Unravelling Cotton Nonexpressor of Pathogenesis-Related 1(NPR1)-Like Genes Family: Evolutionary Analysis and Putative Role in Fiber Development and Defense Pathway
Abstract
1. Introduction
2. Results
2.1. Identification and Classification of NPR Homologs in Cotton
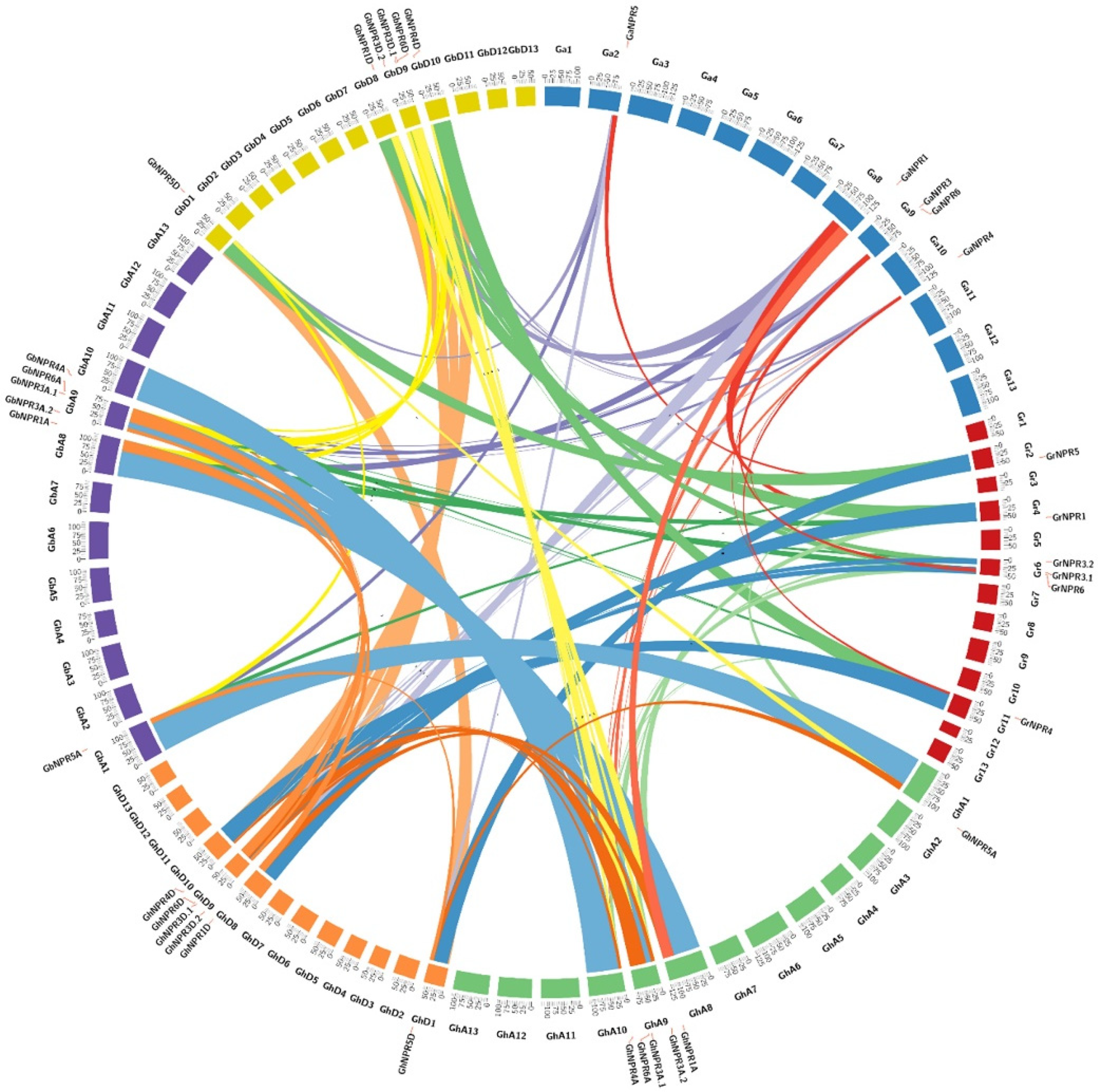
2.2. Phylogenetic and Synteny Analysis of Cotton NPR1-Like Protein Family
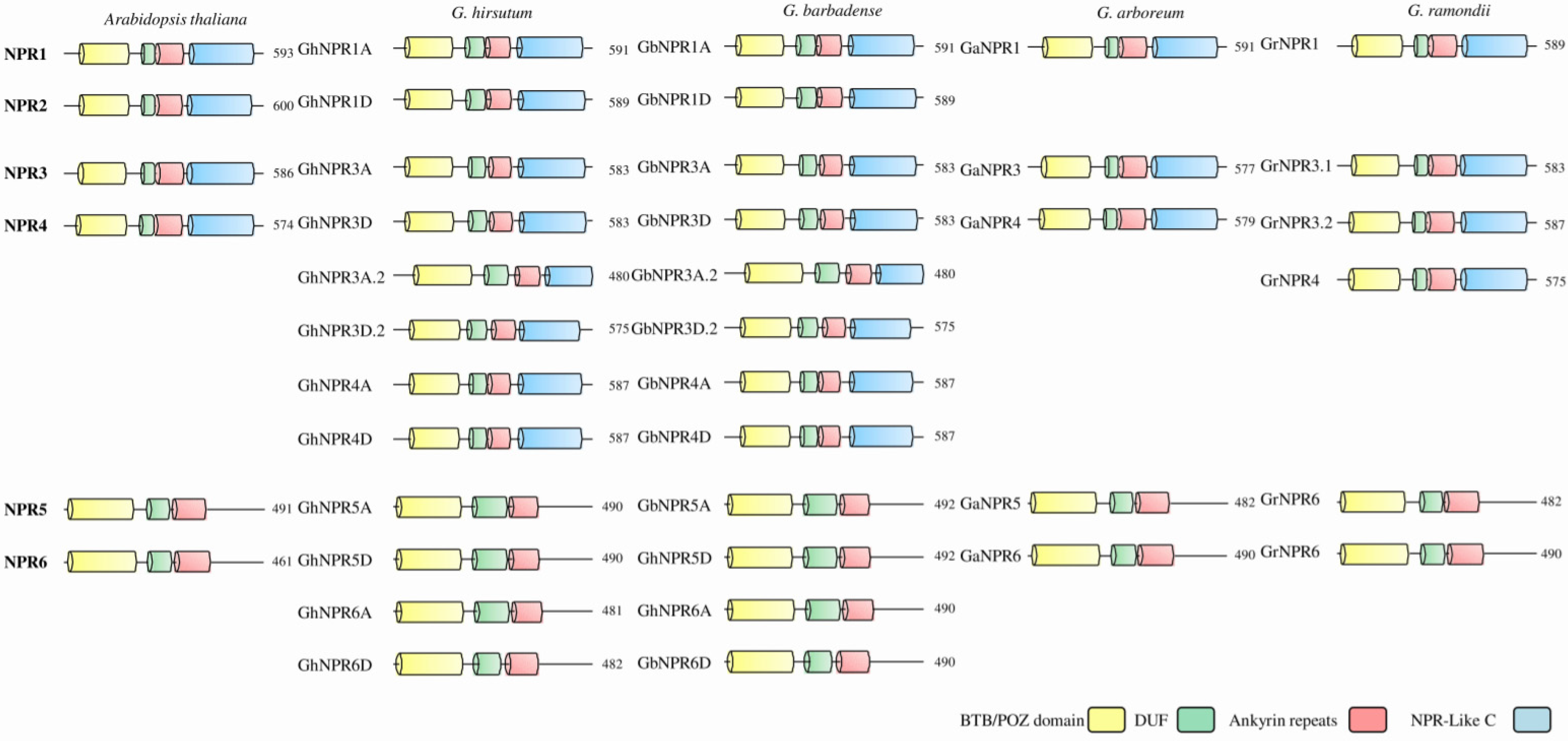
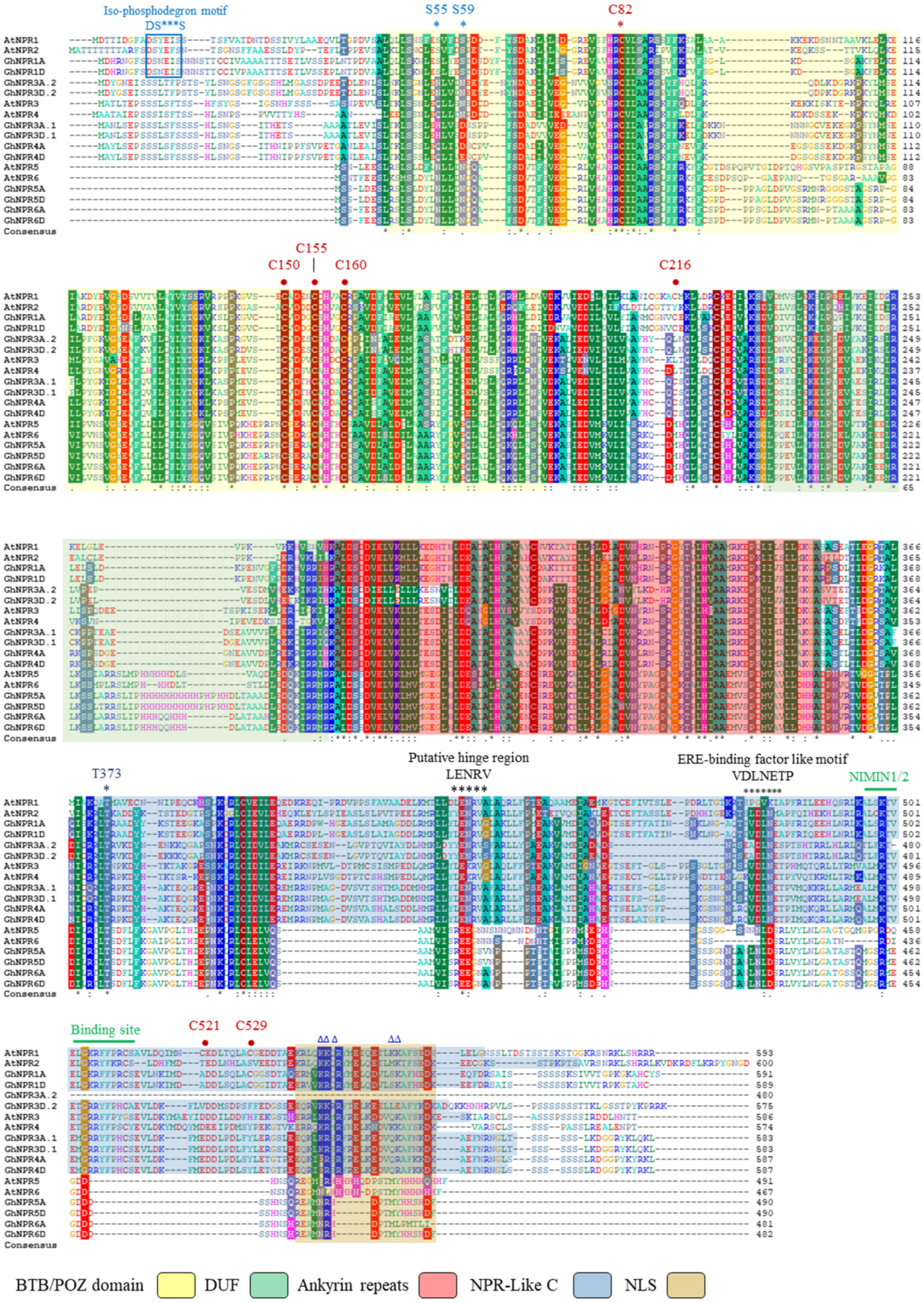
2.3. Cotton NPR1-Like Gene Family Protein and Gene Structural Properties
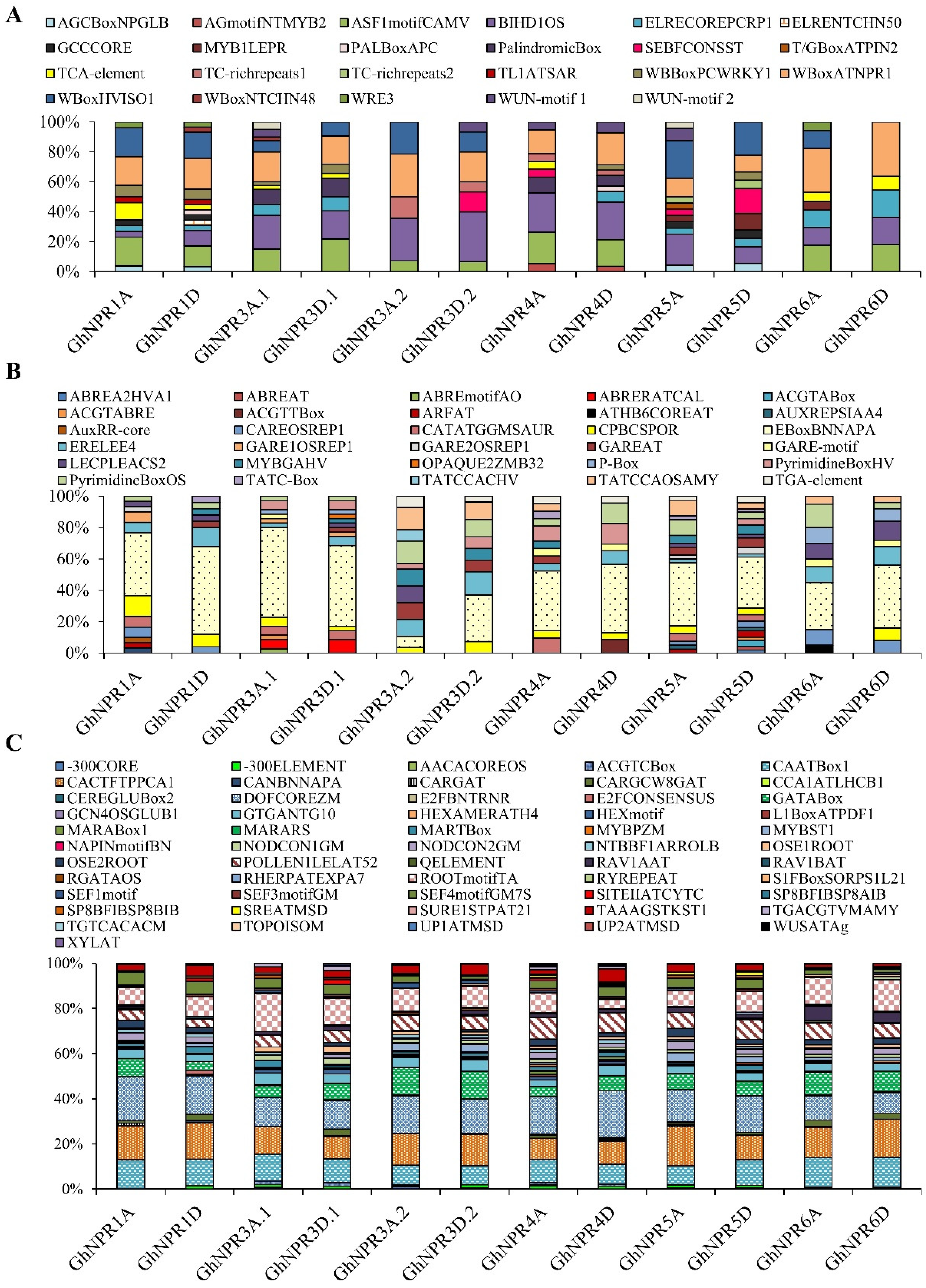
2.4. Prediction of the Cis-Regulatory Elements in Cotton NPR1-Like Gene Family Promoter
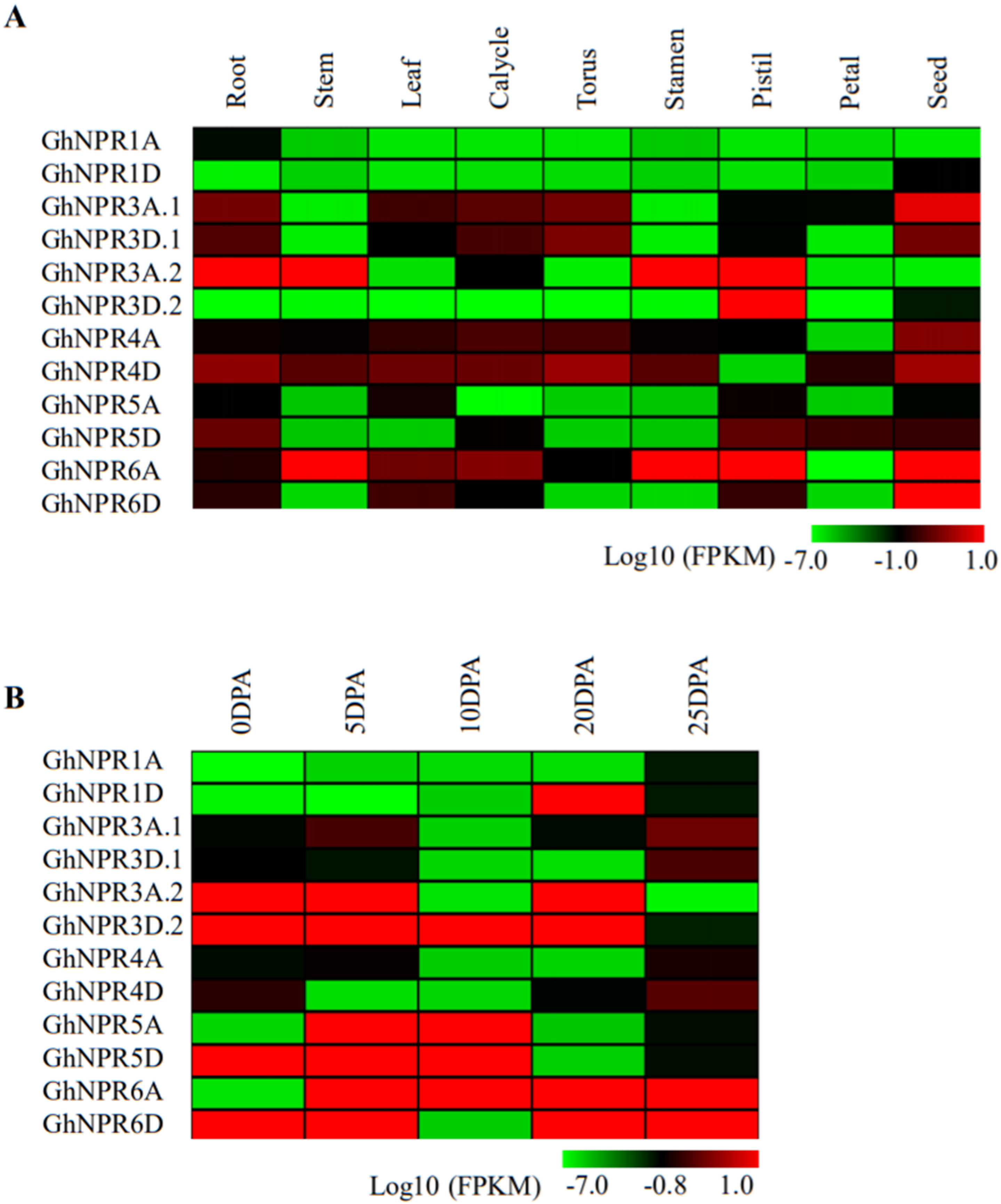
2.5. In Silico Gene Expression Pattern of Cotton NPR1-Like Gene Family
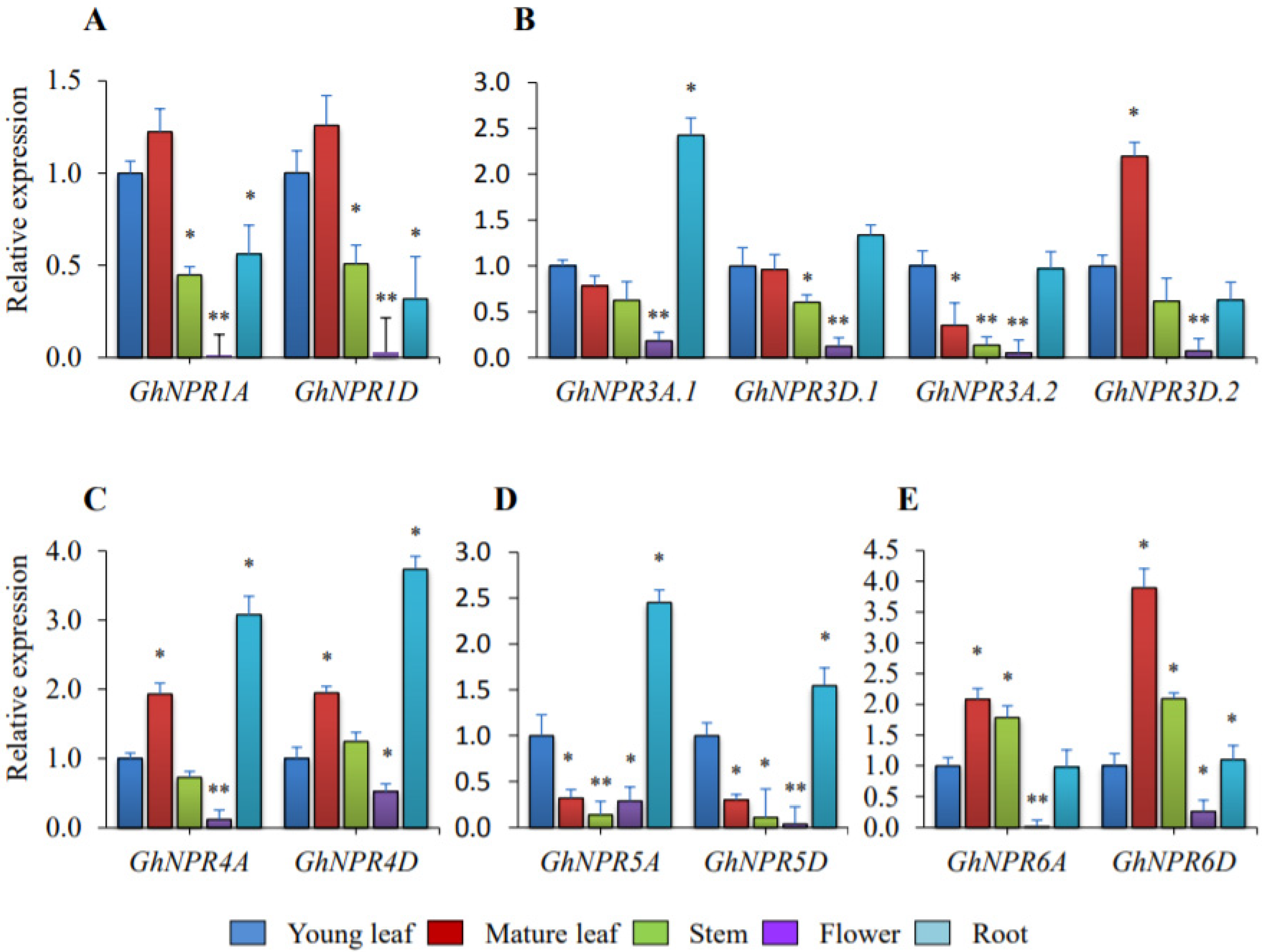
2.6. Expression Analysis of the Cotton NPR1-Like Gene Family in Developmental Tissue
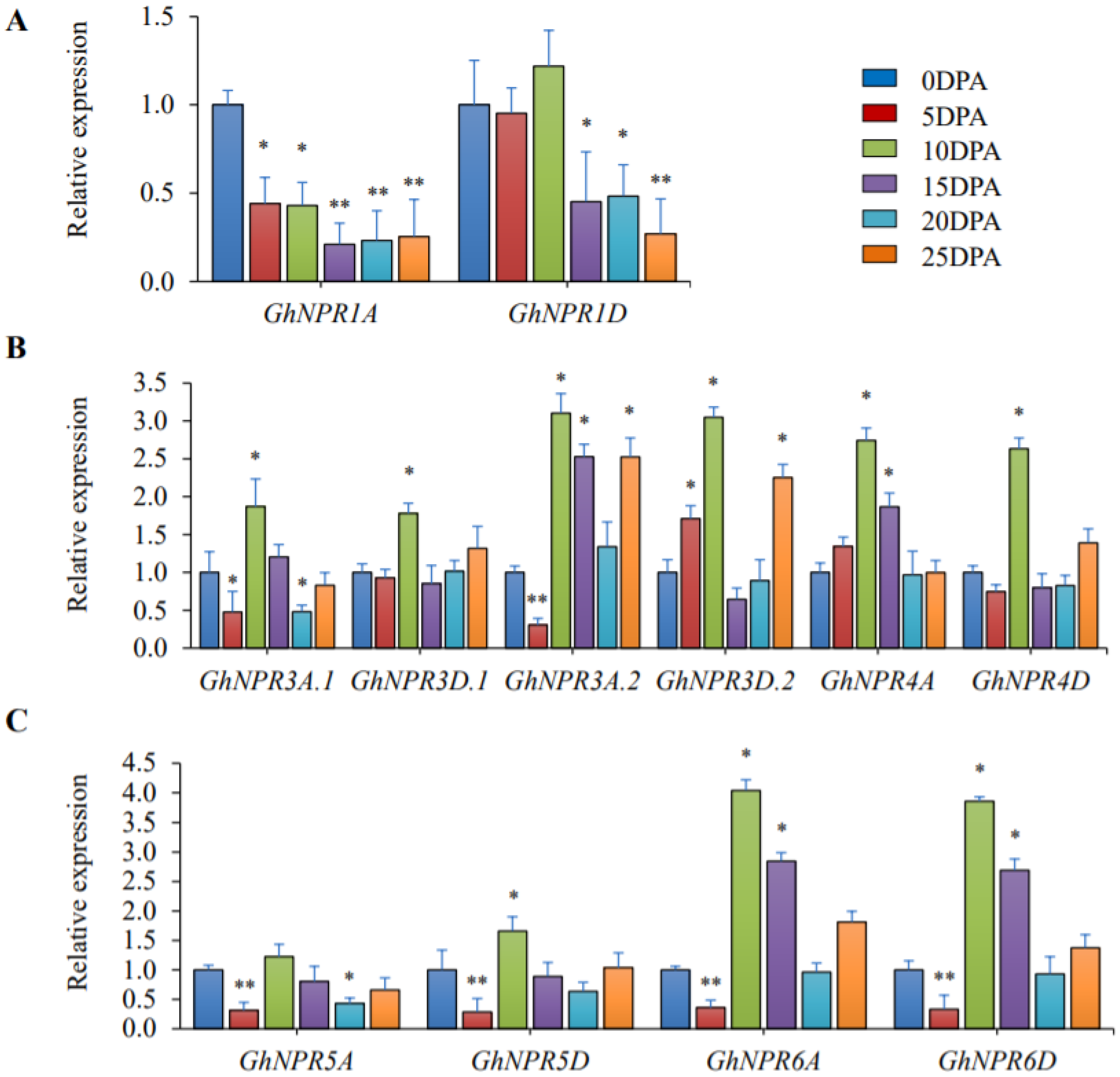
2.7. Expression Analysis of the Cotton NPR1-Like Gene Family in Different Fiber Stages
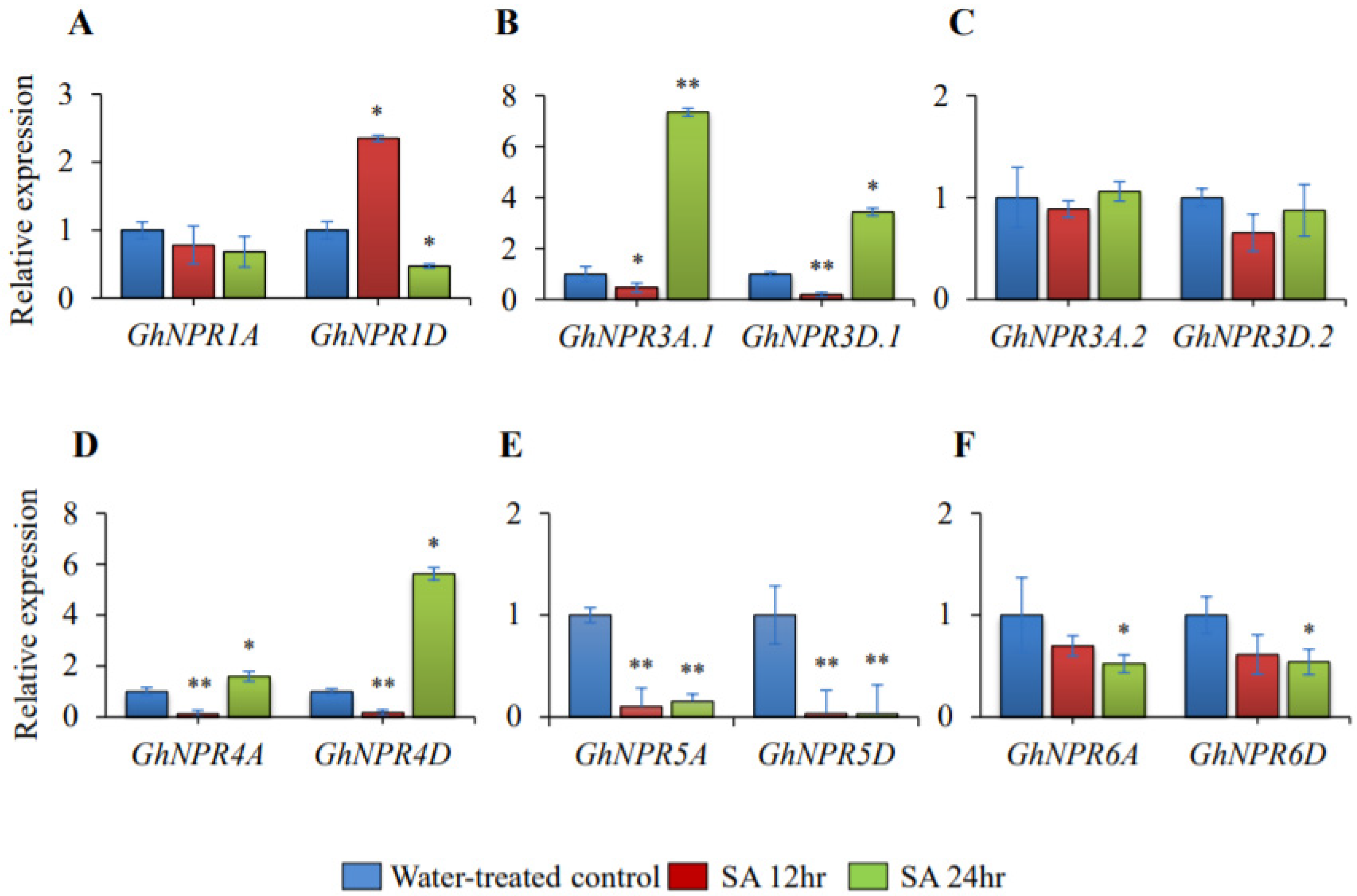
2.8. Expression Analysis of the Cotton NPR1-Like Gene Family During SA-Induction
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of NPR1-Like Genes Family in Cotton
4.3. Protein Structure Analysis and Domain Distribution
4.4. Physical Property Analysis of Cotton NPR Genes
4.5. Analysis of Gene Structure and Chromosomal Localization
4.6. Phylogenetic and Synteny Analysis
4.7. Conserved Cis-Element Analysis in the Promoter
4.8. Analysis of RNA-Seq Data for Putative GhNPR Genes in Different Tissues and Fiber Development Stages
4.9. RNA Extraction and Real-Time PCR Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Mou, Z. Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 2011, 53, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.; Carr, J.P.; Klessig, D.F.; Raskin, I. Salicylic Acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 1990, 250, 1002–1004. [Google Scholar] [CrossRef]
- Cao, H.; Bowling, S.A.; Gordon, A.S.; Dong, X. Characterization of an Arabidopsis Mutant That Is Nonresponsive to Inducers of Systemic Acquired Resistance. Plant Cell 1994, 6, 1583–1592. [Google Scholar] [CrossRef]
