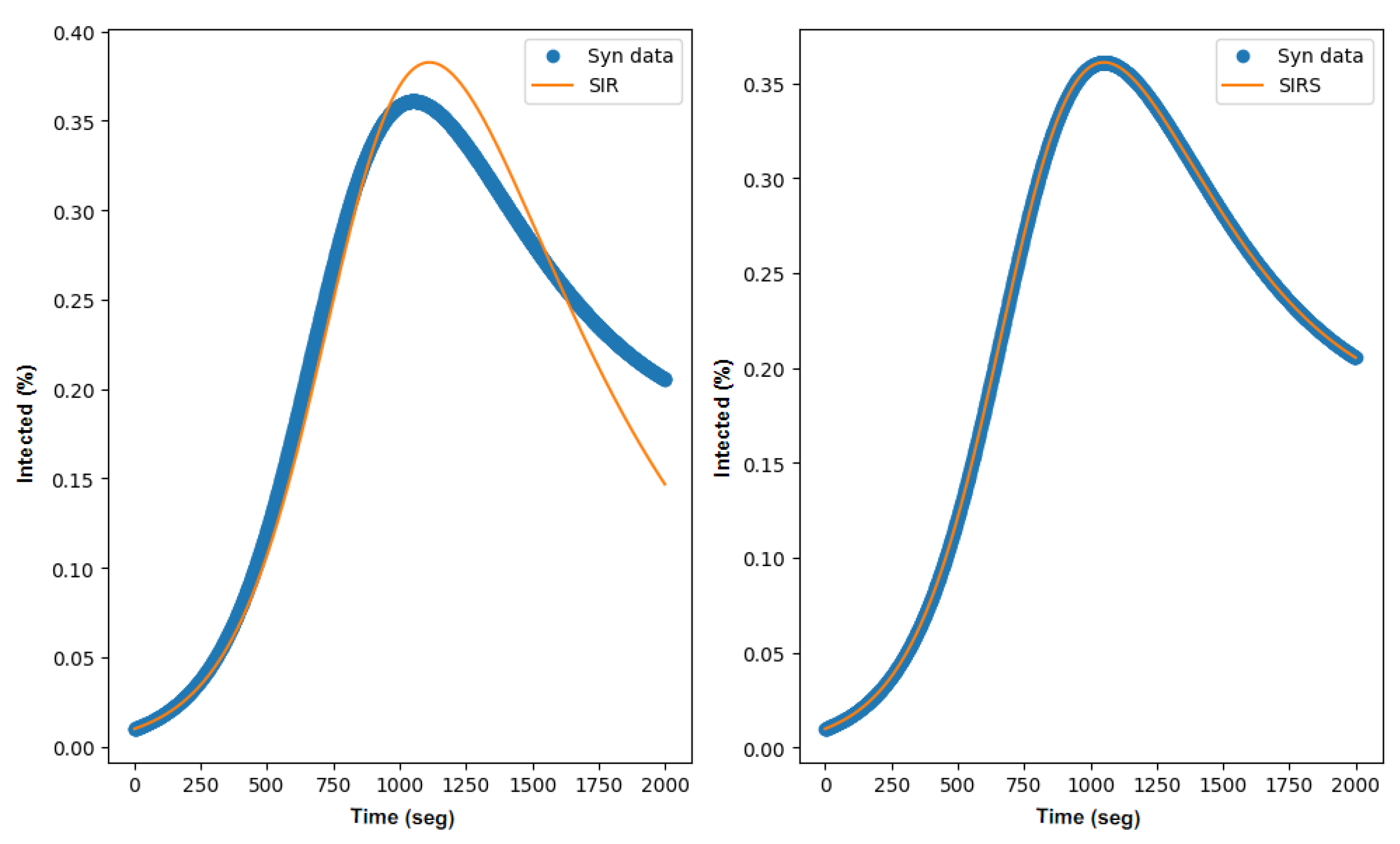
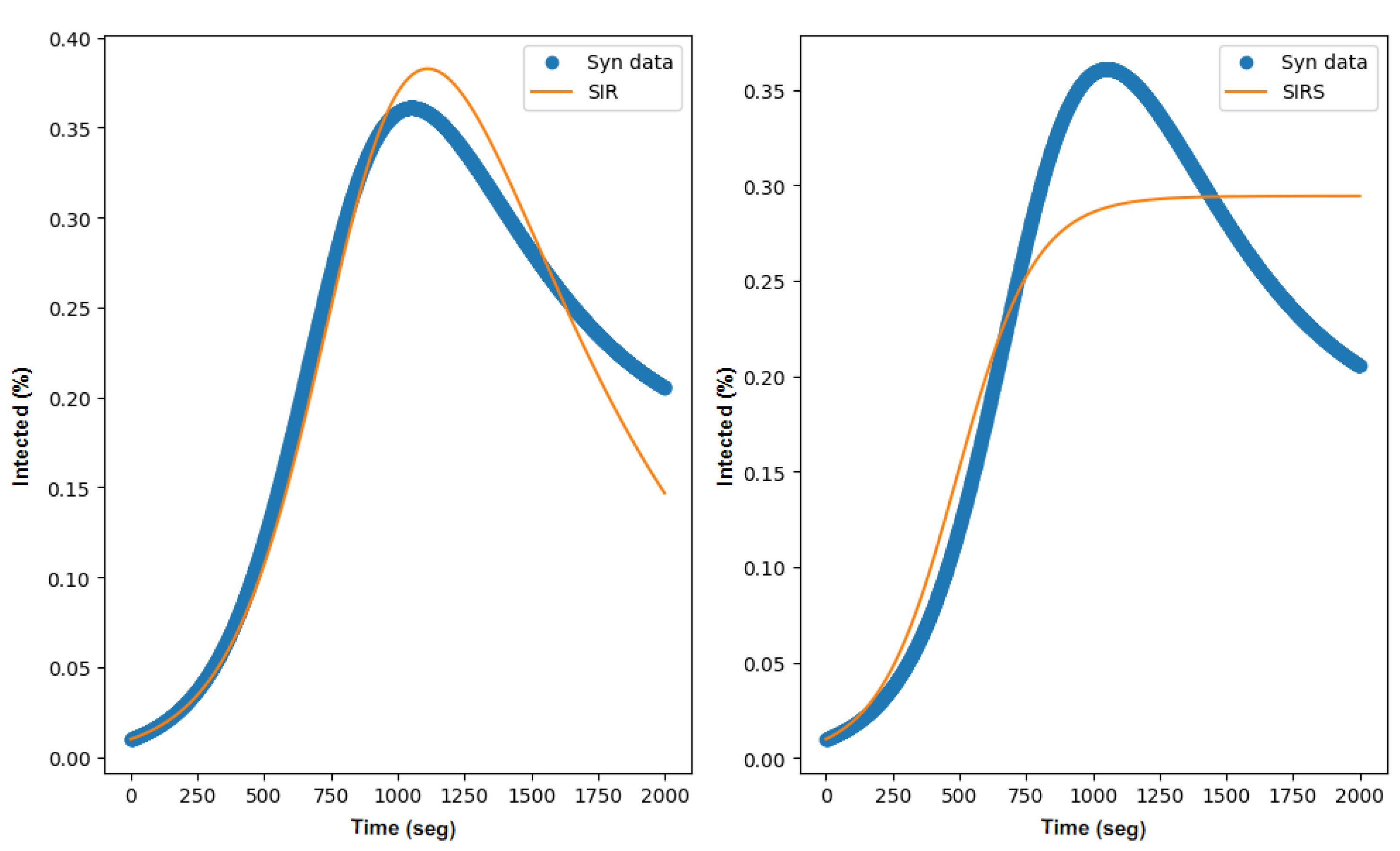
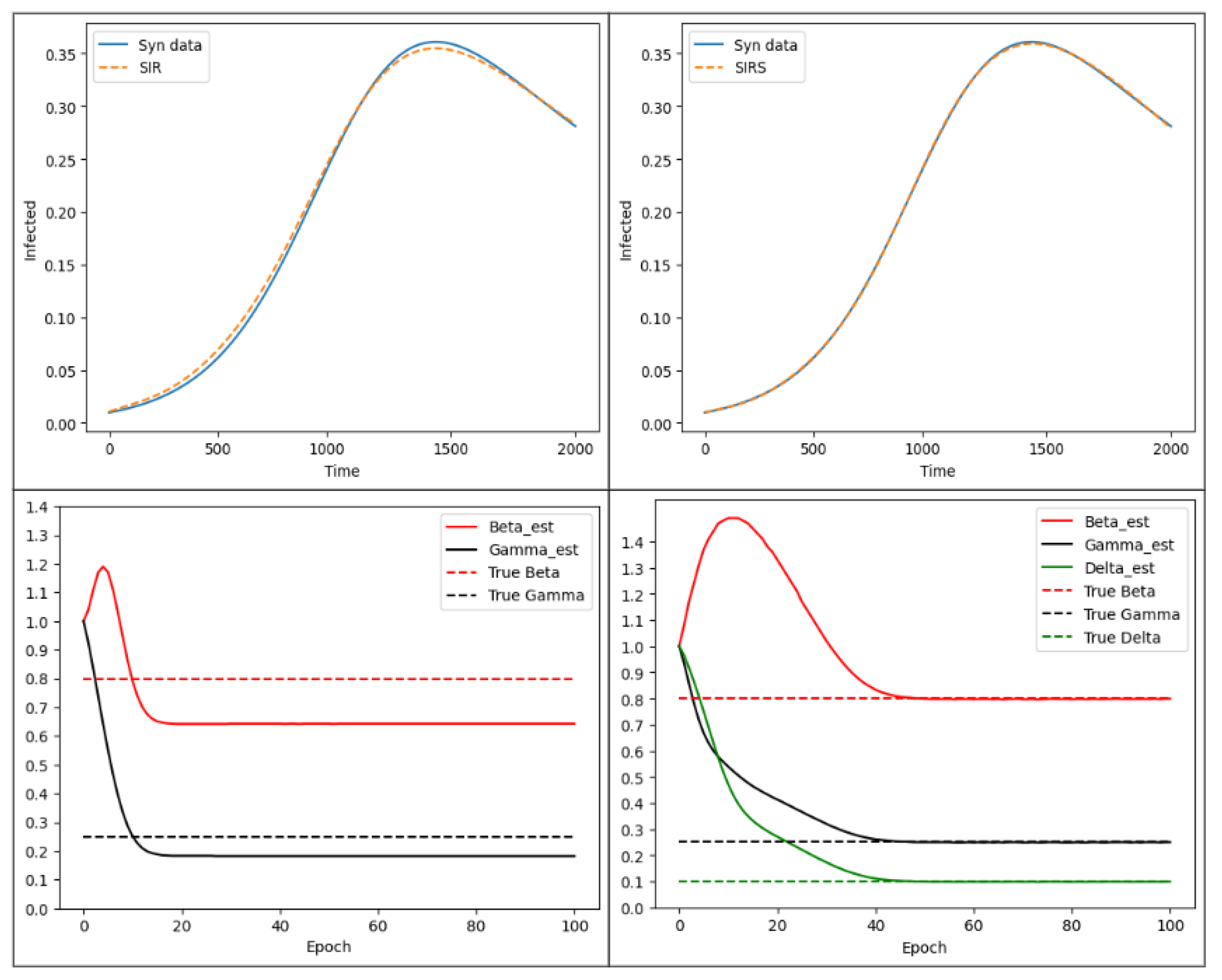
3.2. SIR Parameter Estimation
3.3. SIRS Parameter Estimation
3.4. Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Khan, J.A.; Chowdhury, M.M. Security analysis of 5g network. In Proceedings of the 2021 IEEE International Conference on Electro Information Technology (EIT), Mt. Pleasant, NI, USA, 14–15 May 2021; pp. 001–006. [Google Scholar]
- Wu, H.; Han, H.; Wang, X.; Sun, S. Research on artificial intelligence enhancing internet of things security: A survey. IEEE Access 2020, 8, 153826–153848. [Google Scholar] [CrossRef]
- Sadhu, P.K.; Yanambaka, V.P.; Abdelgawad, A. Internet of things: Security and solutions survey. Sensors 2022, 22, 7433. [Google Scholar] [CrossRef]
- Clim, A.; Toma, A.; Zota, R.D.; Constantinescu, R. The Need for Cybersecurity in Industrial Revolution and Smart Cities. Sensors 2022, 23, 120. [Google Scholar] [CrossRef]
- Aslan, Ö.; Aktuğ, S.S.; Ozkan-Okay, M.; Yilmaz, A.A.; Akin, E. A comprehensive review of cyber security vulnerabilities, threats, attacks, and solutions. Electronics 2023, 12, 1333. [Google Scholar] [CrossRef]
- Mittal, M.; Kumar, K.; Behal, S. Deep learning approaches for detecting DDoS attacks: A systematic review. Soft Comput. 2023, 27, 13039–13075. [Google Scholar] [CrossRef]
- Victoire, T.A.; Vasuki, M.; Karunamurthy, A.; Soundarya, D.; Sarumathi, S. A Survey on Cyber Security Threats and its Impact on Society. Int. J. Res. Eng. Sci. Manag. 2023, 6, 146–152. [Google Scholar]
- del Rey, A.M.; Vara, R.C.; González, S.R. A computational propagation model for malware based on the SIR classic model. Neurocomputing 2022, 484, 161–171. [Google Scholar] [CrossRef]
- Furtado, P. Epidemiology SIR with regression, arima, and Prophet in forecasting COVID-19. Eng. Proc. 2021, 5, 52. [Google Scholar]
- Wong, W.; Juwono, F.H. Estimating effective reproduction number for sir compartmental model: A stochastic evolutionary approach. J. Soc. Comput. 2022, 3, 182–189. [Google Scholar] [CrossRef]
