Seed Transcriptome Annotation Reveals Enhanced Expression of Genes Related to ROS Homeostasis and Ethylene Metabolism at Alternating Temperatures in Wild Cardoon
Abstract
:1. Introduction
2. Materials and Methods
2.1. Achenes Collection
2.2. General Procedures for Germination Tests
2.3. Achenes Treatments and RNA Extraction
2.4. Transcriptome Sequencing, Assembly, and Annotation
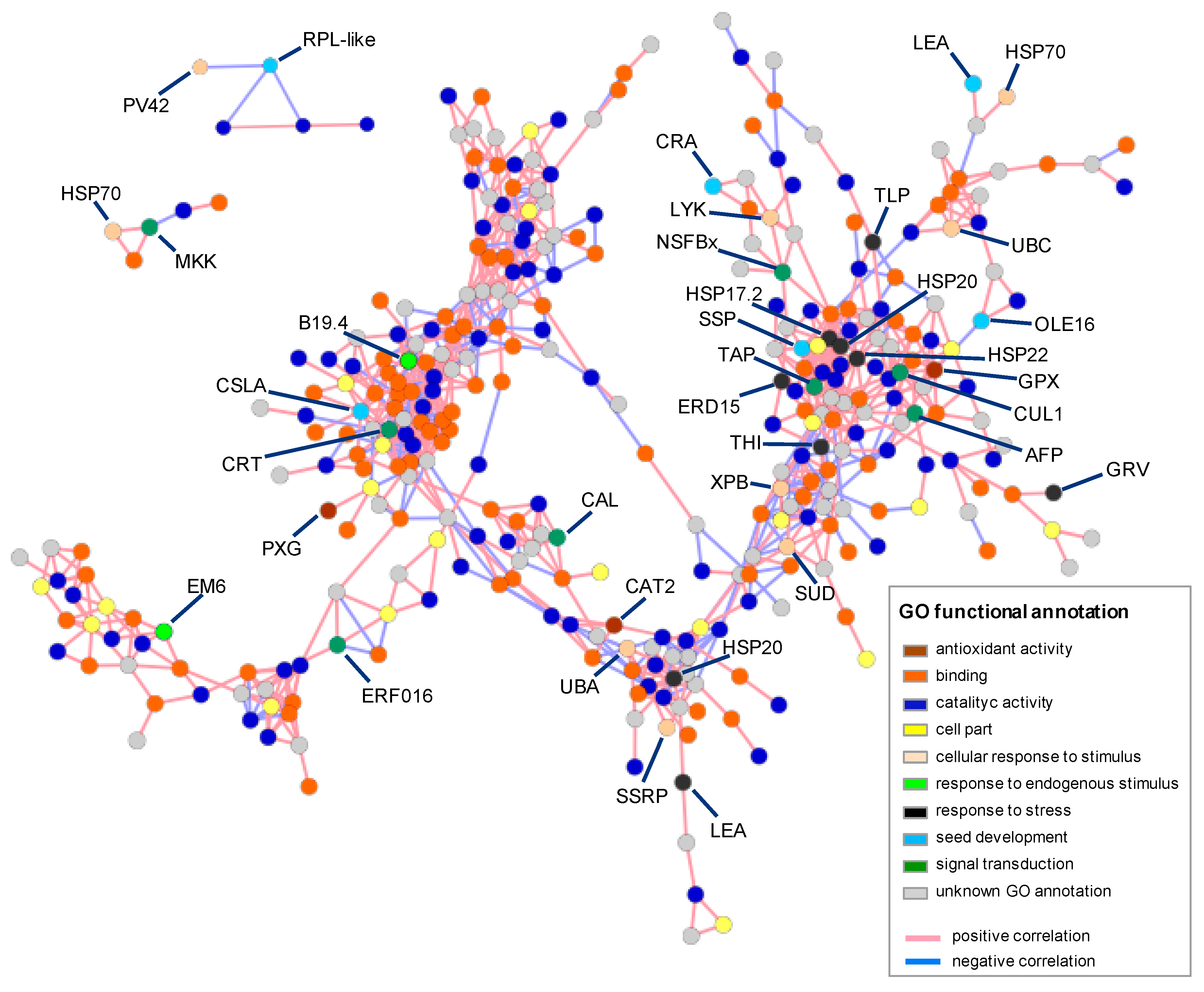
2.5. Differential Gene Expression and Co-Expression Network Analysis
2.6. Analysis of Ethylene and ROS Regulation at Alternating Temperatures
2.7. Ethylene Measurements
3. Results and Discussion
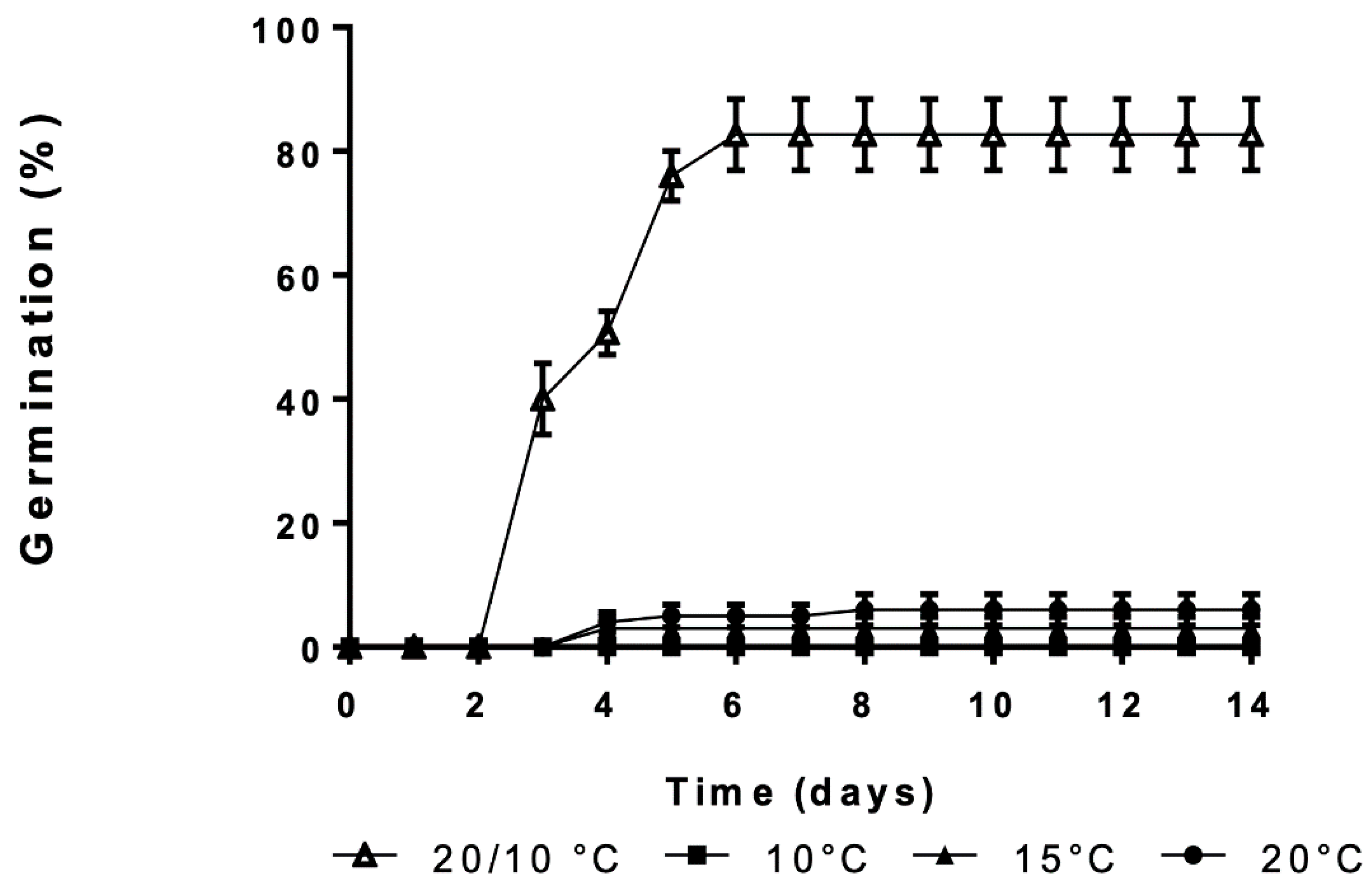
3.1. Effect of Constant and Alternating Temperatures on Germination
3.2. Seed Transcriptome Annotation
3.3. Differential Expression Analysis
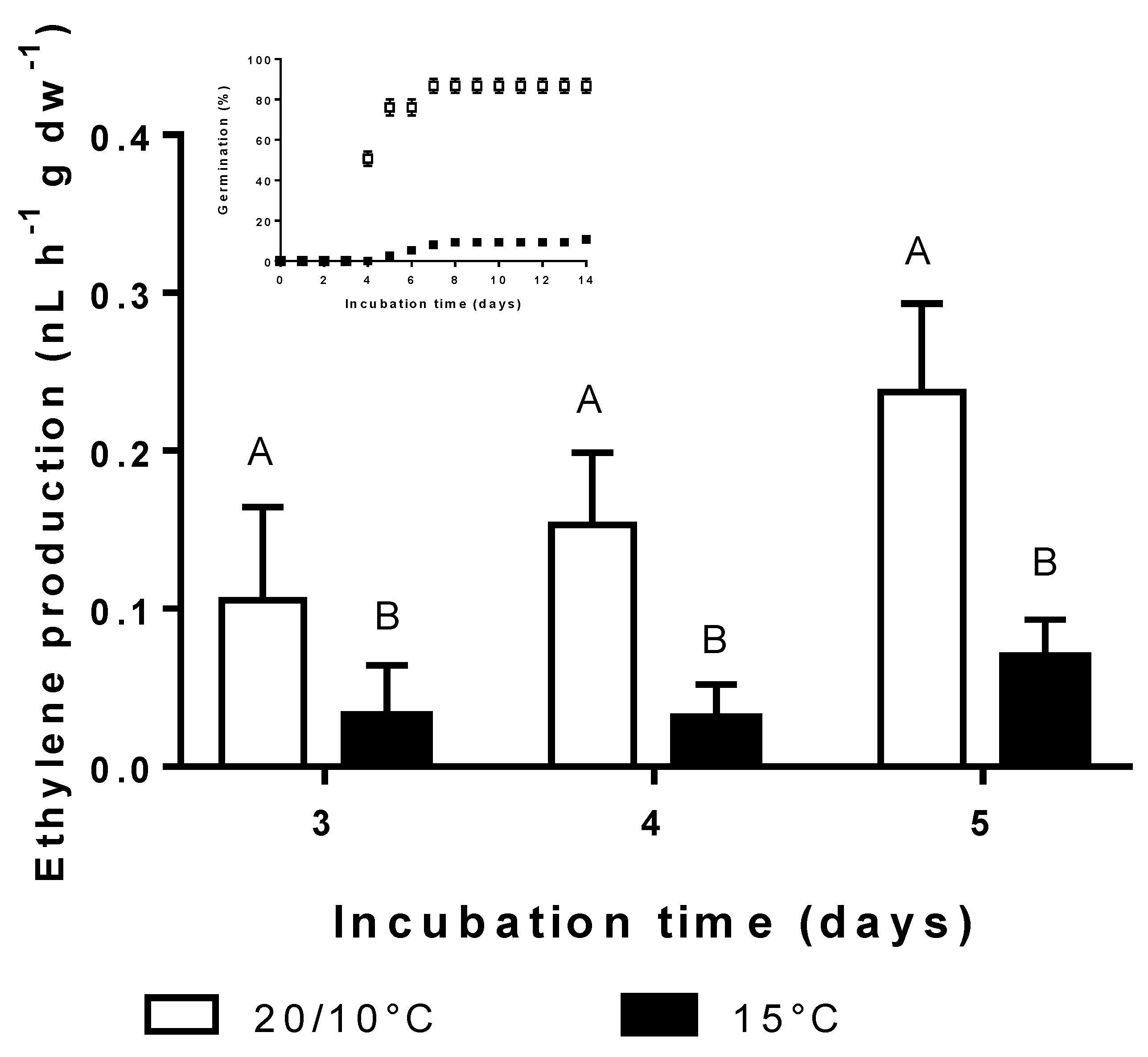
3.4. Effect of Incubation Temperature on Ethylene Synthesis and Germination
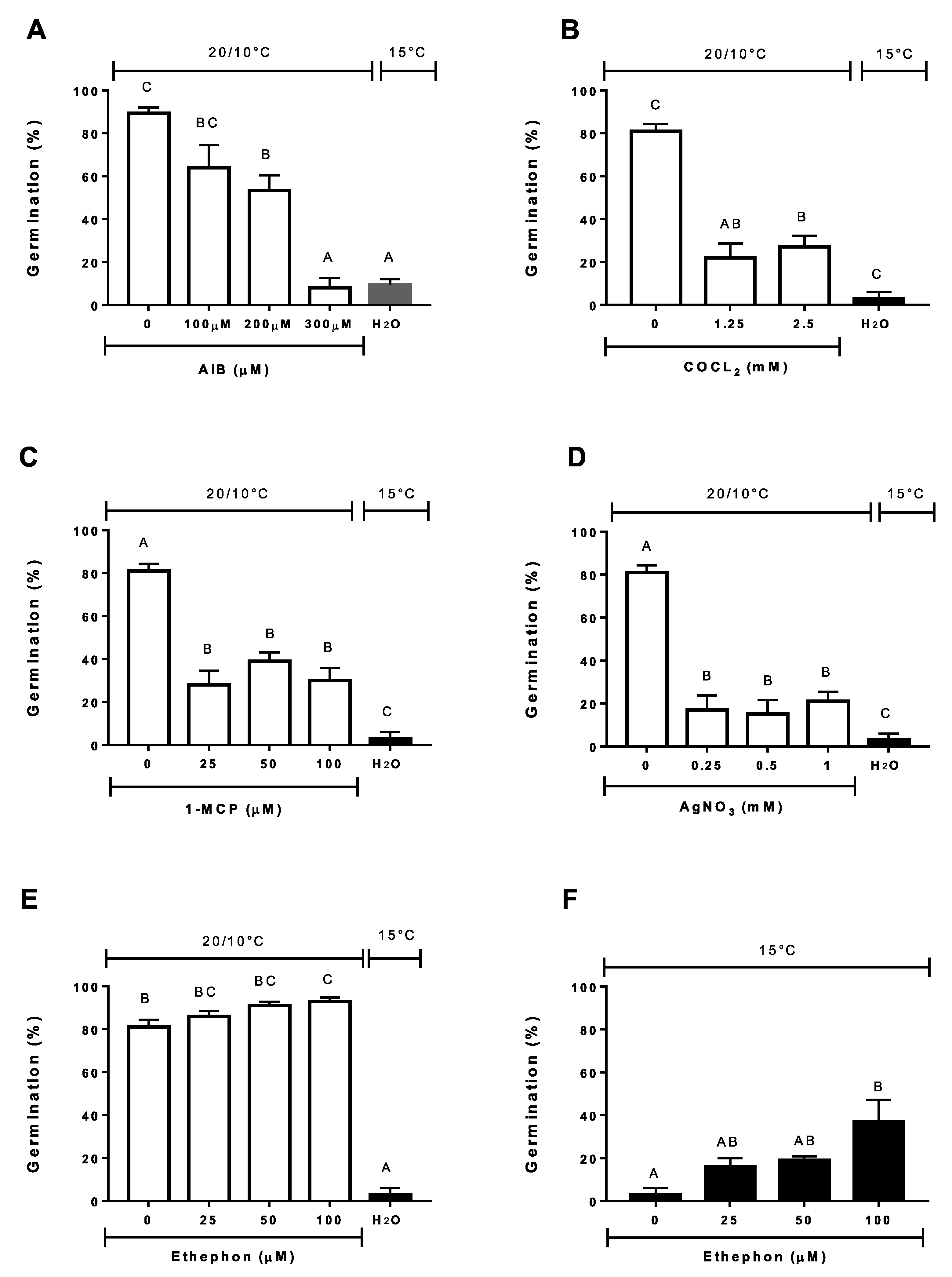
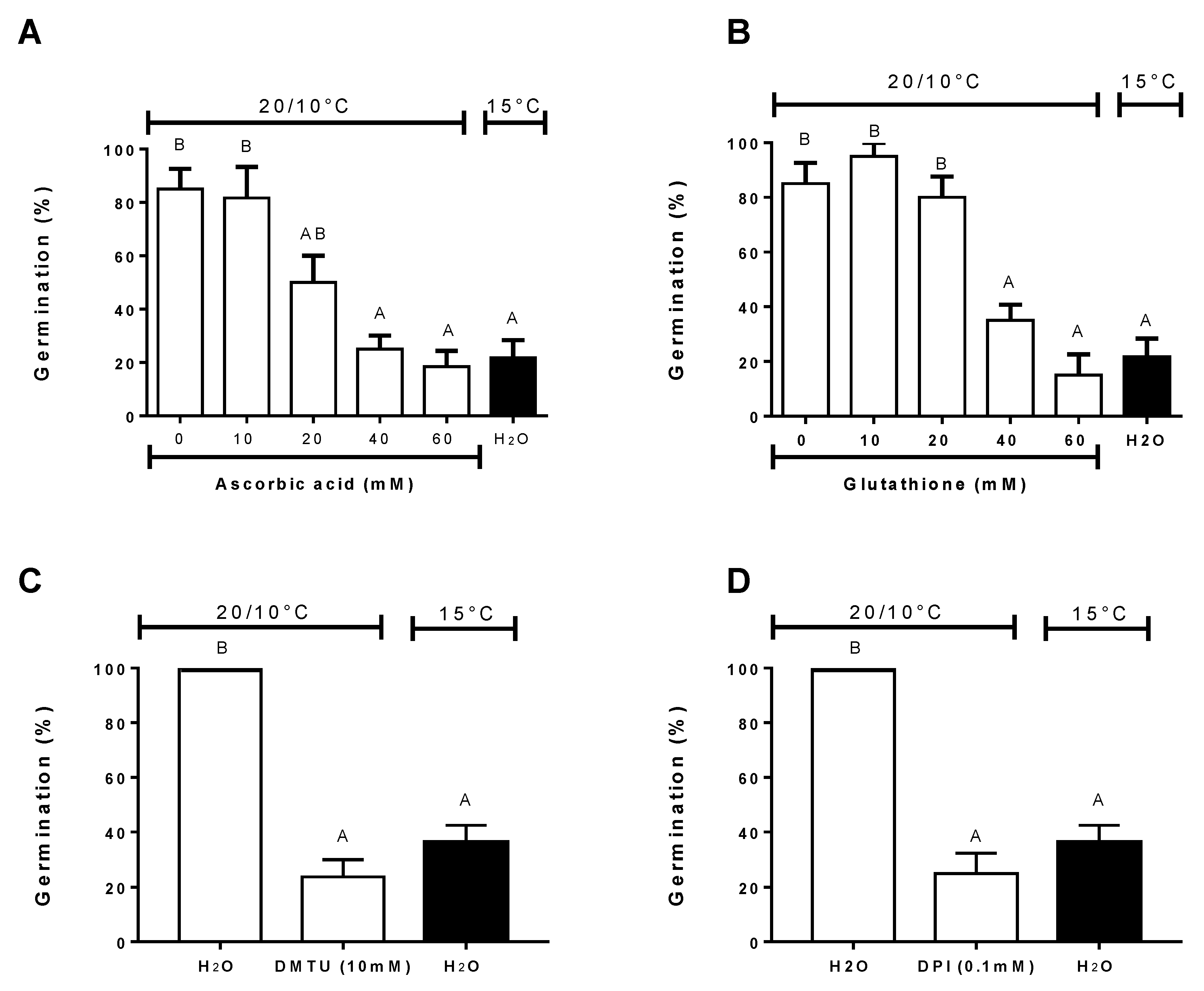
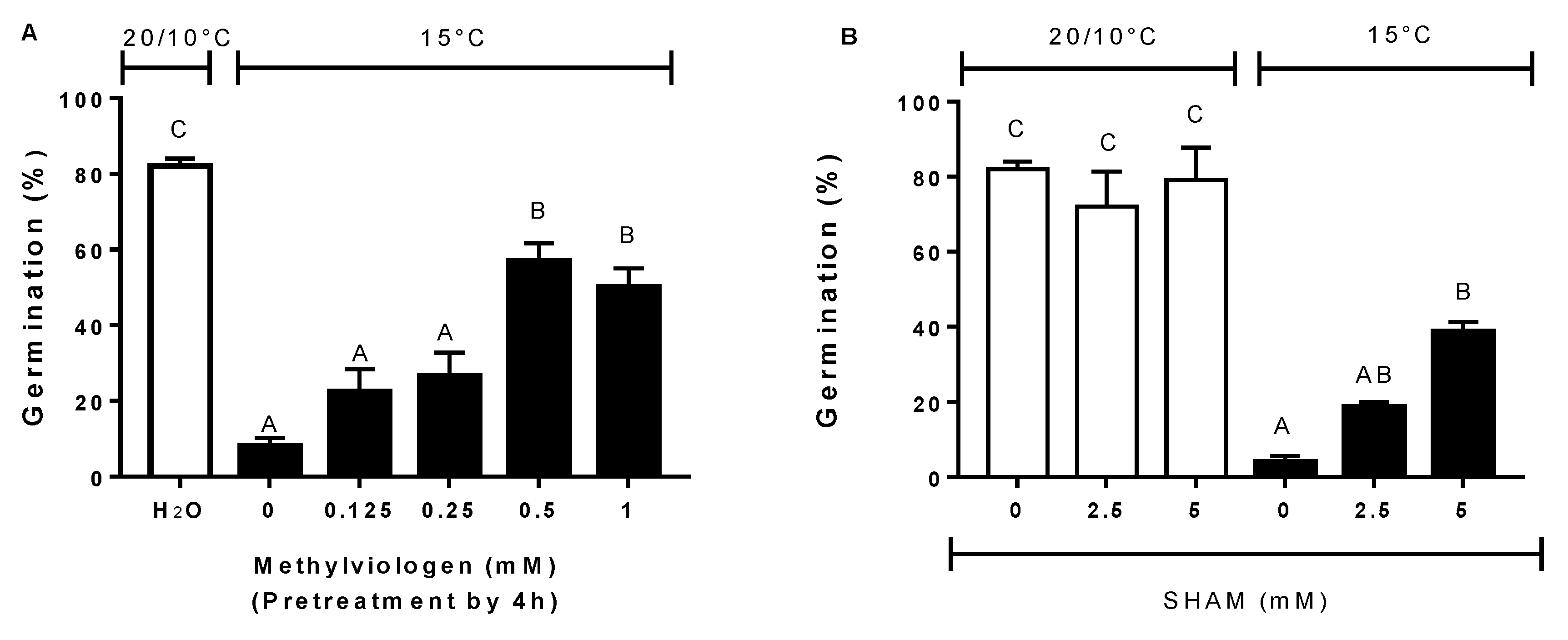
3.5. Effect of Antioxidants, ROS Scavengers, and ROS Donors on Germination at Alternating or Constant Temperatures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Long, R.L.; Gorecki, M.J.; Renton, M.; Scott, J.K.; Colville, L.; Goggin, D.E.; Commander, L.E.; Westcott, D.A.; Cherry, H.; Finch-Savage, W.E. The ecophysiology of seed persistence: A mechanistic view of the journey to germination or demise. Biol. Rev. 2015, 90, 31–59. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Footitt, S. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 2017, 68, 843–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batlla, D.; Benech-Arnold, R.L. A framework for the interpretation of temperature effects on dormancy and germination in seed populations showing dormancy. Seed Sci. Res. 2015, 25, 1–12. [Google Scholar] [CrossRef]
- Chao, W.S.; Foley, M.E.; Dogramaci, M.; Anderson, J.V.; Horvath, D.P. Alternating temperature breaks dormancy in leafy spurge seeds and impacts signaling networks associated with HY5. Funct. Integr. Genom. 2011, 11, 637–649. [Google Scholar] [CrossRef]
- Puglia, G.; Carta, A.; Bizzoca, R.; Toorop, P.; Spampinato, G.; Raccuia, S.A. Seed dormancy and control of germination in Sisymbrella dentata (l.) O.E. Schulz (Brassicaceae). Plant Biol. 2018, 50, 164–170. [Google Scholar] [CrossRef]
- Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.M.; Nonogaki, H. Dormancy and the Control of Germination. In Seeds: Physiology of Development, Germination and Dormancy, 3rd ed.; Springer: New York, NY, USA, 2013; pp. 247–297. ISBN 978-1-4614-4693-4. [Google Scholar]
