Abstract
The pyrabactin resistance (PYR)/PYR1-like (PYL)/regulatory components of ABA receptor (RCAR) (known as PYLs for short) have been identified and characterized as the ABA receptors in some plants. However, little is known about the details regarding PYL family genes in the apple (Malus domestica). In this study, we identified 13 apple PYLs, termed MdPYL1-13, which could be classified into four groups according to structural features of the amino acid sequence. The gene structures and conserved motifs analysis found that the majority of MdPYLs had a similar number of exons and similar conserved motif profile in the same group. In addition, 11 gene pairs were identified to exhibit synteny by synteny analysis between the apple and Arabidopsis. Furthermore, we investigated MdPYLs transcript level in various organs of the red-fleshed apple (Malus sieversii f. Neidzwetzkyana (Dieck) Langenf) ‘Xinjiang No.1’. The results suggested all MdPYLs within group I were expressed at relatively higher levels in all of the organs tested. However, the genes of group IV had little or no variation. Additionally, we found various hormone and stress-related cis-elements in the promoters of MdPYLs by analyzing cis-elements. Therefore, the expression levels of all MdPYLs were further detected under ABA, PEG, salt, and cold stresses in ‘Xinjiang No.1’ seedlings. We found that all MdPYLs except for MdPYL11 were upregulated by ABA treatment, 10 genes were upregulated by PEG treatment, 12 genes were upregulated by NaCl treatment, and six genes were upregulated by cold treatment (4 °C) while seven genes were downregulated. Thus, these MdPYLs might be involved in the defense against abiotic stresses. In addition, the interaction between 13 MdPYLs and two 2C protein phosphatases in the apple (MdPP2C65 and MdPP2C72) was investigated in yeast two-hybrid assays. These results suggested that MdPYLs may bind to MdPP2C65 and MdPP2C72 in different manners and with different intensity. Our studies provide useful information for further investigating and researching the regulatory mechanisms of PYL family genes in response to abiotic stresses in the apple.
1. Introduction
Abscisic acid (ABA) is an important phytohormone involved in plant growth, plant development, and protection of plants against abiotic stresses [1,2]. The core components of ABA signaling included the regulatory component of ABA receptors (PYLs), group A type 2C protein phosphatases (PP2Cs), and the sucrose nonfermenting 1-related protein kinases (SnRKs) [3,4,5,6]. The PYLs interacted with PP2Cs to form PYL-PP2C heterodimer, which inhibited the phosphatase activity of key negative regulators PP2Cs, thereby leading to activation of pivotal positive regulators SnRK2s [7,8,9,10].
PYLs, as ABA receptors, belong to the StAR-related lipid-transfer (START) superfamily containing a ligand-binding pocket enclosed by four conserved loops, CL1-CL4 [4,6], which is important in ABA signal transduction pathway. To date, 14 PYLs containing the conserved structure have been recognized in the Arabidopsis genome [4,6,11], including AtPYR1 (Arabidopsis thaliana pyrabactin resistance1) and 13 AtPYLs (A. thaliana PYR1-like1-13) [12]. Some PYLs, such as AtPYR1, AtPYL1, AtPYL2, and AtPYL3 inhibited PP2Cs in an ABA-dependent manner, while others including AtPYL4, AtPYL5, AtPYL6, AtPYL8, AtPYL9, AtPYL10, and AtPYL13 showed suppression of PP2Cs in the absence of ABA [10,12,13]. In addition, several of these genes were showed having diverse functions in the regulation of stomatal closure [14], drought resistance [5], leaf senescence [15], and root development [16,17,18].
Since the discovery of the PYL family in Arabidopsis, orthologous genes of PYL in other crops have been characterized at genome-wide levels, including six PYLs in sweet orange [19], eight PYLs in grape [20], 13 PYLs in rice [21], 14 PYLs in tomato [22], 14 PYLs in rubber tree [23], 24 PYLs in Brassica rapa [24], 27 PYLs in cotton [25], and 29 PYLs in tobacco [26]. Recently, the study regarding the roles of PYLs in other plant species besides Arabidopsis has begun to be reported, and the various function of the PYLs in plant development and stress resistance was emphasized. For instance, overexpression of rice OsPYL3, cotton GhPYL10/12/26, or maize ZmPYL3/9/10/13 dramatically enhanced the sensitivity to ABA in transgenic Arabidopsis [27,28,29]. Similarly, overexpression of rice OsPYL/RCAR5 or OsPYL3/5/9/11 also increased the sensitivity to ABA and the tolerance to drought stress in transgenic rice [30,31,32]. Overexpressing OsPYL9 significantly enhanced the tolerance to drought stress and leaf senescence induced by drought in transgenic Arabidopsis and rice plants [15]. Moreover, overexpression of Arabidopsis AtPYL4 (A194T), AtPYL5, maize ZmPYL8/9/12, or cotton GhPYL9-11A greatly improved tolerance to drought in transgenic Arabidopsis plants, respectively [5,29,33,34]. In recent years, in addition to herbs, the function of the PYLs in woody and lianas plants has also been reported. Overexpression of Populus trichocarpa PtPYRL1 or PtPYRL5 in both Arabidopsis and poplar also enhanced ABA sensitivity and drought resistance [35]. In grape, VvPYL1 could be combined with ABA and repress the ABI1 phosphatase activity [36]. The VvPYL1 (VvRCAR7) exhibited high expression level in grape leaves treated by drought, salt, and cold stresses [37].
Unfortunately, in spite of an increase in reports in Arabidopsis and other plants such as maize, rice, and grape, the information concerning PYLs remains elusive in the apple. In this study, for the first time, we systematically identified the MdPYLs at the genome level in the M. domestica and performed their exon–intron structure and synteny analysis. Moreover, we investigated the spatiotemporal expression patterns of MdPYLs in various apple organs. Finally, we also examined expression patterns of MdPYLs under various abiotic stresses treatments. Our results will provide some clues for further investigating the function of PYL genes in apple tree growth and development, and resistance to abiotic stress.
2. Results
2.1. Identification and Expansion Patterns of MdPYLs in the M. domestica Genome
We determined that 13 putative MdPYLs were present in the M. domestica genome through BLASTP by using 14 AtPYL sequences as references. Then, we termed MdPYL1 to MdPYL13 based on their chromosomal order (Figure 1). Information regarding MdPYLs was provided in Table 1 consisting of the gene name, gene ID, length of coding DNA sequence (CDS), theoretical isoelectric point (pI), molecular weight (MW), and length of protein. The sequence analysis revealed that the MdPYLs lengths changed obviously in size, ranging from 558 bp (MdPYL5) to 3252 bp (MdPYL1), with one to four exons in each sequence. However, the lengths of CDS and the corresponding MdPYL proteins both had no significant difference, varying from 555 bp (MdPYL1 and MdPYL7) to 777 bp (MdPYL4) and from 184 aa (MdPYL1 and MdPYL7) to 258 aa (MdPYL4), respectively. The predicted MW and pI values of these proteins ranged respectively from 20.13 kDa (MdPYL5) to 27.66 kDa (MdPYL4) and 5.02 (MdPYL10) to 8.30 (MdPYL11) (Table 1).
