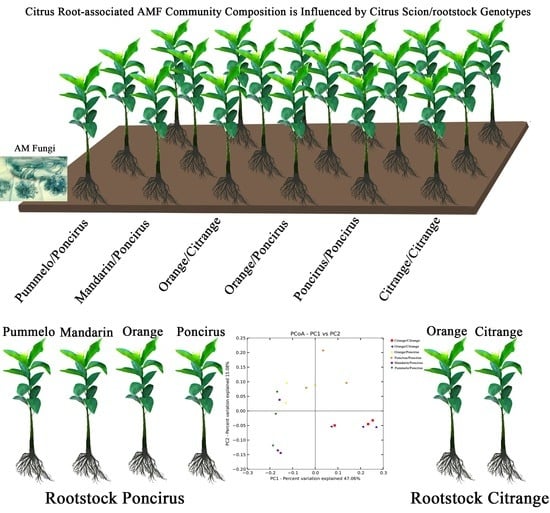
Influence of Citrus Scion/Rootstock Genotypes on Arbuscular Mycorrhizal Community Composition under Controlled Environment Condition
Abstract
1. Introduction
2. Results
2.1. Overall Citrus AMF Taxonomic Richness
2.2. Difference in α Diversity of AMF Community among Different Citrus Genotypes
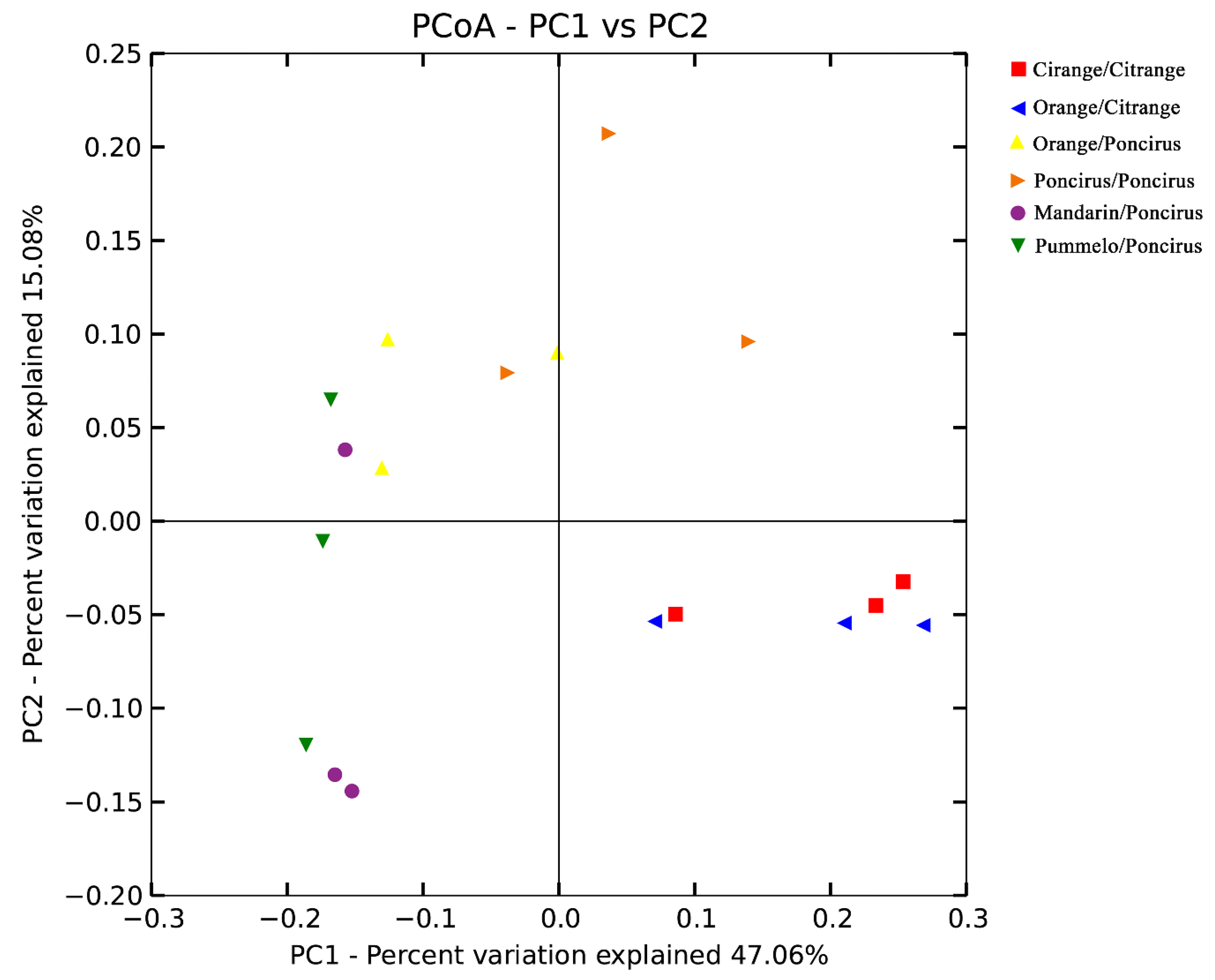
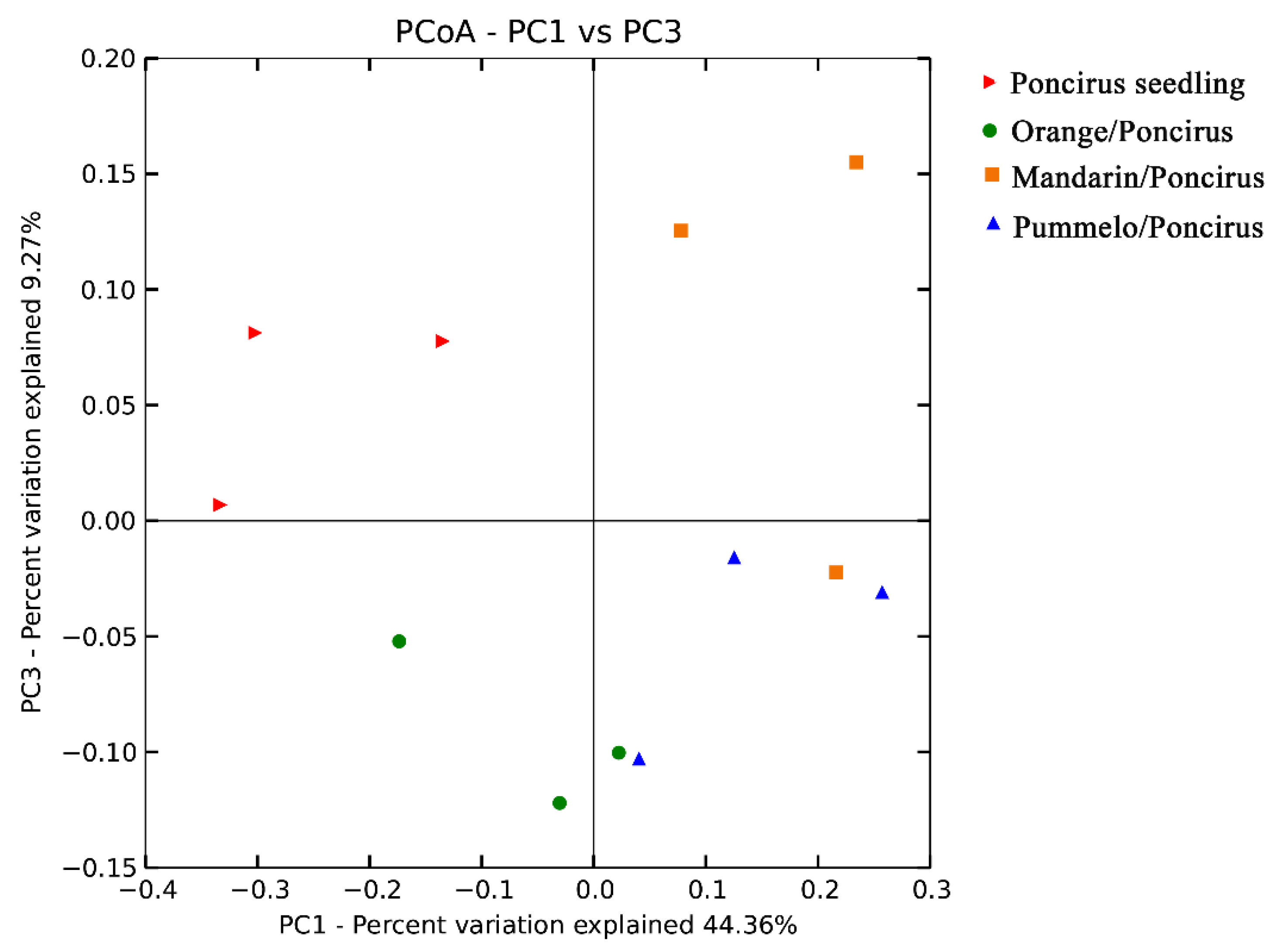
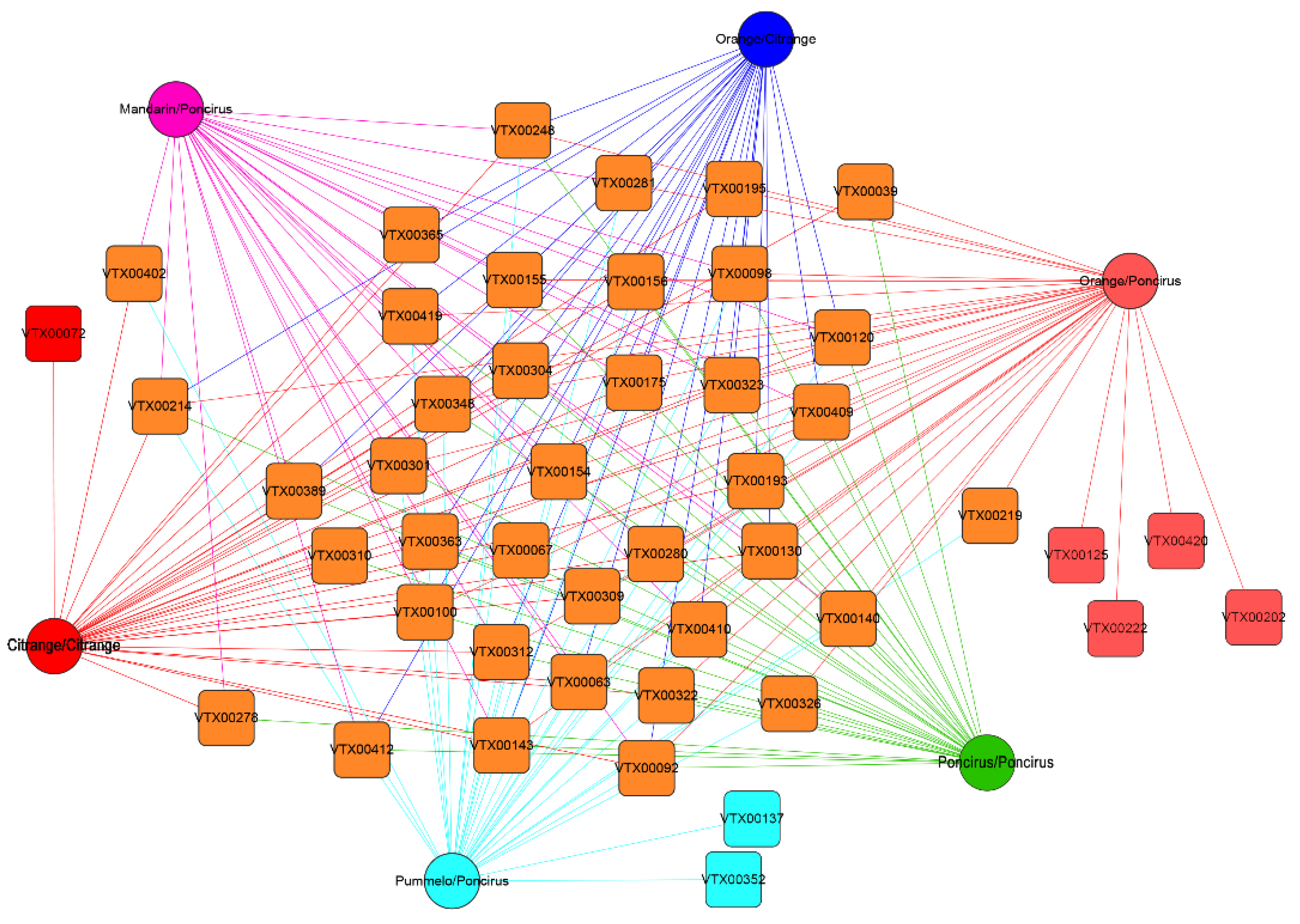
2.3. Citrus Genotypes Shape the AMF Community Composition
2.4. Relative Abundance of AMF Species under Different Citrus Genotypes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials and Sample Preparation
5.2. DNA Extraction, Library Construction and Sequencing
5.3. Data Analysis
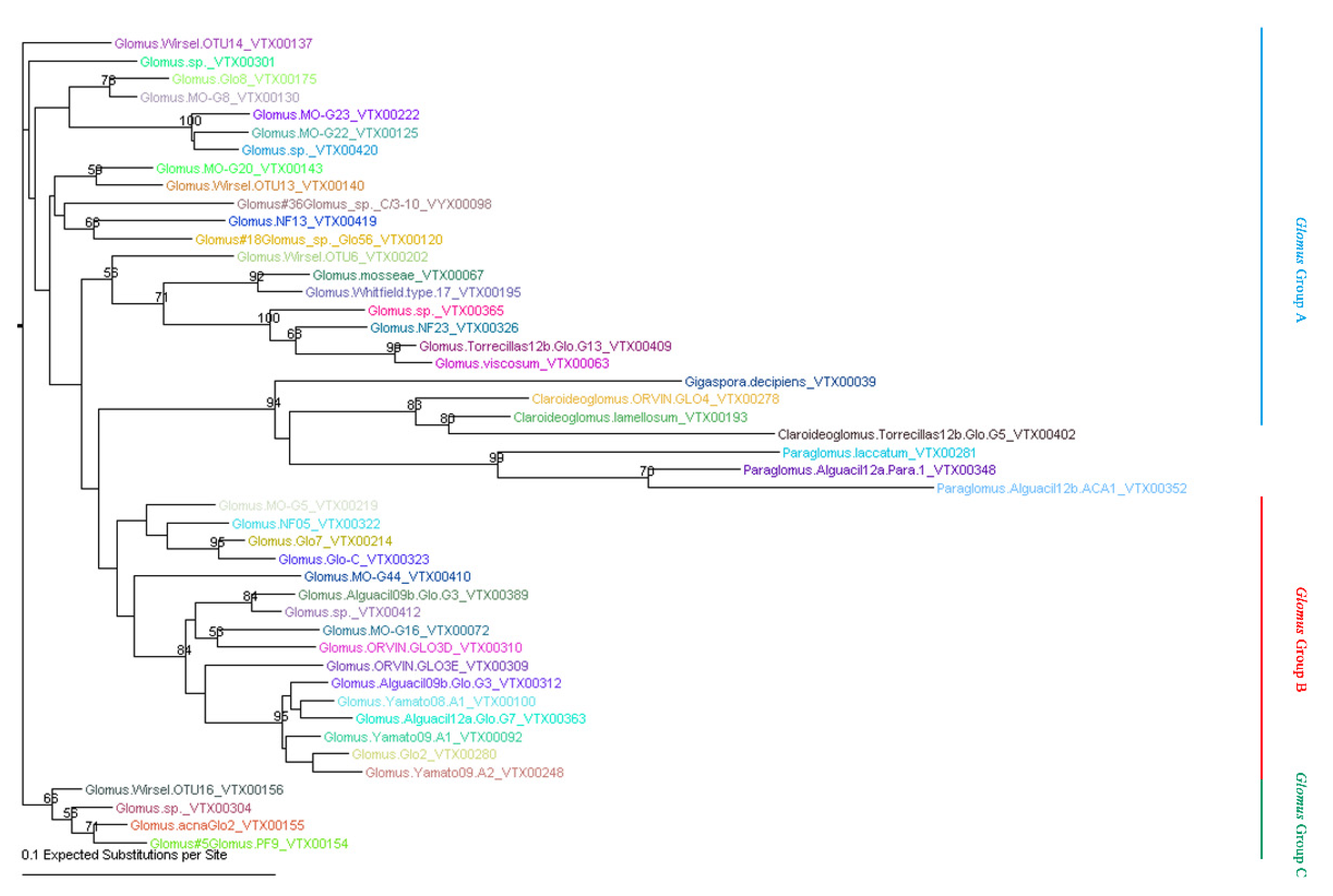
5.4. Phylogenetic Analysis
5.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef]
- Öpik, M.; Davison, J. Uniting species- and community-oriented approaches to understand arbuscular mycorrhizal fungal diversity. Fungal Ecol. 2016, 24, 106–113. [Google Scholar] [CrossRef]