- Vandelook, F.; Newton, R.J.; Carta, A. Photophobia in lilioid monocots: Photoinhibition of seed germination explained by seed traits, habitat adaptation and phylogenetic inertia. Ann. Bot. 2018, 121, 405–413. [Google Scholar] [CrossRef]
- Saatkamp, A.; Affre, L.; Dutoit, T.; Poschlod, P. Germination traits explain soil seed persistence across species: The case of Mediterranean annual plants in cereal fields. Ann. Bot. 2011, 107, 415–426. [Google Scholar] [CrossRef]
- Picciau, R.; Pritchard, H.W.; Mattana, E.; Bacchetta, G. Thermal thresholds for seed germination in Mediterranean species are higher in mountain compared with lowland areas. Seed Sci. Res. 2019, 29, 44–54. [Google Scholar] [CrossRef]
- Arana, M.V.; Tognacca, R.S.; Estravis-Barcalá, M.; Sánchez, R.A.; Botto, J.F. Physiological and molecular mechanisms underlying the integration of light and temperature cues in Arabidopsis thaliana seeds. Plant Cell Environ. 2017, 40, 3113–3121. [Google Scholar] [CrossRef]
- Huarte, H.R.; Luna, V.; Pagano, E.A.; Zavala, J.A.; Benech-Arnold, R.L. Fluctuating temperatures terminate dormancy in Cynara cardunculus seeds by turning off ABA synthesis and reducing ABA signaling, but not stimulating GA synthesis or signaling. Seed Sci. Res. 2014, 24, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Laspina, N.V.; Batlla, D.; Benech-Arnold, R.L. Dormancy cycling in Polygonum aviculare seeds is accompanied by changes in ABA sensitivity. J. Exp. Bot. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ghassemian, M.; Nambara, E.; Cutler, S.; Kawaide, H.; Kamiya, Y.; McCourt, P.; Corbineau, F.; Strnad, M.; Lynn, J.R.; Finch-Savage, W.E.; et al. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 2000, 12, 1117–1126. [Google Scholar] [CrossRef] [Green Version]
- Linkies, A.; Müller, K.; Morris, K.; Turecková, V.; Wenk, M.; Cadman, C.S.C.; Corbineau, F.; Strnad, M.; Lynn, J.R.; Finch-Savage, W.E.; et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 2009, 21, 3803–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucera, B.; Cohn, M.A.; Leubner-Metzger, G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005, 15, 281–307. [Google Scholar] [CrossRef]
- Matilla, A.J.; Matilla-Vázquez, M.A. Involvement of ethylene in seed physiology. Plant Sci. 2008, 175, 87–97. [Google Scholar] [CrossRef]