- Delaney, T.P.; Friedrich, L.; Ryals, J.A. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 1995, 92, 6602–6606. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 Protein Is a Receptor for the Plant Defense Hormone Salicylic Acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Mou, Z. Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis. J. Plant Physiol. 2010, 167, 144–148. [Google Scholar] [CrossRef]
- Rochon, A.; Boyle, P.; Wignes, T.; Fobert, P.R.; Despres, C. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 2006, 18, 3670–3685. [Google Scholar] [CrossRef]
- Spoel, S.H.; Mou, Z.; Tada, Y.; Spivey, N.W.; Genschik, P.; Dong, X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 2009, 137, 860–872. [Google Scholar] [CrossRef]
- Hepworth, S.R.; Zhang, Y.; McKim, S.; Li, X.; Haughn, G.W. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 2005, 17, 1434–1448. [Google Scholar] [CrossRef] [PubMed]
- Makandar, R.; Essig, J.S.; Schapaugh, M.A.; Trick, H.N.; Shah, J. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant Microbe Interact. 2006, 19, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Dutt, M.; Barthe, G.; Irey, M.; Grosser, J. Transgenic Citrus Expressing an Arabidopsis NPR1 Gene Exhibit Enhanced Resistance against Huanglongbing (HLB.; Citrus Greening). PLoS ONE 2015, 10, e0137134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Francis, M.I.; Dawson, W.O.; Graham, J.H.; Orbovic, V.; Triplett, E.W.; Mou, Z.L. Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur. J. Plant Pathol. 2010, 128, 91–100. [Google Scholar] [CrossRef]
- Chern, M.S.; Fitzgerald, H.A.; Yadav, R.C.; Canlas, P.E.; Dong, X.; Ronald, P.C. Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 2001, 27, 101–113. [Google Scholar] [CrossRef]
- Molla, K.A.; Karmakar, S.; Chanda, P.K.; Sarkar, S.N.; Datta, S.K.; Datta, K. Tissue-specific expression of Arabidopsis NPR1 gene in rice for sheath blight resistance without compromising phenotypic cost. Plant Sci. 2016, 250, 105–114. [Google Scholar] [CrossRef]