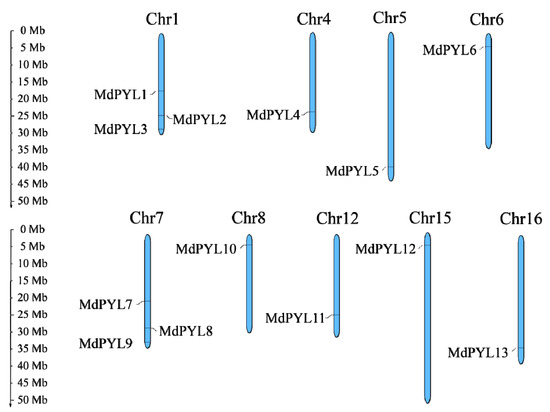
Figure 1.
Chromosomal distribution and localization of MdPYLs. Only nine chromosomes contained MdPYLs are represented in this figure. The chromosome names are shown at the top of each chromosome. The chromosome scale is in millions of bases (Mb) on the left.

Table 1.
Basic informations of 13 MdPYLs identified in apple.
Moreover, according to available annotation information of the GDR database, 13 putative MdPYLs were distributed on nine chromosomes including chromosome 1, 4, 5, 6, 7, 8, 12, 15, and 16 (Figure 1). Three MdPYLs were present on chromosome 1 and 7, respectively, and one MdPYL on the remaining chromosomes (Figure 1).
2.2. Phylogenetic Analysis of the MdPYLs Family
We constructed a phylogenetic tree based on 109 protein sequences of PYLs from seven species, including 14 AtPYLs from A. thaliana, 13 MdPYLs from M. domestica, 7 RcPYLs from R. communis, 5 VvPYLs from V. vinifera, 9 BdPYLs from B. distachyon, 12 OsPYLs from O. sativa, 9 TcPYLs from T. cacao, and 40 GhPYLs from G. hirsutum (Table S1) via MEGA7 software using the neighbor-joining algorithm. In general, the 109 plant PYLs tested were classified into four subgroups and named as group I–IV (Figure 2, Tables S1 and S2). However, the plant PYL family evolutionary relationship was diverse in various species of plants. As expected, MdPYLs from apple generally showed closer genetic relationships to PYLs of dicotyledonous angiosperms (V. vinifera, A. thaliana, G. hirsutum, R. communi, and T. cacao) than monocotyledonous angiosperms (O. sativa and B. distachyon) (Figure 2). The results suggested plant PYLs classifying into the same groups might have similar functions.
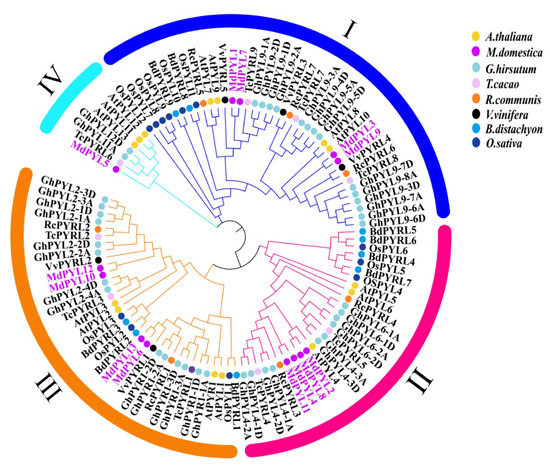
Figure 2.
Phylogenetic analysis of PYLs from seven species. Different groups of MdPYLs are indicated in different colors bars. Circles of different colors denote PYLs from different species, including 14 AtPYLs from A. thaliana (yellow dot), 13 MdPYLs from M. domestica (purple dot), 7 RcPYLs from R. communis (orange dot), 5 VvPYLs from V. vinifera (black dot), 9 BdPYLs from B. distachyon (blue dot), 12 OsPYLs from O. sativa (navy blue dot), 9 TcPYLs from T. cacao (pink dot) and 40 GhPYLs from G. hirsutum (light blue dot). All amino acid sequences used in this analysis are listed in Supplementary Table S1.
2.3. Gene Structure and Duplication Analysis of MdPYLs
To study the structure and expansion mechanisms of MdPYLs gene family, we carried out the analysis of amino acid alignment, phylogenetic tree, exon–intron structures, the conserved motifs, and segmental duplications (Figure 3). In general, 13 MdPYLs protein all contained three α-helixes (α1–α4), seven-stranded β-sheet (β1–β7), and four highly conserved surface loops (CL1–CL4). (Figure 3a). The structure had been well characterized in the AtPYLs gene family, which involved in ABA binding and inhibition of PP2Cs [38]. Moreover, the majority of MdPYLs had a similar profile of exons, and length of ORFs in the same group (Figure 3b,c). For example, all MdPYLs within group I included three exons, while MdPYLs in groups II and III had only one exon (no intron), except MdPYL4 and MdPYL11 both contained two exons (Figure 3b,c). Meanwhile, all of MdPYLs shared highly conserved motif 1, 2, and 3, and MdPYLs belonging to the same group seemed to have a similar conserved motif distribution. For example, in addition to the motif 1, 2, and 3, all of group I, II, and III respectively shared the highly conserved motifs 7 and 10, motif 1, and motif 8 (Figure 3d,e). To further understand the expansion mechanism of the MdPYLs, we also examined segmental and tandem duplications within the apple genome. No tandem duplications were identified and eight segmental duplication were found in 12 pairs of duplicated genomic regions, respectively, including (MdPYL1/MdPYL3, MdPYL1/7, MdPYL2/MdPYL8, MdPYL3/MdPYL7, MdPYL3/MdPYL9, MdPYL4/ MdPYL11, MdPYL6/MdPYL13, MdPYL10/MdPYL12) (Figure 4).
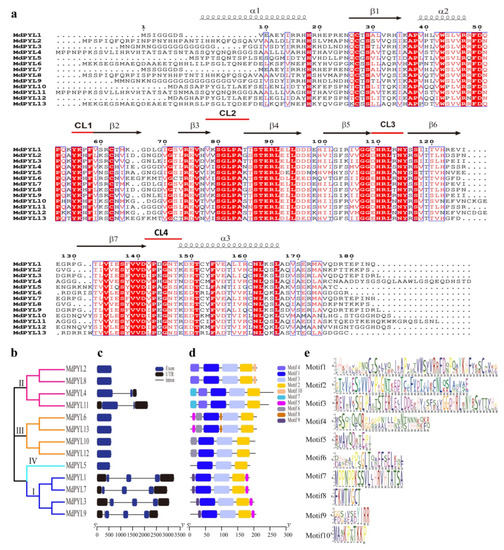
Figure 3.