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 2014, 5, 539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Maarouf-Bouteau, H.; Bailly, C. Oxidative signaling in seed germination and dormancy. Plant Signal. Behav. 2008, 3, 175–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leymarie, J.; Vitkauskaité, G.; Hoang, H.H.; Gendreau, E.; Chazoule, V.; Meimoun, P.; Corbineau, F.; El-Maarouf-Bouteau, H.; Bailly, C. Role of reactive oxygen species in the regulation of Arabidopsis seed dormancy. Plant Cell Physiol. 2012, 53, 96–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailly, C. The signaling role of ROS in the regulation of seed germination and dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef]
- Oracz, K.; El-Maarouf-Bouteau, H.; Kranner, I.; Bogatek, R.; Corbineau, F.; Bailly, C. The mechanisms involved in seed dormancy alleviation by hydrogen cyanide unravel the role of reactive oxygen species as key factors of cellular signaling during germination. Plant Physiol. 2009, 150, 494–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Maarouf-Bouteau, H.; Sajjad, Y.; Bazin, J.; Langlade, N.; Cristescu, S.M.; Balzergue, S.; Baudouin, E.; Bailly, C. Reactive oxygen species, abscisic acid and ethylene interact to regulate sunflower seed germination. Plant Cell Environ. 2015, 38, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Huarte, H.R.; Borlandelli, F.; Varisco, D.; Batlla, D. Understanding dormancy breakage and germination ecology of Cynara cardunculus (Asteraceae). Weed Res. 2018, 58, 450–462. [Google Scholar] [CrossRef]
- Huarte, H.R.; Puglia, G.D.; Varisco, D.; Pappalardo, H.; Calderaro, P.; Toscano, V.; Raccuia, S.A. Effect of reactive oxygen species on germination of Cynara cardunculus (L.) cultivars. Acta Hort. 2020, 2, 33–40. [Google Scholar] [CrossRef]
- Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. From intracellular signaling networks to cell death: The dual role of reactive oxygen species in seed physiology. Comptes Rendus Biol. 2008, 331, 806–814. [Google Scholar] [CrossRef]
- Bazin, J.; Langlade, N.; Vincourt, P.; Arribat, S.; Balzergue, S.; El-Maarouf-Bouteau, H.; Bailly, C. Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 2011, 23, 2196–2208. [Google Scholar] [CrossRef] [Green Version]
- Layat, E.; Leymarie, J.; El-Maarouf-Bouteau, H.; Caius, J.; Langlade, N.; Bailly, C. Translatome profiling in dormant and nondormant sunflower (Helianthus annuus) seeds highlights post-transcriptional regulation of germination. New Phytol. 2014, 204, 864–872. [Google Scholar] [CrossRef]
- Han, Z.; Wang, B.; Tian, L.; Wang, S.; Zhang, J.; Guo, S.L.; Zhang, H.; Xu, L.; Chen, Y. Comprehensive dynamic transcriptome analysis at two seed germination stages in maize (Zea mays L.). Physiol. Plant 2020, 168, 205–217. [Google Scholar] [CrossRef]