- Lin, W.C.; Lu, C.F.; Wu, J.W.; Cheng, M.L.; Lin, Y.M.; Yang, N.S.; Black, L.; Green, S.K.; Wang, J.F.; Cheng, C.P. Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 2004, 13, 567–581. [Google Scholar] [CrossRef]
- Potlakayala, S.D.; DeLong, C.; Sharpe, A.; Fobert, P.R. Conservation of NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES1 function between Arabidopsis thaliana and Brassica napus. Physiol. Mol. Plant Pathol. 2007, 71, 174–183. [Google Scholar] [CrossRef]
- Meur, G.; Budatha, M.; Srinivasan, T.; Rajesh Kumar, K.R.; Dutta Gupta, A.; Kirti, P.B. Constitutive expression of Arabidopsis NPR1 confers enhanced resistance to the early instars of Spodoptera litura in transgenic tobacco. Physiol. Plant 2008, 133, 765–775. [Google Scholar] [CrossRef]
- Wally, O.; Jayaraj, J.; Punja, Z.K. Broad-spectrum disease resistance to necrotrophic and biotrophic pathogens in transgenic carrots (Daucus carota L.) expressing an Arabidopsis NPR1 gene. Planta 2009, 231, 131–141. [Google Scholar] [CrossRef]
- Kumar, V.; Joshi, S.G.; Bell, A.A.; Rathore, K.S. Enhanced resistance against Thielaviopsis basicola in transgenic cotton plants expressing Arabidopsis NPR1 gene. Transgenic Res. 2013, 22, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Parkhi, V.; Kumar, V.; Campbell, L.M.; Bell, A.A.; Shah, J.; Rathore, K.S. Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res. 2010, 19, 959–975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Cheng, C.; Gao, Q.; Liu, J.; Guo, X. Molecular cloning and characterization of GhNPR1, a gene implicated in pathogen responses from cotton (Gossypium hirsutum L.). Biosci. Rep. 2008, 28, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Mahomed, W.; Reeksting, B.J.; Engelbrecht, J.; Ibarra-Laclette, E.; van den Berg, N. Phylogenetic and expression analysis of the NPR1-like gene family from Persea americana (Mill.). Front. Plant Sci. 2015, 6, 300. [Google Scholar] [CrossRef]