Amino acid sequence alignment, phylogenetic relationships, gene structure, and conserved motifs of MdPYLs. (a) Amino acid sequence alignment and secondary structural elements of the 13 MdPYLs. CL1–CL4 shown with red lines represent four conserved ABA receptor regions. (b) The phylogenetic tree of MdPYLs. Bars of different colors represent different groups of PYLs. (c) Exon–intron structure of apple MdPYLs. Black boxes denote untranslated regions; blue boxes denote exons; black lines denote introns. (d) Distributions of ten conserved motifs in MdPYLs. (e) The sequence logos of ten conserved motifs.
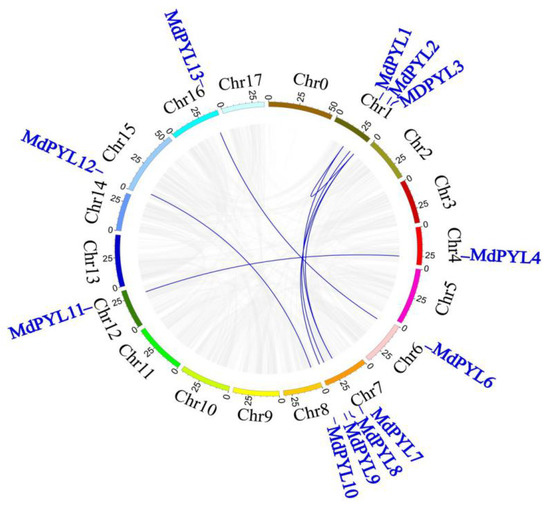
Figure 4.
The synteny analysis of MdPYLs in the apple. Gray lines denote all synteny blocks in the apple genome, and the blue lines denote MdPYL duplicated gene pairs.
2.4. Evolutionary Relationship of PYLs Between Apple and Arabidopsis
Comparative genomic analysis between the apple and Arabidopsis genomes found the syntenies were clear and included the following ortholog pairs: MdPYL1, 7-AtPYL9; MdPYL2, 8-AtPYL5, 6; MdPYL5-AtPYL13; MdPYL6, 13-AtPYL1; MdPYL11-AtPYL4 and MdPYL12-AtPYL2 (Figure 5). Furthermore, AtPYLs and MdPYLs from the same group had similar exon–intron structures (Figure 6a,b). Meanwhile, all of the AtPYLs and MdPYLs shared highly conserved motifs 1, 2, and 3, except AtPYL11, 12, and 13 (Figure 6c). Significantly, many ortholog pairs between the apple and Arabidopsis genomes included nearly the same structure and conserved motif. For example, in MdPYL6, 13-AtPYL1 gene pairs belonging to group III all had only one exon and conserved motif 6. MdPYL1,7-AtPYL9 gene pairs belonging to group I all had three exons and conserved motif 8 (Figure 5,6). The result will provide further insight into the functions of apple MdPYLs.
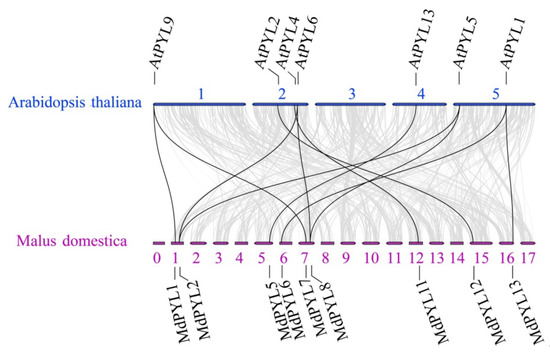
Figure 5.
Synteny analysis of PYLs between apple and A. thaliana. Gray lines denote the collinear blocks between apple and A. thaliana genomes and the black lines denote the syntenic gene pairs of PYLs. Blue and purple lines represent respectively the apple chromosomes (0–17) and Arabidopsis chromosomes (1–5).
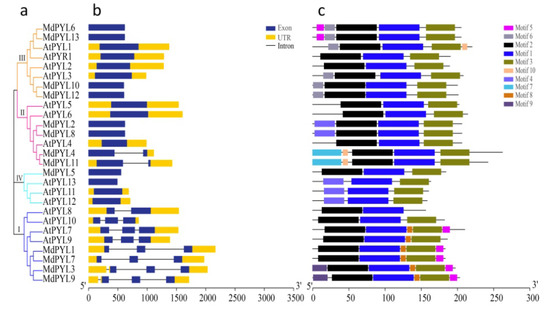
Figure 6.
Phylogenetic relationships, gene structure, and conserved motifs of MdPYLs and AtPYLs. (a). The phylogenetic tree of PYLs from the apple and Arabidopsis. Bars of different colors represent different groups of PYLs. (b). Exon–intron structure of MdPYLs and AtPYLs. Yellow boxes denote untranslated regions; blue boxes denote exons; black lines denote introns. (c). Distributions of ten conserved motifs in MdPYLs and AtPYLs.
2.5. Cis-Element Analysis of the MdPYLs Promoter in Apple
Previous works have indicated that many MdPYLs involved in the response to different abiotic stresses. To further forecast the potential functions of MdPYLs, we analyzed the cis-acting elements involved in abiotic stresses responses in the MdPYLs promoter regions using PlantCARE software (Table S3,S4). In total, three abiotic stress-related and four hormone-related elements were discovered in the promoter of 13 MdPYLs, including drought-inducibility elements (MBS), defense and stress-responsive elements (TC-rich repeats), dehydration reaction elements (MYC), low-temperature-responsive elements (LTR), abscisic acid-responsive elements (ABRE), salicylic acid-responsive elements (TCA-element), ethylene-responsive elements (ERE), and MeJA-responsive elements (CGTCA-motif, TGACG-motif) (Figure 7). All MdPYLs promoter regions contained at least three cis-elements (Figure 7). For example, the MdPYL7 promoter had seven stress-responsive elements including ABRE, MBS, MYC, LTR, TC-rich repeats, CGTCA, and TCA-element (Figure 7). The MdPYL13 promoter region contained six stress-responsive elements including ABRE, MBS, MYC, LTR, ERE, and CGTCA-element. Moreover, the MdPYL2, 5, 8 promoter included three, the MdPYL1, 6, 7, 10, 11 promoter comprised four, and the MdPYL3, 4, 12 promoter contained five stress-responsive elements. Furthermore, the MdPYLs promoter had two tissue-specifc cis-elements (root-specific regulatory element and seed-specific regulation element) (Figure 7). The results concluded that the expression of MdPYLs with different cis-elements may differ in response to various abiotic stresses.
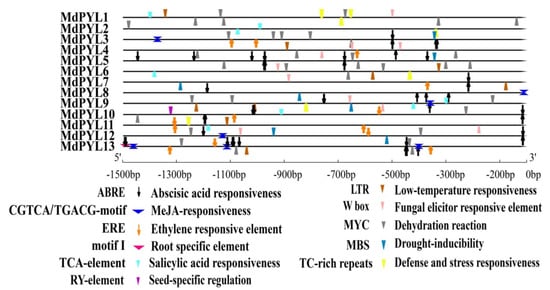
Figure 7.