- Liao, D.; Zhu, J.; Zhang, M.; Li, X.; Sun, P.; Wei, J.; Qi, J. Comparative transcriptomic analysis reveals genes regulating the germination of morphophysiologically dormant Paris polyphylla seeds during a warm stratification. PLoS ONE 2019, 14, e0212514. [Google Scholar] [CrossRef] [Green Version]
- Visscher, A.M.; Yeo, M.; Castillo-Lorenzo, E.; Toorop, P.E.; Junio da Silva, L.; Yeo, M.; Pritchard, H.W. Pseudophoenix ekmanii (Arecaceae) seeds at suboptimal temperature show reduced imbibition rates and enhanced expression of genes related to germination inhibition. Plant Biol. 2020. [Google Scholar] [CrossRef]
- Foley, M.E.; Anderson, J.V.; Chao, W.S.; Doğramacı, M.; Horvath, D.P. Initial changes in the transcriptome of Euphorbia esula seeds induced to germinate with a combination of constant and diurnal alternating temperatures. Plant Mol. Biol. 2010, 73, 131–142. [Google Scholar] [CrossRef]
- Ma, W.; Guan, X.; Li, J.; Pan, R.; Wang, L.; Liu, F.; Ma, H.; Zhu, S.; Hu, J.; Ruan, Y.L.; et al. Mitochondrial small heat shock protein mediates seed germination via thermal sensing. Proc. Natl. Acad. Sci. USA 2019, 116, 4716–4721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raccuia, S.A.; Melilli, M.G. Biomass and grain oil yields in Cynara cardunculus L. genotypes grown in a Mediterranean environment. Field Crop Res. 2007, 101, 187–197. [Google Scholar] [CrossRef]
- Pappalardo, H.D.; Toscano, V.; Puglia, G.D.; Genovese, C.; Raccuia, S.A. Cynara cardunculus L. as a multipurpose crop for plant secondary metabolites production in marginal stressed lands. Front. Plant Sci. 2020, 11, 240. [Google Scholar] [CrossRef] [PubMed]
- Raccuia, S.A.; Mainolfi, A.; Mandolino, G.; Melilli, M.G. Genetic diversity in Cynara cardunculus revealed by AFLP markers: Comparison between cultivars and wild types from Sicily. Plant Breed. 2004, 123, 280–284. [Google Scholar] [CrossRef]
- Huarte, H.R.; Benech-Arnold, R.L. Hormonal nature of seed responses to fluctuating temperatures in Cynara cardunculus (L.). Seed Sci. Res. 2010, 20, 39–45. [Google Scholar] [CrossRef]
- Scaglione, D.; Reyes-Chin-Wo, S.; Acquadro, A.; Froenicke, L.; Portis, E.; Beitel, C.; Tirone, M.; Mauro, R.; Lo Monaco, A.; Mauromicale, G.; et al. The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 2016, 6, 19427. [Google Scholar] [CrossRef] [Green Version]