- Govindarajulu, M.; Pfeffer, P.E.; Jin, H.; Abubaker, J.; Douds, D.D.; Allen, J.W.; Bucking, H.; Lammers, P.J.; Shachar-Hill, Y. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 2005, 435, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J.; Dewbre, G.R.; Liu, J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002, 14, 2413–2429. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D.; Leyser, O. A plant’s diet, surviving in a variable nutrient environment. Science 2020, 368. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Lozano, J.M.; Aroca, R.; Zamarreno, A.M.; Molina, S.; Andreo-Jimenez, B.; Porcel, R.; Garcia-Mina, J.M.; Ruyter-Spira, C.; Lopez-Raez, J.A. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Env. 2016, 39, 441–452. [Google Scholar] [CrossRef]
- Santander, C.; Aroca, R.; Ruiz-Lozano, J.M.; Olave, J.; Cartes, P.; Borie, F.; Cornejo, P. Arbuscular mycorrhiza effects on plant performance under osmotic stress. Mycorrhiza 2017, 27, 639–657. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, W.; Xie, Q.; Liu, N.; Liu, L.; Wang, D.; Zhang, X.; Yang, C.; Chen, X.; Tang, D.; et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017, 356, 1172–1175. [Google Scholar] [CrossRef]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef]
- Smith, S.; Read, D. Mycorrhizal Symbiosis; Academic Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Davies, F.; Albrigo, L. Citrus; Acribia, S.A.: Zaragoza, Spain, 1999. [Google Scholar]
- Rillig, M.C.; Aguilar-Trigueros, C.A.; Camenzind, T.; Cavagnaro, T.R.; Degrune, F.; Hohmann, P.; Lammel, D.R.; Roy, J.; van der Heijden, M.G.; Yang, G. Why farmers should manage the arbuscular mycorrhizal symbiosis. New Phytol. 2019, 222, 1–5. [Google Scholar] [CrossRef]
- Srivastava, A.; Singh, S.; Marathe, R. Organic citrus: Soil fertility and plant nutrition. J. Sustain. Agric. 2002, 19, 5–29. [Google Scholar] [CrossRef]