- Cantó, B.; Coll, C.; Sánchez, E. Estimation of parameters in a structured SIR model. Adv. Differ. Equ. 2017, 2017, 33. [Google Scholar] [CrossRef]
- Marinov, T.T.; Marinova, R.S.; Omojola, J.; Jackson, M. Inverse problem for coefficient identification in SIR epidemic models. Comput. Math. Appl. 2014, 67, 2218–2227. [Google Scholar] [CrossRef]
- Zang, W.; Zhang, P.; Zhou, C.; Guo, L. Locating multiple sources in social networks under the SIR model: A divide-and-conquer approach. J. Comput. Sci. 2015, 10, 278–287. [Google Scholar] [CrossRef]
- Piazzola, C.; Tamellini, L.; Tempone, R. A note on tools for prediction under uncertainty and identifiability of SIR-like dynamical systems for epidemiology. Math. Biosci. 2021, 332, 108514. [Google Scholar] [CrossRef] [PubMed]
- Taddy, M.A.; Lee, H.K.; Sansó, B. Fast inference for statistical inverse problems. Inverse Probl. 2009, 25, 085001. [Google Scholar] [CrossRef]
- da Silva, W.B.; Dutra, J.C.; Knupp, D.C.; Abreu, L.A.; Silva Neto, A.J. Estimation of timewise varying boundary heat flux via Bayesian filters and Markov Chain Monte Carlo method. In Computational Intelligence in Emerging Technologies for Engineering Applications; Springer: Cham, Switzerland, 2020; pp. 137–153. [Google Scholar]
- Schiassi, E.; De Florio, M.; D’Ambrosio, A.; Mortari, D.; Furfaro, R. Physics-informed neural networks and functional interpolation for data-driven parameters discovery of epidemiological compartmental models. Mathematics 2021, 9, 2069. [Google Scholar] [CrossRef]
- Grimm, V.; Heinlein, A.; Klawonn, A.; Lanser, M.; Weber, J. Estimating the time-dependent contact rate of SIR and SEIR models in mathematical epidemiology using physics-informed neural networks. Electron. Trans. Numer. Anal. 2022, 56, 1–27. [Google Scholar] [CrossRef]
- Ferrández, M.R.; Ivorra, B.; Redondo, J.L.; Ramos del Olmo, Á.M.; Ortigosa, P.M. A multi-objective approach to estimate parameters of compartmental epidemiological models. Application to Ebola Virus Disease epidemics. Ene 2021, 12, 42. [Google Scholar]