The cis-elements analysis in MdPYL promoters. The 1500 bp promoter sequences of 13 MdPYLs were analyzed by PlantCARE. The up and down directions respectively denote the cis-elements existing in the plus or minus strand of MdPYL promoters.
2.6. Expression Profiles of Apple MdPYLs in Diverse Organs
To further understand the expression profiles of MdPYLs in apple development, we detected their expression levels in different tissues and organs from Chinese wild apple clone ‘Xinjiang No.1’ using qRT-PCR. In general, most MdPYLs were expressed differently in various organs, showing they might have different functions (Figure 8). Notably, MdPYL5 (group IV) had little or almost no expression in the organs tested. In contrast, the expression levels of all MdPYLs within group I, including MdPYL1, MdPYL3, MdPYL7, and MdPYL9, were higher in all of the organs tested, and were more than 20 times of that in group IV. Furthermore, MdPYLs of group II, including MdPYL2, MdPYL4, MdPYL8, and MdPYL11, were preferentially expressed in the root and their expression level were more than fivefold of the other organs. At the same time, we found MdPYLs within group III expression exhibited the relatively higher levels than MdPYLs of group II in all tissue analyzed except for root. For example, the expression of MdPYL10 and MdPYL12 in peel, sarcocarp, and young fruit were over fivefold of all genes within group II (Figure 8). These results suggested MdPYLs might have a distinct effect on growth and development of the apple tree.
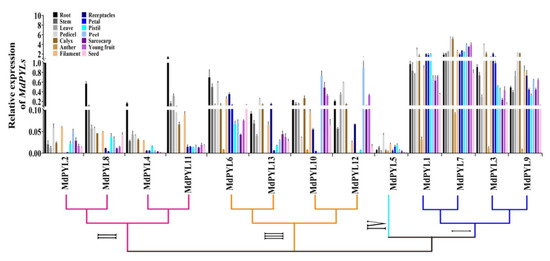
Figure 8.
Tissue-specific expression of MdPYLs. Quantitative RT-PCR was performed on roots, stems, leaves, pedicels, calyxes, anthers, filaments, receptacles, petals, pistils, peels, sarcocarps, young fruits, and seeds. The expression of MdPYL2 in roots was set to 1 and MdActin was used as an internal control. The relative expression levels of genes were calculated based on the 2-ΔΔCt method, and values are the mean ± SD obtained from three biological replicates.
2.7. Expression Profiles of MdPYLs in Response to Various Abiotic Stresses
PYLs, as ABA receptors, might play a vital function in abiotic stress signaling pathways. Therefore, we detected the expression of all 13 apple MdPYLs in Chinese wild apple clone ‘Xinjiang No.1’ tissue-cultured seedlings upon various abiotic stress treatments (including ABA, NaCl, PEG, cold) by carrying out qRT-PCR. Genes with expression levels that were increased or decreased more than two-fold, were used for the following analyses (Figure 9).
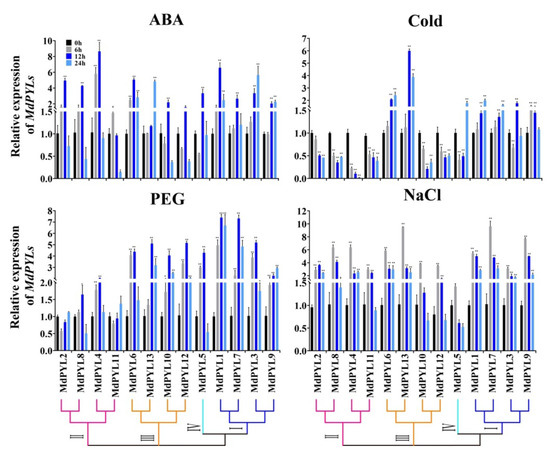
Figure 9.
Expression of the apple MdPYLs under ABA, PEG, cold (4 °C), and salt treatment in the tissue-cultured apple seedlings (Xinjiang No.1). The gene relative expression was calculated using the 2-ΔΔCt method with MdActin as an internal control, and value represents mean ± SD of three biological replicates. Asterisks indicated values that are significantly different from CK (0 h) (* p < 0.05, ** p < 0.01, one-way ANOVA).
In our study, we found that the expression of almost all of MdPYLs were significantly changed by ABA, PEG, NaCl, and cold. For example, the expression of all MdPYLs except for MdPYL11 exhibited significant increases by ABA treatment (Figure 9). Nine genes (MdPYL1, MdPYL2, MdPYL4, MdPYL5, MdPYL6, MdPYL7, MdPYL8, MdPYL10, and MdPYL12) reached the peak at 12 h after ABA treatment, while three genes (MdPYL3, MdPYL9, and MdPYL13) reached the peak at 24 h. Notably, MdPYL1, MdPYL2, MdPYL3, MdPYL4, MdPYL6, MdPYL8, and MdPYL13 all were upregulated more than fourfold after 12 h or 24 treatment, respectively. Similarly, following with PEG treatment, 10 genes (MdPYL1, MdPYL2, MdPYL3, MdPYL5, MdPYL6, MdPYL7, MdPYL9, MdPYL10, MdPYL12, and MdPYL13) were sharply upregulated in expression after 6 h of treatment and reached the maximum at 12 h, while the expression of remaining three apple MdPYLs had no obvious changes (Figure 9). The expression of all MdPYLs within group I, group III, and group IV were over fourfold higher than CK (0 h) at 12 h after PEG treatment. After NaCl treatment, the expression of all MdPYLs except for MdPYL5 showed significant increases and surged to the peak at 6 h (more than threefold of CK) (Figure 9). Following cold treatment (4 °C), all MdPYLs within group I, and MdPYL6, 13 within group III demonstrated increased expression. More remarkably, MdPYL13 was upregulated more than sixfold after 6 h treatment. In contrast, all MdPYLs within group II, and MdPYL10, 12 within group III were significantly downregulated (Figure 9).