- Peraza-Echeverria, S.; Santamaría, J.M.; Fuentes, G.; de los Ángeles Menéndez-Cerón, M.; Vallejo-Reyna, M.Á.; Herrera-Valencia, V.A. The NPR1 family of transcription cofactors in papaya: Insights into its structure, phylogeny and expression. Genes Genom. 2012, 34, 379–390. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Niu, X.; Xu, Q.; Yang, L. Genome-Wide Identification and Analysis of the NPR1-Like Gene Family in Bread Wheat and Its Relatives. Int. J. Mol. Sci. 2019, 20, 5974. [Google Scholar] [CrossRef]
- Zhang, J.; Jiao, P.; Zhang, C.; Tong, X.; Wei, Q.; Xu, L. Apple NPR1 homologs and their alternative splicing forms may contribute to SA and disease responses. Tree Genet. Genomes 2016, 12, 92. [Google Scholar] [CrossRef]
- Shu, L.J.; Liao, J.Y.; Lin, N.C.; Chung, C.L. Identification of a strawberry NPR-like gene involved in negative regulation of the salicylic acid-mediated defense pathway. PLoS ONE 2018, 13, e0205790. [Google Scholar] [CrossRef]
- Phillips, A.Z.; Berry, J.C.; Wilson, M.C.; Vijayaraghavan, A.; Burke, J.; Bunn, J.I.; Allen, T.W.; Wheeler, T.; Bart, R.S. Genomics-enabled analysis of the emergent disease cotton bacterial blight. PLoS Genet. 2017, 13, e1007003. [Google Scholar] [CrossRef]
- Dixit, G.; Srivastava, A.; Rai, K.M.; Dubey, R.S.; Srivastava, R.; Verma, P.C. Distinct defensive activity of phenolics and phenylpropanoid pathway genes in different cotton varieties toward chewing pests. Plant Signal. Behav. 2020, 15, 1747689. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fan, G.; Wang, K.; Sun, F.; Yuan, Y.; Song, G.; Li, Q.; Ma, Z.; Lu, C.; Zou, C.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Backer, R.; Naidoo, S.; van den Berg, N. The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and Related Family: Mechanistic Insights in Plant Disease Resistance. Front. Plant Sci. 2019, 10, 102. [Google Scholar] [CrossRef]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Kinkema, M.; Fan, W.; Dong, X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 2000, 12, 2339–2350. [Google Scholar] [CrossRef]
- Castelló, M.J.; Medina-Puche, L.; Lamilla, J.; Tornero, P. NPR1 paralogs of Arabidopsis and their role in salicylic acid perception. PLoS ONE 2018, 13, e0209835. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, P.; Luo, X.; Yang, C.; Tang, Y.; Wang, Z.; Hu, G.; Ge, X.; Xia, G.; Wu, J. Cotton plant defence against a fungal pathogen is enhanced by expanding BLADE-ON-PETIOLE1 expression beyond lateral-organ boundaries. Commun. Biol. 2019, 2, 238. [Google Scholar] [CrossRef]