- Shandilya, S.K.; Upadhyay, S.; Kumar, A.; Nagar, A.K. AI-assisted Computer Network Operations testbed for Nature-Inspired Cyber Security based adaptive defense simulation and analysis. Future Gener. Comput. Syst. 2022, 127, 297–308. [Google Scholar] [CrossRef]
- Barlas, Y. Formal aspects of model validity and validation in system dynamics. Syst. Dyn. Rev. J. Syst. Dyn. Soc. 1996, 12, 183–210. [Google Scholar] [CrossRef]
- Lee, G.; Kim, W.; Oh, H.; Youn, B.D.; Kim, N.H. Review of statistical model calibration and validation—from the perspective of uncertainty structures. Struct. Multidiscip. Optim. 2019, 60, 1619–1644. [Google Scholar] [CrossRef]
- Savage, L.J. The Foundations of Statistics; Courier Corporation: New York, NY, USA, 1972. [Google Scholar]
- Bar-Yam, Y. Dynamics of Complex Systems; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Mingers, J. A critique of statistical modelling in management science from a critical realist perspective: Its role within multimethodology. J. Oper. Res. Soc. 2006, 57, 202–219. [Google Scholar] [CrossRef]
- Kaniadakis, G.; Baldi, M.M.; Deisboeck, T.S.; Grisolia, G.; Hristopulos, D.T.; Scarfone, A.M.; Sparavigna, A.; Wada, T.; Lucia, U. The κ-statistics approach to epidemiology. Sci. Rep. 2020, 10, 19949. [Google Scholar] [CrossRef] [PubMed]
- Andrade, J.; Duggan, J. An evaluation of Hamiltonian Monte Carlo performance to calibrate age-structured compartmental SEIR models to incidence data. Epidemics 2020, 33, 100415. [Google Scholar] [CrossRef] [PubMed]
- Hattaf, K.; Yousfi, N.; Tridane, A. Mathematical analysis of a virus dynamics model with general incidence rate and cure rate. Nonlinear Anal. Real World Appl. 2012, 13, 1866–1872. [Google Scholar] [CrossRef]
- Miao, H.; Xia, X.; Perelson, A.S.; Wu, H. On identifiability of nonlinear ODE models and applications in viral dynamics. SIAM Rev. 2011, 53, 3–39. [Google Scholar] [CrossRef]