- Puglia, G.D.; Prjibelski, A.D.; Vitale, D.; Bushmanova, E.; Schmid, K.J.; Raccuia, S.A. Hybrid transcriptome sequencing approach improved assembly and gene annotation in Cynara cardunculus (L.). BMC Genom. 2020, 21, 317. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Rau, A.; Maugis-Rabusseau, C. Transformation and model choice for RNA-seq co-expression analysis. Brief. Bioinform. 2018, 19, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Raccuia, S.A.; Puglia, G.; Pappalardo, H.; Argento, S.; Leonardi, C.; Calderaro, P.; Melilli, M.G. Dormancy-related genes isolation in Cynara cardunculus var. sylvestris. Acta Hortic. 2016, 1147, 315–322. [Google Scholar] [CrossRef]
- Passaia, G.; Queval, G.; Bai, J.; Margis-Pinheiro, M.; Foyer, C.H. The effects of redox controls mediated by glutathione peroxidases on root architecture in Arabidopsis. J. Exp. Bot. 2014, 65, 1403–1413. [Google Scholar] [CrossRef] [Green Version]
- Bailly, C. Active oxygen species and antioxidants in seed biology. Seed Sci. Res. 2004, 14, 93–107. [Google Scholar] [CrossRef]
- Rahman, F.; Hassan, M.; Rosli, R.; Almousally, I.; Hanano, A.; Murphy, D.J. Evolutionary and genomic analysis of the caleosin/peroxygenase (CLO/PXG) gene/ protein families in the Viridiplantae. PLoS ONE 2018, 13, e0196669. [Google Scholar] [CrossRef] [Green Version]
- Jin, D.; Wu, M.; Li, B.; Bücker, B.; Keil, P.; Zhang, S.; Li, J.; Kang, D.; Liu, J.; Dong, J.; et al. The COP9 Signalosome regulates seed germination by facilitating protein degradation of RGL2 and ABI5. PLOS Genet. 2018, 14, e1007237. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Cao, H.; Sun, Y.; Li, X.; Chen, F.; Carles, A.; Li, Y.; Ding, M.; Zhang, C.; Deng, X.; et al. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid–ethylene antagonism mediated by histone deacetylation. Plant Cell 2013, 25, 149–166. [Google Scholar] [CrossRef] [Green Version]
- Paik, I.; Chen, F.; Ngoc Pham, V.; Zhu, L.; Kim, J.I.; Huq, E. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 2019, 10, 4216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.; Lei, L.; Yang, H.; Liu, G.; Ren, D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, R.; Zhu, Y.; Shen, G.; Zhang, H. TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiol. 2014, 164, 721–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.E.; Lynch, T.; Peeters, J.; Snowden, C.; Finkelstein, R. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 2008, 67, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Maia, J.; Dekkers, B.J.W.; Provart, N.J.; Ligterink, W.; Hilhorst, H.W.M. The re-establishment of desiccation tolerance in germinated Arabidopsis thaliana seeds and its associated transcriptome. PLoS ONE 2011, 6, e29123. [Google Scholar] [CrossRef] [Green Version]