2.8. Interaction between MdPYL and MdPP2C Proteins
PYLs can interact with PP2Cs to form PYL-ABA-PP2C triple complexes promoting the ABA signals transmission [1]. However, other studies suggested that PYLs can bind PP2Cs in an ABA-dependent or ABA-independent manner in yeast two-hybrid (Y2H) assays [4,12]. In this study, 13 MdPYLs and 2 MdPP2Cs were isolated (Figure S1) to detect their potential interactions in the yeast two-hybrid system. The full coding sequence of all MdPYLs were respectively fused to the DNA-binding domain (BD) of pGBKT7 vector, while two MdPP2Cs were respectively fused with activation domain (AD) of pGADT7 vector. After co-transformed into the Y2H Gold yeast strain, all of yeast cells were capable of growth on SD-Leu/-Trp medium in the absence or presence of ABA. However, the co-transformed yeast cells grew faster in medium with ABA (Figure 10). Interaction between 13 MdPYLs and two MdPP2Cs was determined by growth assay on SD-Leu/-Trp/-His/X-α-Gal/3-AT medium with and without 10 μM ABA. In this experiment, all MdPYLs interacted with MdPP2C65 in an ABA-dependent manner, and these interactions were considerably enhanced in presence of ABA. However, MdPYLs differentially interacted with MdPP2C72. MdPYL1, MdPYL3, MdPYL6, MdPYL10, MdPYL11, and MdPYL12 interacted with MdPP2C72 in an ABA-dependent manner. MdPYL4, MdPYL7, MdPYL8, and MdPYL9 showed weak interaction signals with MdPP2C72 in the absence of ABA. Furthermore, we found that MdPYL2,5,13 showed stronger interaction signals activating the reporter gene X-α-Gal with MdPP2C72 in an ABA-independent manner (Figure 10). In addition, all MdPYLs and MdPP2Cs had no obvious self-activation (Figure S2). These results suggested that MdPYLs may bind to MdPP2C65 and MdPP2C72 in different manners and with different affinities.

Figure 10.
Interactions between MdPYLs and MdPP2Cs in yeast. Co-transformed positive yeast cells dilutions (10−1, 10−2, and 10−3) were spotted onto the plates. Interaction was determined by growth assay on SD-Leu/-Trp/-His/X-α-Gal/3-AT medium in the presence and absence of 10 μM ABA.
3. Discussion
3.1. Gene Functional Diversification of Apple MdPYLs
PYLs encoding an ABA receptor family contained a similar ligand-binding pocket embraced by four conserved surface loops (CL1–CL4). In the study, 13 MdPYLs shared the typical structure were identified in the apple, with an almost equal number of AtPYLs from Arabidopsis (Figure 2 and Figure 3a). Comparative genomic analysis found that nine MdPYLs (MdPYL1, 2, 5, 6, 7, 8, 11, 12, 13) were positioned in syntenic regions of the apple and Arabidopsis genomes (Figure 5), and the other four MdPYLs (MdPYL3, 4, 6, 9) were formed by segment duplication events in the apple genome (Figure 4). Meanwhile, almost all of the ortholog pairs between apple and Arabidopsis genomes also contained similar exon–intron structures and conserved motifs. The results suggested that the function of PYLs gene might be relatively conserved between the apple and Arabidopsis. To date, the functions of many AtPYLs have been reported in Arabidopsis [15,16,17,18,22,39]. According to MdPYLs expression data (Figure 8 and Figure 9) and the information known concerning their Arabidopsis counterparts, we will better predict the probable functions of apple MdPYLs.
3.2. Expression Profiles of Apple PYLs and Their Potential Functions in Various Apple Organs
ABA is an important phytohormone for plant growth and development. PYL family members, as ABA receptors, have been identified and characterized in many plants including Arabidopsis, maize, rice, grape, rubber tree, and so on. To date, their expression profiles in various tissues have been revealed substantial differences among PYL genes for many plants. For example, some PYLs respectively displayed relatively higher levels in seeds of soybean, the callus of B. rapa, and the latex of rubber tree than other tissues [23,24,40,41]. Moreover, most OsPYLs were expressed in all organs of rice, OsPYL3 and OsPYL5 were predominantly expressed in leaves, and OsPYL1 in roots [42]. Similarly, in maize, ZmPYL11 was upregulated in leaves and ZmPYL6 and ZmPYL10 in roots [43]. In tomato, PYR/PYL/RCAR genes (10g085310 and 3g095780) had high levels of expression in root, and 1g095700 showed predominant expression in leaf, 12g055990 and 8g082180 in fruit tissues [22]. In grape, most of the VyPYLs genes, especially VyPYL1, VyPYL4a, and VyPYL7-9 were expressed relatively strongly in roots, stems, and tendrils [44]. These reports indicated that PYLs play an important role in plant growth and development. The function of PYLs in Arabidopsis has been studied to support this idea. For example, overexpression of RCAR11, RCAR12, RCAR13, or RCAR14 inhibited the germination and root growth of transgenic Arabidopsis in an ABA-treated condition [45]. AtPYL9 promoted ABA-induced leaf senescence in transgenic Arabidopsis [15]. AtPYL8 promoted lateral root growth by interacting with AtMYB77 and enhancing its transcriptional activity [18]. However, MdPYLs expression patterns and functions are still unclear in apple. Therefore, we speculated the roles of the apple MdPYLs based on qRT-PCR results in different tissues, and the information known of Arabidopsis counterparts (Figure 5) and homology genes of other species (Figure 2).
In general, the apple MdPYLs exhibited qualitatively and quantitatively distinct expression profiles. However, MdPYLs clustered to the same group, especially the eight pairs of genes in duplicated genomic regions, showed similar expression patterns, respectively (Figure 8), suggesting that they might have similar functions. For example, all MdPYLs within group I, including MdPYL1, MdPYL3, MdPYL7, and MdPYL9, exhibited relatively higher expression levels in all of the tissues tested compared to PYLs in other groups (Figure 8). Thus, we speculated that MdPYLs of the group I might function in the regulation of plant different growing and developmental stages. The function of homologous genes in other species has been studied to support this hypothesis. For instance, AtPYL8 and AtPYL9, which are orthologs of MdPYL1, 7, are involved in controlling root sensitivity to ABA [16]. OsPYL8 and OsPYL9, belonging to group I, were specifically expressed in the rice endosperms and positively regulated by ABA during seed germination [46]. Furthermore, MdPYLs of group II, including MdPYL2, MdPYL4, MdPYL8, and MdPYL11 were preferentially expressed in the root (Figure 8), indicating that these genes might play a role in root development. Indeed, the AtPYL5/AtPYL6 gene, as the ortholog of MdPYL2/MdPYL8, was expressed at the higher levels in root and might play a critical role for the regulation of root growth and root system architecture [5]. In short, these results indicated the diverse biological functions of different PYLs in plant growth and development.