- Figueira, A.; Vaz, B. Survey on synthetic data generation, evaluation methods and GANs. Mathematics 2022, 10, 2733. [Google Scholar] [CrossRef]
- Diekmann, O.; Heesterbeek, J.A.P. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation; John Wiley & Sons: Hoboken, NJ, USA, 2000; Volume 5. [Google Scholar]
- Brauer, F.; Van den Driessche, P.; Wu, J.; Allen, L.J. Mathematical Epidemiology; Springer: Berlin/Heidelberg, Germany, 2008; Volume 1945. [Google Scholar]
- Kwok, K.O.; Tang, A.; Wei, V.W.; Park, W.H.; Yeoh, E.K.; Riley, S. Epidemic models of contact tracing: Systematic review of transmission studies of severe acute respiratory syndrome and middle east respiratory syndrome. Comput. Struct. Biotechnol. J. 2019, 17, 186–194. [Google Scholar] [CrossRef]
- del Rey, A.M. Mathematical modeling of the propagation of malware: A review. Secur. Commun. Netw. 2015, 8, 2561–2579. [Google Scholar] [CrossRef]
- Reis, W.P.N.d.; Silva, G.J.d.; Junior, O.M.; Vivaldini, K.C.T. An extended analysis on tuning the parameters of Adaptive Monte Carlo Localization ROS package in an automated guided vehicle. Int. J. Adv. Manuf. Technol. 2021, 117, 1975–1995. [Google Scholar] [CrossRef]
- Driggers, J.C.; Vitale, S.; Lundgren, A.; Evans, M.; Kawabe, K.; Dwyer, S.; Izumi, K.; Schofield, R.; Effler, A.; Sigg, D.; et al. Improving astrophysical parameter estimation via offline noise subtraction for Advanced LIGO. Phys. Rev. D 2019, 99, 042001. [Google Scholar] [CrossRef]
- Reis, M.d.; Yang, Z. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol. Biol. Evol. 2011, 28, 2161–2172. [Google Scholar] [CrossRef] [PubMed]
- Kanaan, M.; Farrington, C. Matrix models for childhood infections: A Bayesian approach with applications to rubella and mumps. Epidemiol. Infect. 2005, 133, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Dangerfield, B.; Duggan, J. Optimization of system dynamics models. In System Dynamics: Theory and Applications; Springer: New York, NY, USA, 2020; pp. 139–152. [Google Scholar]