- Footitt, S.; Douterelo-Soler, I.; Clay, H.; Finch-Savage, W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 20236–20241. [Google Scholar] [CrossRef] [Green Version]
- Zhiguo, E.; Yuping, Z.; Tingting, L.; Lei, W.; Heming, Z. Characterization of the ubiquitin-conjugating enzyme gene family in rice and evaluation of expression profiles under abiotic stresses and hormone treatments. PLoS ONE 2015, 10, e0122621. [Google Scholar]
- Michl-Holzinger, P.; Mortensen, S.A.; Grasser, K.D. The SSRP1 subunit of the histone chaperone FACT is required for seed dormancy in Arabidopsis. J. Plant Physiol. 2019, 236, 105–108. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Bray, C.M.; West, C.E. Seeds and the art of genome maintenance. Front. Plant Sci. 2019, 10, 706. [Google Scholar] [CrossRef] [Green Version]
- Kariola, T.; Brader, G.; Helenius, E.; Li, J.; Heino, P.; Palva, E.T. Early responsive to dehydration 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol. 2006, 142, 1559–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toorop, P.E.; Barroco, R.M.; Engler, G.; Groot, S.P.C.; Hilhorst, H.W.M. Differentially expressed genes associated with dormancy or germination of Arabidopsis thaliana seeds. Planta 2005, 221, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.S.; Kavi Kishor, P.B.; Seiler, C.; Kuhlmann, M.; Eschen-Lippold, L.; Lee, J.; Reddy, M.K.; Sreenivasulu, N. Unraveling regulation of the small heat shock proteins by the heat shock factor HvHsfB2c in barley: Its implications in drought stress response and seed development. PLoS ONE 2014, 9, e89125. [Google Scholar] [CrossRef] [Green Version]
- Sall, K.; Dekkers, B.J.W.; Nonogaki, M.; Katsuragawa, Y.; Koyari, R.; Hendrix, D.; Willems, L.A.J.; Bentsink, L.; Nonogaki, H. DELAY OF GERMINATION 1-LIKE 4 acts as an inducer of seed reserve accumulation. Plant J. 2019, 100, 7–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linkies, A.; Leubner-Metzger, G. Beyond gibberellins and abscisic acid: How ethylene and jasmonates control seed germination. Plant Cell Rep. 2012, 31, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Kępczyński, J.; Sznigir, P. Participation of GA3, ethylene, NO and HCN in germination of Amaranthus retroflexus L. seeds with various dormancy levels. Acta Physiol. Plant 2014, 36, 1463–1472. [Google Scholar] [CrossRef] [Green Version]
- Oracz, K.; Bouteau, H.E.M.; Farrant, J.M.; Cooper, K.; Belghazi, M.; Job, C.; Job, D.; Corbineau, F.; Bailly, C. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007, 50, 452–465. [Google Scholar] [CrossRef] [Green Version]