3.3. Apple MdPYLs Were Responsive to Various Abiotic Stresses
To date, PYLs, as important ABA receptors, have been found to play crucial roles in responding to various abiotic stresses in plants. Overexpressing pRD29A::PYL9 dramatically promoted drought resistance and leaf senescence induced by drought in Arabidopsis and rice plants [15]. Similarly, overexpression of AtPYL4, 5, 7, 8, and 13 also respectively enhanced drought tolerance and showed hypersensitivity to ABA during early seedling development in transgenic Arabidopsis [47]. Furthermore, overexpressing ABA receptors (RCAR11-14) also increased drought resistance and accelerated stress-responsive gene expression in transgenic Arabidopsis [45]. With the deepening study in Arabidopsis, the functions of orthologous PYL proteins’ responses to stresses were reported in several plants, such as rice, cotton, tomato, and grape. In rice, ectopic expression of OsPYL3 improved cold and drought tolerance in transgenic Arabidopsis [31]. Constitutive expression of OsPYL/RCAR5 increased drought and salt stress tolerance in rice [30]. In Gossypium, the expression level of many GhPYLs were downregulated by ABA treatment and upregulated by osmotic stress [27,34]. Overexpression of GhPYL9-11A, GhPYL10, GhPYL12, and GhPYL26 improved the tolerance to drought stress in transgenic Arabidopsis [27,34]. Heterologous expression of tomato ABA receptors (6g050500 or 3g007310, or 8g076960) and Vitis yeshanensis VyPYL9 all enhanced transgenic Arabidopsis drought resistance [22,44]. However, there is only a handful information concerning these genes response to stress in apple. Thus, we investigated the response of all 13 MdPYLs to various abiotic stress conditions in the study (Figure 9), and found that 12 MdPYLs (except MdPYL11), 12 MdPYLs (except MdPYL5), and 10 MdPYLs (except MdPYL4, MdPYL8, and MdPYL11) had significant expression changes when treated with ABA, salinity, and PEG, respectively (Figure 9). The results were consistent with the function of PYLs in Arabidopsis and rice, which indicated they may involve in drought and salinity stresses response. In addition, MdPYL6 and MdPYL13, which were located in duplicated genomic regions, exhibited significant increases in expression levels following cold treatment (4 °C) (Figure 9). Meanwhile, the promoter regions of MdPYL6 and MdPYL13 both contained low-temperature-responsive elements (LTR) (Figure 7, Tables S3,S4). This result insinuated that these two genes will be the key candidate ABA receptors responding to cold stresses in apple. Indeed, the rice OsPYL3 and OsPYL9 gene, which the ortholog of MdPYL6/13 (Figure 2 and Table S2), was reported to be involved in enhancing cold tolerance in transgenic Arabidopsis and rice [28,42].
Furthermore, many evidences have shown that PYLs involve in plant response to abiotic stress by inhibiting PP2Cs in an ABA-dependent manner [3,5,6,48]. Different works have been reported that PYLs interacted with PP2Cs in an ABA-dependent or ABA-independent manner [5,8]. For instance, AtPYL8/RCAR3 interacted with Fagus sylvatica FsPP2C1 in an ABA-independent manner and positively regulates ABA signaling during abiotic stress responses [48]. AtPYL5 enhanced transgenic Arabidopsis resistance to drought by inhibited HAB1 phosphatase activity in an ABA-dependent manner [5]. In this work, we evaluated the interactions between 13 MdPYLs and two MdPP2Cs. Our results indicated that 13 MdPYLs selectively interacted with MdPP2Cs in ABA-dependent or ABA-independent manner (Figure 10), implying the function diversity among different MdPYLs. In conclusion, it seems that PYLs have various functions in the apple and play a key role in abiotic stress induced by ABA molecular signals.
4. Materials and Methods
4.1. Identification of MdPYL Gene Family and Chromosomal Location in Apple Genome
We used the protein sequence of 14 AtPYLs from the Arabidopsis genome to search for MdPYLs of the apple genome. The BLAST program was set to the default value (e-value < e−10) [49]. Manual reanalysis was performed for suspicious genes with a PYL structure and low E-value. The amino acid number, isoelectric point (pI), and molecular weight (MW) were analyzed by using the ExPASy website. The localization of MdPYLs on the chromosome were mapped by using the Gene Structure Display Server.
4.2. Phylogenetic Analysis of PYL Gene Family
The phylogenetic tree was constructed using the PYLs amino acid sequences from M. domestica, A. thaliana, B. distachyon, O. sativa, R. communis, T. cacao, V. vinifera, and G. hirsutum. Multiple alignments of protein sequences were analyzed by the neighbor-join (NJ) algorithm from 1000 repeated by MEGA 5.2 software [50].
4.3. Co-linear Analysis of Apple MdPYLs
The homologous genes were searched by MCScanx software [51], and the collinearity of the MdPYLs was obtained using the Circos program [52].
4.4. Gene Structure and Motif Composition of Apple MdPYLs
The MdPYLs protein sequences were found from the apple genome, and their intron–exon structures were analyzed by GSDS [53]. The MdPYL protein motifs were analyzed by Multiple EM for Motif Elicitation version 4.11.4 (MEME) [54]. The maximum number of motifs was set to 10, and the motif length was set to 6–200 amino acids.
4.5. Analysis of Cis-Acting Elements in the Promoter of MdPYLs
The cis-acting elements were detected using the PLACE database in the 1500 base pair (bp) upstream of the gene initiation codon (ATG) of the MdPYLs (Tables S3 and S4).
4.6. Plant Materials and Treatments
The red-fleshed apple (Malus sieversii f. Neidzwetzkyana (Dieck) Langenf) ‘Xinjiang No.1’, a wild apple resource with resistance to abiotic stress, was from Xinjiang of China.
The apple ‘Xinjiang No.1’ tissue culture seedlings used for stress treatment was cultured in a specific medium with Murashigeand Skoog (MS) medium, 0.8% agar, 0.5 mmol L−1 indole-3-butytric acid (IBA) and 0.7 mmol L−1 6-benzylaminopurine (6-BA). After 30 days of growth, the ‘Xinjiang No.1’ seedlings were transferred to the medium, which supplied with 10% polyethylene glycol (PEG) 6000 or 100 mmol L−1 abscisic acid (ABA) or 100 mmol L−1 NaCl to induce different abiotic stress, respectively. Furthermore, the temperature for apple tissue culture seedlings cultured in a specific medium was adjusted to 4 °C to induce cold stress. The stress-treated seedlings were harvested at 0, 6, 12, and 24 h, then immediately frozen with liquid nitrogen and stored in a −80 °C freezer for follow-up studies.
The different apple organs, including roots, stems, leaves, pedicels, calyxes, anthers, filaments, receptacles, petals, pistils, sarcocarps, peels, young fruits (20 days after flowering), and seeds were collected from seven-year old ‘Xinjiang No.1’, which was obtained from tissue culture seedlings transplanted to the Jiao Zhou Experimental Farm (longitude 120°39′ E, latitude 36°27′ N) of Qing Dao Agricultural University.
4.7. Quantitative Real-Time RT-PCR Analysis
RNA was extracted from the collected samples using the EASYspin Plant RNA Rapid Extraction Kit (YPHBIO, Beijing, China). The RNA concentration was measured with a NanoDrop 2000 (Gene Company Limited, Hong Kong, China) instrument. The cDNA was obtained according to a reverse transcription kit (Takara, Dalian, China). Specific primers were designed on NCBI-BLAST (Table S5). According to the manufacturer’s instructions, quantitative real-time RT-PCR (qRT-PCR) was performed on a LightCycler® 480 real-time PCR instrument (Roche, Shanghai, China). The PCR program was as followed: 95 °C for 2 min, 40 cycles of 95 °C for 30 s, 56 °C for 30 s and 72 °C for 30 s with a final dissociation stage. Actin of apple was used as an internal control for normalized gene expression levels. Repeat at least three times for each sample. Gene relative expression levels were calculated using GraphPad.Prism.5.0 software.