- Raissi, M.; Perdikaris, P.; Karniadakis, G.E. Physics-informed neural networks: A deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J. Comput. Phys. 2019, 378, 686–707. [Google Scholar] [CrossRef]
- Berkhahn, S.; Ehrhardt, M. A physics-informed neural network to model COVID-19 infection and hospitalization scenarios. Adv. Contin. Discret. Model. 2022, 2022, 61. [Google Scholar] [CrossRef] [PubMed]
- Tartakovsky, A.M.; Marrero, C.O.; Perdikaris, P.; Tartakovsky, G.D.; Barajas-Solano, D. Physics-informed deep neural networks for learning parameters and constitutive relationships in subsurface flow problems. Water Resour. Res. 2020, 56, e2019WR026731. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, D.; Chen, X.; Zhang, M. Physics-Informed Neural Network for Optical Fiber Parameter Estimation From the Nonlinear Schrödinger Equation. J. Light. Technol. 2022, 40, 7095–7105. [Google Scholar] [CrossRef]
- Zhao, S.; Peng, Y.; Zhang, Y.; Wang, H. Parameter estimation of power electronic converters with physics-informed machine learning. IEEE Trans. Power Electron. 2022, 37, 11567–11578. [Google Scholar] [CrossRef]
- Robert, C.P.; Casella, G.; Casella, G. Introducing Monte Carlo Methods with r; Springer: Berlin/Heidelberg, Germany, 2010; Volume 18. [Google Scholar]
- Jin, Y.F.; Yin, Z.Y.; Zhou, W.H.; Horpibulsuk, S. Identifying parameters of advanced soil models using an enhanced transitional Markov chain Monte Carlo method. Acta Geotech. 2019, 14, 1925–1947. [Google Scholar] [CrossRef]
- Durmus, A.; Moulines, É.; Pereyra, M. A Proximal Markov Chain Monte Carlo Method for Bayesian Inference in Imaging Inverse Problems: When Langevin Meets Moreau. SIAM Rev. 2022, 64, 991–1028. [Google Scholar] [CrossRef]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Baydin, A.G.; Pearlmutter, B.A.; Radul, A.A.; Siskind, J.M. Automatic differentiation in machine learning: A survey. J. Marchine Learn. Res. 2018, 18, 1–43. [Google Scholar]