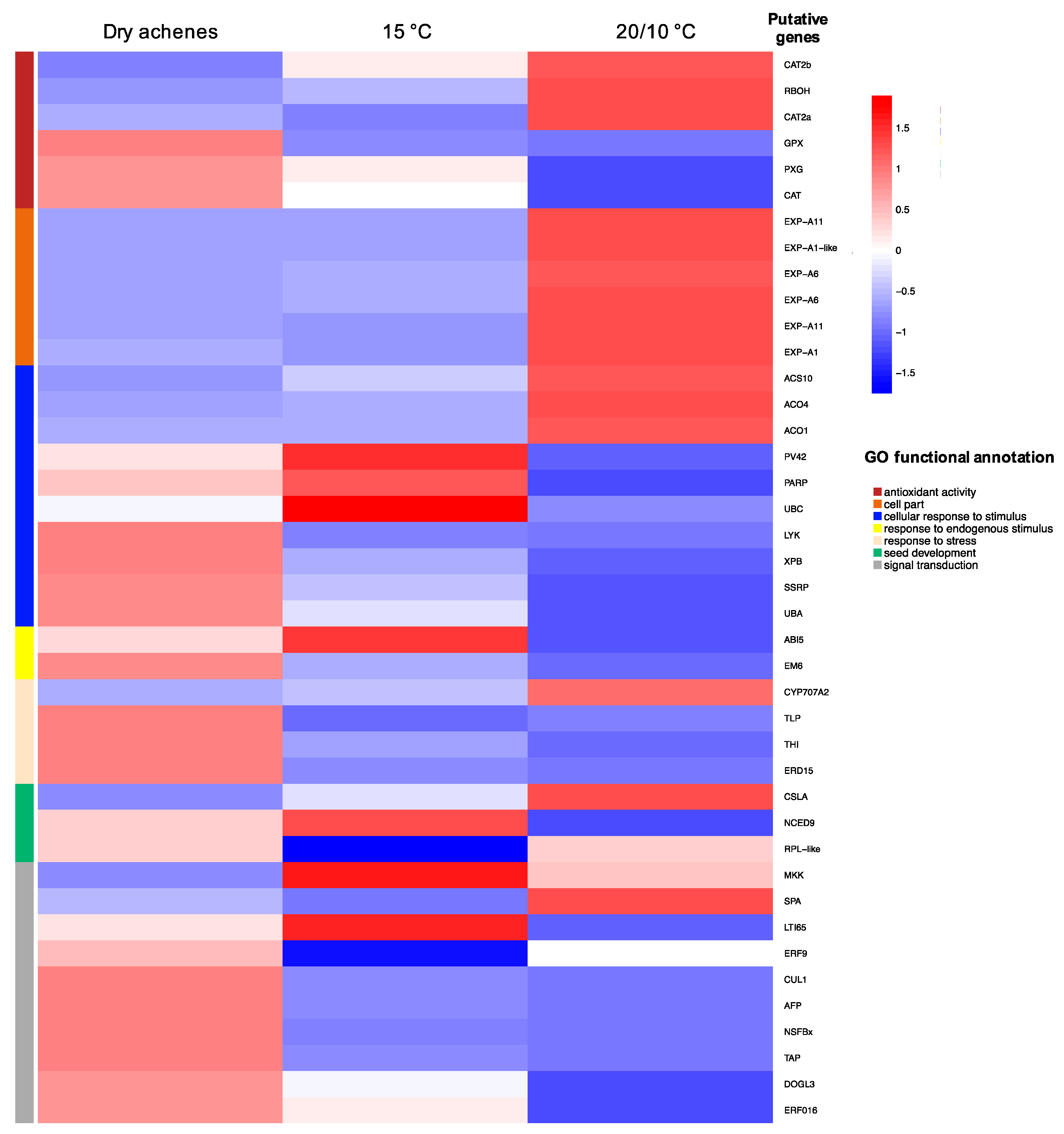
| Gene Identifier | Gene Description | Effect on Dormancy Regulation |
|---|---|---|
| Ccrd_v2_22156_g15 | GPX—phospholipid hydroperoxide glutathione peroxidase | Reduction of H2O2 or organic hydroperoxides [47] |
| Ccrd_v2_22208_g15 | RBOH—Respiratory burst oxidase protein | Biosynthesis of superoxide/Dormancy alleviation factor [20] |
| split_gene_Ccrd_v2_02613_g01-g146 | CAT2—Catalase-like isoform X2 | Protect from H2O2 and lipid peroxidation [48] |
| Ccrd_v2_14857_g10 | PXG—Plant seed peroxygenase | Protect from dehydration stress [49] |
| Ccrd_v2_04883_g02 | DOGL3—protein DOG1-like 3 isoform | Effect unclear |
| Ccrd_v2_02779_g02 | CUL1—Cullin-1-like isoform X1 | Control of ABA biosynthesis [50] |
| Ccrd_v2_12828_g08 | ERF9—ETHYLENE RESPONSE FACTOR9 | Ethylene signaling [51] |
| Ccrd_v2_23452_g16 | SPA—protein SUPPRESSOR OF PHYA-105 1-like isoform | Regulates circadian rhythms/germination enhancer [52] |
| Ccrd_v2_21316_g15 | MKK—Mitogen-activated protein kinase 9-like | Induces the synthesis of ethylene [53] |
| Ccrd_v2_08680_g05 | NSFBx—Probable F-box protein (At5g04010) | Effect unknown |
| Ccrd_v2_00002_g01 | TAP—2A phosphatase associated protein | Dormancy enhancer [54] |
| Ccrd_v2_09661_g05 | AFP—ninja-family protein AFP3-like | Control of ABA biosynthesis [55] |
| Ccrd_v2_03046_g02 | ERF016—Ethylene-responsive transcription factor ERF016-like | Effect unclear |
| novel_gene_1_5b8548b9 | LTI65—low-temperature-induced 65 kDa protein-like | Responsive to ABA [56] |
| Ccrd_v2_06887_g03 | PV42—SNF1-related protein kinase regulatory subunit gamma-like PV42a | Dormancy enhancer [57] |
| Ccrd_v2_15915_g11 | PARP—putative Poly [ADP-ribose] polymerase 3 | DNA protection system |
| Ccrd_v2_01121_g01 | UBC—Ubiquitin-conjugating enzyme E2 2 | Induced by ABA [58] |
| Ccrd_v2_24782_g17 | ACO1—1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE1 | Ethylene biosynthesis [51] |
| Ccrd_v2_23833_g17 | ACO4—1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE4 | Ethylene biosynthesis [51] |
| Ccrd_v2_16461_g11 | ACS10—ACC synthase10 | Ethylene biosynthesis [51] |
| Ccrd_v2_19164_g13 | LYK—LYSIN MOTIF RECEPTOR KINASE | Effect unknown |
| Ccrd_v2_01115_g01 | SSRP—FACT complex subunit SSRP1-like isoform X1 | Dormancy enhancer [59] |
| Ccrd_v2_14002_g09 | UBA—ubiquitin-activating enzyme E1 1-like isoform X | Effect unknown |
| Ccrd_v2_22183_g15 | XPB—general transcription and DNA repair factor IIH helicase subunit XPB1 | DNA repair [60] |