4.8. Yeast Two-Hybrid Assay
Full-length sequences of 13 MdPYLs and two MdPP2Cs (MdPP2C72: XM_008359137.2 and MdPP2C65: XM_008371984.3) were respectively amplified by PCR from ‘Xinjiang No.1’ leaf using the specific primers with enzyme digestion sites (Table S5). The full coding sequence of all MdPYLs were respectively fused to the DNA-binding domain (BD) of pGBKT7 vector, while two MdPP2Cs were respectively fused with activation domain (AD) of pGADT7 vector via double digestion technique. Combinations of pGADT7-T with pGBKT7-53 and pGBKT7-Lam were used as positive and negative controls, respectively. Combinations of empty pGADT7 with pGBKT7-MdPYLs, and empty pGBKT7 with pGADT7-MdPP2Cs were used to investigate whether MdPYLs or MdPP2Cs protein had self-activation activity. Combinations of pGBKT7-MdPYLwith pGADT7-MdPP2Cs were used to determine the potential interactions between MdPYLs and MdPP2Cs.
After co-transformation into the Y2H Gold yeast strain, the yeasts were plated on a SD/-Leu/-Trp media. Then, the positive yeasts were grown in liquid SD-Leu/Trp medium for 12 h at 30 °C. Finally, 3 μL of the co-transformed yeast cells dilutions of 10−1, 10−2, and 10−3 were spread on plates containing SD/-Leu/-Trp, SD/-His/-Leu/-Trp/X-α-Gal medium with 10 mM 3-AT (3-amino-1, 2, 4-triazole) and SD/-His/-Leu/-Trp/X-α-Gal medium with 10 mM 3-AT (3-amino-1, 2, 4-triazole) and 10 μM ABA. The plates were photographed after four days of incubation at 30 °C. The experiments were repeated three times.
5. Conclusions
Thirteen MdPYLs were identified in M. domestica genome. Phylogenetic reconstruction and gene structure analysis demonstrated that MdPYLs could be divided into four groups, and that they had similar gene structures and high conserved motifs in the same group. Moreover, comparative genomic analysis showed that homologs of nine MdPYLs were located in corresponding syntenic regions of Arabidopsis. The MdPYLs expression analysis in various organs revealed distinct spatiotemporal patterns. Furthermore, gene expression analysis showed that MdPYLs are possibly involved in multiple abiotic stress responses (ABA, salt, PEG, and cold). The results presented here call for further research aiming at revealing the potential important functions of these genes in the apple.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2223-7747/9/8/1028/s1. Table S1: Amino acid sequences of the plant PYLs. Table S2: The clustered groups of PYL proteins from eight species. Table S3: The 1500 bp promoter sequences of the 13 MdPYLs. Table S4: Cis-element analysisof the MdPYLs promoter regions in the apple. Table S5: Primers used in this study. Figure S1: The results of agarose gel electrophoresis test of 13 MdPYLs and two MdPP2Cs. Figure S2: Self activation analysis of 13 MdPYLs and two MdPP2Cs fusions.
Author Contributions
Y.Z. and H.H. (Hongmin Hou) designed the study. H.H. (Hongmin Hou), L.L., and Y.Z. performed data analysis. Y.Z. provided guidance on the whole study. H.H. (Hongmin Hou), L.L., H.D., H.H. (Heqiang Huo), and Y.Z. wrote and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31801841); China Agriculture Research System Foundation (Grant No. CARS-27); Natural Science Foundation of Shandong (ZR2019MC003); Qingdao Scientific Research Foundation (Grant No. 19-6-1-60-nsh); Breeding Plan of Shandong Provincial Qingchuang Research Team (2019).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant. Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant. Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.Y.; Márquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant. J. 2009, 60, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Miyazono, K.; Miyakawa, T.; Sawano, Y.; Kubota, K.; Kang, H.J.; Asano, A.; Miyauchi, Y.; Takahashi, M.; Zhi, Y.; Fujita, Y.; et al. Structural basis of abscisic acid signalling. Nature 2009, 462, 609–614. [Google Scholar] [CrossRef]
- Nishimura, N.; Hitomi, K.; Arvai, A.S.; Rambo, R.P.; Hitomi, C.; Cutler, S.R.; Schroeder, J.I.; Getzoff, E.D. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 2009, 326, 1373–1379. [Google Scholar] [CrossRef]
- Peterson, F.C.; Burgie, E.S.; Park, S.Y.; Jensen, D.R.; Weiner, J.J.; Bingman, C.A.; Chang, C.E.; Cutler, S.R.; Phillips, G.N., Jr.; Volkman, B.F. Structural basis for selective activation of ABA receptors. Nat. Struct. Mol. Biol. 2010, 17, 1109–1113. [Google Scholar] [CrossRef]
- Miyakawa, T.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Tanokura, M. Structure and function of abscisic acid receptors. Trends. Plant. Sci. 2013, 18, 259–266. [Google Scholar] [CrossRef]
- Santiago, J.; Dupeux, F.; Betz, K.; Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Márquez, J.A.; Rodriguez, P.L. Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs. Plant. Sci. 2012, 182, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Yin, P.; Li, W.; Wang, L.; Yan, C.; Lin, Z.; Wu, J.Z.; Wang, J.; Yan, S.F.; Yan, N. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell. 2011, 42, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, L.; Sheng, X.; Yan, C.; Zhou, R.; Hang, J.; Yin, P.; Yan, N. Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13. Cell Res. 2013, 23, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernandez, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant. Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Peirats-Llobet, M.; Pizzio, G.A.; Fernandez, M.A.; De Winne, N.; De Jaeger, G.; Dietrich, D.; Bennett, M.J.; et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant. Physiol. 2013, 161, 931–941. [Google Scholar] [CrossRef]
- Xing, L.; Zhao, Y.; Gao, J.; Xiang, C.; Zhu, J.K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 2016, 6, 27177. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.J.; Gao, J.; Wang, P.; Duan, C.G.; Zhu, X.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Romero, P.; Lafuente, M.T.; Rodrigo, M.J. The Citrus ABA signalosome: Identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. J. Exp. Bot. 2012, 63, 4931–4945. [Google Scholar] [CrossRef]
- Boneh, U.; Biton, I.; Zheng, C.; Schwartz, A.; Ben-Ari, G. Characterization of potential ABA receptors in Vitis vinifera. Plant. Cell Rep. 2012, 31, 311–321. [Google Scholar] [CrossRef]