- Pajerowska-Mukhtar, K.M.; Emerine, D.K.; Mukhtar, M.S. Tell me more: Roles of NPRs in plant immunity. Trends Plant Sci. 2013, 18, 402–411. [Google Scholar] [CrossRef]
- Ha, C.M.; Jun, J.H.; Nam, H.G.; Fletcher, J.C. BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, Y.T.; Qu, N.; Zhao, Q.; Bi, D.; Li, X. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 2006, 48, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Salasini, B.C.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, S.R. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate BLADE-ON-PETIOLE-Dependent Regulation of Development. Plant Physiol. 2019, 180, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Innes, R. The Positives and Negatives of NPR: A Unifying Model for Salicylic Acid Signaling in Plants. Cell 2018, 173, 1314–1315. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Withers, J.; Mohan, R.; Marqués, J.; Gu, Y.; Yan, S.; Zavaliev, R.; Nomoto, M.; Tada, Y.; Dong, X. Posttranslational Modifications of the Master Transcriptional Regulator NPR1 Enable Dynamic but Tight Control of Plant Immune Responses. Cell Host Microbe 2015, 18, 169–182. [Google Scholar] [CrossRef]
- Withers, J.; Dong, X. Posttranslational Modifications of NPR1: A Single Protein Playing Multiple Roles in Plant Immunity and Physiology. PLoS Pathog. 2016, 12, e1005707. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef]
- Lee, H.J.; Park, Y.J.; Seo, P.J.; Kim, J.H.; Sim, H.J.; Kim, S.G.; Park, C.M. Systemic Immunity Requires SnRK2.8-Mediated Nuclear Import of NPR1 in Arabidopsis. Plant Cell 2015, 27, 3425–3438. [Google Scholar] [CrossRef]
- Maier, F.; Zwicker, S.; Hückelhoven, A.; Meissner, M.; Funk, J.; Pfitzner, A.J.P.; Pfitzner, U.M. NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1) and some NPR1-related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 2011, 12, 73–91. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Pandey, B.; Singh, S.P.; Sawant, S.V. Spt-Ada-Gcn5-Acetyltransferase (SAGA) Complex in Plants: Genome Wide Identification, Evolutionary Conservation and Functional Determination. PLoS ONE 2015, 10, e0134709. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Srivastava, M.; Kumar, V.; Pandey, B.; Singh, S.P.; Bag, S.K.; Singh, B.D.; Tuli, R.; Sawant, S.V. Distinct Role of Core Promoter Architecture in Regulation of Light Mediated Responses in Plant Genes. Mol. Plant 2014, 7, 626–641. [Google Scholar] [CrossRef]
- Biłas, R.; Szafran, K.; Hnatuszko-Konka, K.; Kononowicz, A.K. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cul. (PCTOC) 2016, 127, 269–287. [Google Scholar] [CrossRef]
- Pandey, B.; Prakash, P.; Verma, P.C.; Srivastava, R. Regulated gene expression by synthetic modulation of the promoter architecture in plants. In Current Developments in Biotechnology and Bioengineering: Synthetic Biology, Cell Engineering and Bioprocessing Technologies; Elsevier: Amsterdam, The Netherlands, 2019; pp. 235–255. [Google Scholar]
- Srivastava, R.; Srivastava, R.; Singh, U.M. Understanding the patterns of gene expression during climate change. In Climate Change Effect on Crop Productivity; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2014; pp. 279–328. ISBN 978-1-4822-2920-2. [Google Scholar]
- Liu, W.; Stewart, C.N. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Rai, K.M.; Srivastava, R. Plant Biosynthetic Engineering Through Transcription Regulation: An Insight into Molecular Mechanisms During Environmental Stress. In Biosynthetic Technology and Environmental Challenges; Springer Nature Singapore Pte Ltd.: Singapore, 2018; pp. 51–72. ISBN 9789811074332. ISBN 9789811074349 (online). [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E. Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1998, 1, 311–315. [Google Scholar] [CrossRef]
- Chen, J.; Mohan, R.; Zhang, Y.; Li, M.; Chen, H.; Palmer, I.A.; Chang, M.; Qi, G.; Spoel, S.H.; Mengiste, T.; et al. NPR1 Promotes Its Own and Target Gene Expression in Plant Defense by Recruiting CDK8. Plant Physiol. 2019, 181, 289–304. [Google Scholar] [CrossRef]