- Kermack, W.O.; McKendrick, A.G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. Ser. Contain. Pap. Math. Phys. Character 1927, 115, 700–721. [Google Scholar]
- Lu, L.; Meng, X.; Mao, Z.; Karniadakis, G.E. DeepXDE: A deep learning library for solving differential equations. SIAM Rev. 2021, 63, 208–228. [Google Scholar] [CrossRef]
- Heldmann, F.; Berkhahn, S.; Ehrhardt, M.; Klamroth, K. PINN training using biobjective optimization: The trade-off between data loss and residual loss. J. Comput. Phys. 2023, 488, 112211. [Google Scholar] [CrossRef]


| Method | SIR | SIRS |
|---|---|---|
| MC MSE | = −1.49 | |
| = −1.49 | ||
| = 1.46 | ||
| MC Log Square | = −18.71 | |
| = −18.71 | ||
| = 18.69 | ||
| PINN | ||
| Method | SIR | SIRS |
|---|---|---|
| MC MSE | ||
| = 0.187 | = 0.25 | |
| = 0.1 | ||
| MC Log Square | = 0.7 | = 1.9 |
| = 0.18 | = 1.22 | |
| = 5.83 | ||
| PINN | = 0.7 | = 0.8 |
| = 0.187 | = 0.25 | |
| = 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Severt, M.; Casado-Vara, R.; Martín del Rey, A. A Comparison of Monte Carlo-Based and PINN Parameter Estimation Methods for Malware Identification in IoT Networks. Technologies 2023, 11, 133. https://doi.org/10.3390/technologies11050133
Severt M, Casado-Vara R, Martín del Rey A. A Comparison of Monte Carlo-Based and PINN Parameter Estimation Methods for Malware Identification in IoT Networks. Technologies. 2023; 11(5):133. https://doi.org/10.3390/technologies11050133
Chicago/Turabian StyleSevert, Marcos, Roberto Casado-Vara, and Angel Martín del Rey. 2023. "A Comparison of Monte Carlo-Based and PINN Parameter Estimation Methods for Malware Identification in IoT Networks" Technologies 11, no. 5: 133. https://doi.org/10.3390/technologies11050133
APA StyleSevert, M., Casado-Vara, R., & Martín del Rey, A. (2023). A Comparison of Monte Carlo-Based and PINN Parameter Estimation Methods for Malware Identification in IoT Networks. Technologies, 11(5), 133. https://doi.org/10.3390/technologies11050133