- He, Y.; Hao, Q.; Li, W.; Yan, C.; Yan, N.; Yin, P. Identification and characterization of ABA receptors in Oryza sativa. PLoS ONE 2014, 9, e95246. [Google Scholar] [CrossRef] [PubMed]
- González-Guzmán, M.; Rodríguez, L.; Lorenzo-Orts, L.; Pons, C.; Sarrión-Perdigones, A.; Fernández, M.A.; Peirats-Llobet, M.; Forment, J.; Moreno-Alvero, M.; Cutler, S.R.; et al. Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 2014, 65, 4451–4464. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Zhou, Y.; Li, H.L.; Zhu, J.H.; Wang, Y.; Chen, X.T.; Peng, S.Q. Identification and characterization of the abscisic acid (ABA) receptor gene family and its expression in response to hormones in the rubber tree. Sci. Rep. 2017, 7, 45157. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, D.; Sun, C.; Hu, X.; Mu, X.; Hu, J.; Yang, Y.; Zhang, Y.; Xie, C.G.; Zhou, X. Molecular characterization of an AtPYL1-like protein, BrPYL1, as a putative ABA receptor in Brassica rapa. Biochem. Biophys. Res. Commun. 2017, 487, 684–689. [Google Scholar] [CrossRef]
- Zhang, G.; Lu, T.; Miao, W.; Sun, L.; Tian, M.; Wang, J.; Hao, F. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. Peer J. 2017, 5, e4126. [Google Scholar] [CrossRef]
- Bai, G.; Xie, H.; Yao, H.; Li, F.; Chen, X.; Zhang, Y.; Xiao, B.; Yang, J.; Li, Y.; Yang, D.H. Genome-wide identification and characterization of ABA receptor PYL/RCAR gene family reveals evolution and roles in drought stress in Nicotiana tabacum. BMC Genom. 2019, 20, 575. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, L.; Wei, N.; Liu, Z.H.; Hu, S.; Li, X.B. Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant. Physiol. Biochem. 2017, 115, 229–238. [Google Scholar] [CrossRef]
- Lenka, S.K.; Muthusamy, S.K.; Chinnusamy, V.; Bansal, K.C. Ectopic Expression of Rice PYL3 Enhances Cold and Drought Tolerance in Arabidopsis thaliana. Mol. Biotechnol. 2018, 60, 350–361. [Google Scholar] [CrossRef]
- He, Z.; Zhong, J.; Sun, X.; Wang, B.; Terzaghi, W.; Dai, M. The Maize ABA Receptors ZmPYL8, 9, and 12 Facilitate Plant Drought Resistance. Front. Plant. Sci. 2018, 9, 422. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef]
- Kim, H.; Lee, K.; Hwang, H.; Bhatnagar, N.; Kim, D.Y.; Yoon, I.S.; Byun, M.O.; Kim, S.T.; Jung, K.H.; Kim, B.G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014, 65, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef] [PubMed]
- Pizzio, G.A.; Rodriguez, L.; Antoni, R.; Gonzalez-Guzman, M.; Yunta, C.; Merilo, E.; Kollist, H.; Albert, A.; Rodriguez, P.L. The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant. Physiol. 2013, 163, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Liu, Y.; Li, Y.; Meng, Z.; Yan, R.; Zhu, T.; Wang, Y.; Kang, S.; Ali Abid, M.; Malik, W.; et al. Activation of ABA Receptors Gene GhPYL9-11A Is Positively Correlated with Cotton Drought Tolerance in Transgenic Arabidopsis. Front. Plant. Sci. 2017, 8, 1453. [Google Scholar] [CrossRef]
- Yu, J.; Yang, L.; Liu, X.; Tang, R.; Wang, Y.; Ge, H.; Wu, M.; Zhang, J.; Zhao, F.; Luan, S.; et al. Overexpression of Poplar Pyrabactin Resistance-Like Abscisic Acid Receptors Promotes Abscisic Acid Sensitivity and Drought Resistance in Transgenic Arabidopsis. PLoS ONE 2016, 11, e0168040. [Google Scholar] [CrossRef]
- Li, G.; Xin, H.; Zheng, X.F.; Li, S.; Hu, Z. Identification of the abscisic acid receptor VvPYL1 in Vitis vinifera. Plant. Biol. (Stuttg) 2012, 14, 244–248. [Google Scholar] [CrossRef]
- Boneh, U.; Biton, I.; Schwartz, A.; Ben-Ari, G. Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant. Sci. 2012, 187, 89–96. [Google Scholar] [CrossRef]
- Yin, P.; Fan, H.; Hao, Q.; Yuan, X.; Wu, D.; Pang, Y.; Yan, C.; Li, W.; Wang, J.; Yan, N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 2009, 16, 1230–1236. [Google Scholar] [CrossRef]
- Fuchs, S.; Tischer, S.V.; Wunschel, C.; Christmann, A.; Grill, E. Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc. Natl. Acad. Sci. USA 2014, 111, 5741–5746. [Google Scholar] [CrossRef]
- Bai, G.; Yang, D.H.; Zhao, Y.; Ha, S.; Yang, F.; Ma, J.; Gao, X.S.; Wang, Z.M.; Zhu, J.K. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant. Mol. Biol. 2013, 83, 651–664. [Google Scholar] [CrossRef]
- Di, F.; Jian, H.; Wang, T.; Chen, X.; Ding, Y.; Du, H.; Lu, K.; Li, J.; Liu, L. Genome-Wide Analysis of the PYL Gene Family and Identification of PYL Genes That Respond to Abiotic Stress in Brassica napus. Genes 2018, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and Functional Analysis of Pyrabactin Resistance-Like Abscisic Acid Receptor Family in Rice. Rice 2015, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Zhao, M.; Li, S.; Bai, X.; Li, J.; Meng, H.; Mu, Z. Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant. Biol. 2016, 16, 99. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, F.L.; Guo, Y.; Fan, X.C.; Wang, Y.j.; Wen, Y.Q. The ABA receptor-like gene VyPYL9 from drought-resistance wild grapevine confers drought tolerance and ABA hypersensitivity in Arabidopsis. Plant. Cell Tissue Organ. Cult. 2019, 138, 543–558. [Google Scholar] [CrossRef]
- Li, X.; Li, G.; Li, Y.; Kong, X.; Zhang, L.; Wang, J.; Li, X.; Yang, Y. ABA Receptor Subfamily III Enhances Abscisic Acid Sensitivity and Improves the Drought Tolerance of Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1938. [Google Scholar] [CrossRef]
- Chen, Z.; Kong, L.; Zhou, Y.; Chen, Z.; Tian, D.; Lin, Y.; Wang, F.; Chen, S. Endosperm-specific OsPYL8 and OsPYL9 act as positive regulators of the ABA signaling pathway in rice seed germination. Funct. Plant. Biol. 2017, 44, 635–645. [Google Scholar] [CrossRef]
- Han, Y.L.; Jang, G.; Um, T.; Kim, J.K.; Lee, J.S.; Yang, D.C. The soluble ABA receptor PYL8 regulates drought resistance by controlling ABA signaling in Arabidopsis. Plant. Biotechnol. Rep. 2015, 9, 319–330. [Google Scholar]
- Saavedra, X.; Modrego, A.; Rodríguez, D.; González-García, M.P.; Sanz, L.; Nicolás, G.; Lorenzo, O. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant. Physiol. 2010, 152, 133–150. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).