- Srivastava, R.; Ahn, S.H. Modifications of RNA polymerase II CTD: Connections to the histone code and cellular function. Biotechnol. Adv. 2015, 33, 856–872. [Google Scholar] [CrossRef]
- Zhong, X.; Xi, L.; Lian, Q.; Luo, X.; Wu, Z.; Seng, S.; Yuan, X.; Yi, M. The NPR1 homolog GhNPR1 plays an important role in the defense response of Gladiolus hybridus. Plant Cell Rep. 2015, 34, 1063–1074. [Google Scholar] [CrossRef]
- Shi, Z.; Maximova, S.N.; Liu, Y.; Verica, J.; Guiltinan, M.J. Functional analysis of the Theobroma cacao NPR1 gene in arabidopsis. BMC Plant Biol. 2010, 10, 248. [Google Scholar] [CrossRef]
- Sandhu, D.; Tasma, I.M.; Frasch, R.; Bhattacharyya, M.K. Systemic acquired resistance in soybean is regulated by two proteins, Orthologous to Arabidopsis NPR1. BMC Plant Biol. 2009, 9, 105. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. Mev: Multiexperiment viewer. In Biomedical Informatics for Cancer Research; Springer: Berlin/Heidelberg, Germany, 2010; pp. 267–277. [Google Scholar]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Lodhi, N.; Ranjan, A.; Singh, M.; Srivastava, R.; Singh, S.P.; Chaturvedi, C.P.; Ansari, S.A.; Sawant, S.V.; Tuli, R. Interactions between upstream and core promoter sequences determine gene expression and nucleosome positioning in tobacco PR-1a promoter. Biochim. Biophys. Acta 2008, 1779, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Artico, S.; Nardeli, S.M.; Brilhante, O.; Grossi-de-Sa, M.F.; Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed]


| Arabidopsis | Tetraploid Cotton | Diploid Cotton | |||||||
|---|---|---|---|---|---|---|---|---|---|
| G. hirsutum | G. barbadense | G. arboreum | G. raimondii | ||||||
| Gene Name | Gene ID | Gene Name | Gene ID | Gene Name | Gene ID | Gene Name | Gene ID | Gene Name | Gene ID |
| NPR1 | AT1G64280 | GhNPR1A | GH_A08G2791 | GbNPR1A | GB_A08G2903 | GaNPR1 | Ga08G2883 | GrNPR1 | Gorai.004G284300 |
| GhNPR1D | GH_D08G2784 | GbNPR1D | GB_D08G2893 | - | - | - | - | ||
| NPR2 | AT4G26120 | - | - | - | - | - | - | - | - |
| NPR3 | AT5G45110 | GhNPR3A.1 | GH_A09G0955 | GbNPR3A.1 | GB_A09G1067 | GaNPR3 | Ga09G0926 | GrNPR3.1 | Gorai.006G091900 |