| Ccrd_v2_00258_g01 | ERD15—protein EARLY RESPONSIVE TO DEHYDRATION 15-like | Induced by dehydration stress/Modulates ABA response [61] |
| Ccrd_v2_22449_g15 | CYP707A2—Cytochrome P450, Family 707, Subfamily A, Polypeptide2 | Reduced dormancy [51] |
| Ccrd_v2_15609_g11 | NCED9—NINE-cis-EPOXYCAROTENOID DIOXYGENASE -9 | Dormancy enhancer [51] |
| Ccrd_v2_13522_g09 | ABI5—protein ABSCISIC ACID-INSENSITIVE 5 | Dormancy enhancer [55] |
| Ccrd_v2_00305_g01 | EM6—em-like protein GEA6 | Effect unclear |
| Ccrd_v2_02516_g01 | TLP—thaumatin-like protein 1b | Effect unknown |
| Ccrd_v2_05395_g03 | THI—Gamma thionin | Effect unknown |
| Ccrd_v2_00955_g01 | CSLA—CELLULOSE SYNTHASE-LIKE (CSL) | Effect unknown |
| Ccrd_v2_08837_g05 | RPL-like—Ribosomal Protein-like | Reduced dormancy [62] |
| Ccrd_v2_19371_g13 | EXPA6—Expansin A6 | Cell wall loosening/Ethylene signaling [51] |
| Ccrd_v2_05454_g03 | EXPA11—Expansin A11 | Cell wall loosening/Ethylene signaling [51] |
| Ccrd_v2_19070_g13 | EXPA11—Expansin A11 | Cell wall loosening/Ethylene signaling [51] |
| Ccrd_v2_21520_g15 | EXPA1—Expansin A1 | Cell wall loosening/Ethylene signaling [51] |
| Ccrd_v2_01833_g01 | EXPA1-like—Expansin A11-like | Cell wall loosening/Ethylene signaling [51] |
| Ccrd_v2_00414_g01 | EXPA6—Expansin A6 | Cell wall loosening/Ethylene signaling [51] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huarte, H.R.; Puglia, G.D.; Prjibelski, A.D.; Raccuia, S.A. Seed Transcriptome Annotation Reveals Enhanced Expression of Genes Related to ROS Homeostasis and Ethylene Metabolism at Alternating Temperatures in Wild Cardoon. Plants 2020, 9, 1225. https://doi.org/10.3390/plants9091225
Huarte HR, Puglia GD, Prjibelski AD, Raccuia SA. Seed Transcriptome Annotation Reveals Enhanced Expression of Genes Related to ROS Homeostasis and Ethylene Metabolism at Alternating Temperatures in Wild Cardoon. Plants. 2020; 9(9):1225. https://doi.org/10.3390/plants9091225
Chicago/Turabian StyleHuarte, Hector R., Giuseppe. D. Puglia, Andrey D. Prjibelski, and Salvatore A. Raccuia. 2020. "Seed Transcriptome Annotation Reveals Enhanced Expression of Genes Related to ROS Homeostasis and Ethylene Metabolism at Alternating Temperatures in Wild Cardoon" Plants 9, no. 9: 1225. https://doi.org/10.3390/plants9091225
APA StyleHuarte, H. R., Puglia, G. D., Prjibelski, A. D., & Raccuia, S. A. (2020). Seed Transcriptome Annotation Reveals Enhanced Expression of Genes Related to ROS Homeostasis and Ethylene Metabolism at Alternating Temperatures in Wild Cardoon. Plants, 9(9), 1225. https://doi.org/10.3390/plants9091225