| GhNPR3D.1 | GH_D09G0911 | GbNPR3D.1 | GB_D09G0919 | - | - | GrNPR3.2 | Gorai.006G009000 | ||
| GhNPR3A.2 | GH_A09G0085 | GbNPR3A.2 | GB_A09G0108 | - | - | - | - | ||
| GhNPR3D.2 | GH_D09G0089 | GbNPR3D.2 | GB_D09G0086 | - | - | - | - | ||
| NPR4 | AT4G19660 | GhNPR4A | GH_A10G0470 | GbNPR4A | GB_A10G0469 | GaNPR4 | Ga10G2594 | GrNPR4 | Gorai.011G050200 |
| GhNPR4D | GH_D10G0496 | GbNPR4D | GB_D10G0482 | - | - | - | - | ||
| NPR5 | AT2G41370 | GhNPR5A | GH_A01G2100 | GbNPR5A | GB_A01G2205 | GaNPR5 | Ga02G1425 | GrNPR5 | Gorai.002G226700 |
| GhNPR5D | GH_D01G2194 | GbNPR5D | GB_D01G2289 | - | - | - | - | ||
| NPR6 | AT3G57130 | GhNPR6A | GH_A09G1355 | GbNPR6A | GB_A09G1470 | GaNPR6 | Ga09G1363 | GrNPR6 | Gorai.006G133600 |
| GhNPR6D | GH_D09G1306 | GbNPR6D | GB_D09G1311 | - | - | - | - | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agarwal, N.; Srivastava, R.; Verma, A.; Rai, K.M.; Singh, B.; Verma, P.C. Unravelling Cotton Nonexpressor of Pathogenesis-Related 1(NPR1)-Like Genes Family: Evolutionary Analysis and Putative Role in Fiber Development and Defense Pathway. Plants 2020, 9, 999. https://doi.org/10.3390/plants9080999
Agarwal N, Srivastava R, Verma A, Rai KM, Singh B, Verma PC. Unravelling Cotton Nonexpressor of Pathogenesis-Related 1(NPR1)-Like Genes Family: Evolutionary Analysis and Putative Role in Fiber Development and Defense Pathway. Plants. 2020; 9(8):999. https://doi.org/10.3390/plants9080999
Chicago/Turabian StyleAgarwal, Neha, Rakesh Srivastava, Akash Verma, Krishan Mohan Rai, Babita Singh, and Praveen Chandra Verma. 2020. "Unravelling Cotton Nonexpressor of Pathogenesis-Related 1(NPR1)-Like Genes Family: Evolutionary Analysis and Putative Role in Fiber Development and Defense Pathway" Plants 9, no. 8: 999. https://doi.org/10.3390/plants9080999
APA StyleAgarwal, N., Srivastava, R., Verma, A., Rai, K. M., Singh, B., & Verma, P. C. (2020). Unravelling Cotton Nonexpressor of Pathogenesis-Related 1(NPR1)-Like Genes Family: Evolutionary Analysis and Putative Role in Fiber Development and Defense Pathway. Plants, 9(8), 999. https://doi.org/10.3390/plants9080999