- Liu, L.; Hart, M.M.; Zhang, J.; Cai, X.; Gai, J.; Christie, P.; Li, X.; Klironomos, J.N. Altitudinal distribution patterns of AM fungal assemblages in a Tibetan alpine grassland. Fems Microbiol Ecol 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Chen, H.; Shi, Z.; Liu, S.; Cao, X.; Zhang, M.; Chen, M.; Chen, J.; Xiong, K.; Yang, H. Mycorrhizal and rhizospheric fungal community assembly differs during subalpine forest restoration on the eastern Qinghai-Tibetan Plateau. Plant Soil 2019, 1–15. [Google Scholar] [CrossRef]
- Sheldrake, M.; Rosenstock, N.P.; Revillini, D.; Olsson, P.A.; Mangan, S.; Sayer, E.J.; Wallander, H.; Turner, B.L.; Tanner, E.V. Arbuscular mycorrhizal fungal community composition is altered by long-term litter removal but not litter addition in a lowland tropical forest. New Phytol. 2017, 214, 455–467. [Google Scholar] [CrossRef]
- Van Geel, M.; Jacquemyn, H.; Plue, J.; Saar, L.; Kasari, L.; Peeters, G.; van Acker, K.; Honnay, O.; Ceulemans, T. Abiotic rather than biotic filtering shapes the arbuscular mycorrhizal fungal communities of European seminatural grasslands. New Phytol. 2018, 220, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Qiang, W.; He, X.; Wang, J.; Zhao, L. Temporal and spatial variation of arbuscular mycorrhizal fungi under the canopy of Hedysarum scoparium in the northern desert, China. Appl. Soil Ecol. 2019, 136, 139–147. [Google Scholar] [CrossRef]
- Smilauer, P.; Kosnar, J.; Kotilinek, M.; Smilauerova, M. Contrasting effects of host identity, plant community, and local species pool on the composition and colonization levels of arbuscular mycorrhizal fungal community in a temperate grassland. New Phytol. 2020, 225, 461–473. [Google Scholar] [CrossRef]
- Williams, A.; Manoharan, L.; Rosenstock, N.P.; Olsson, P.A.; Hedlund, K. Long-term agricultural fertilization alters arbuscular mycorrhizal fungal community composition and barley (H ordeum vulgare) mycorrhizal carbon and phosphorus exchange. New Phytol. 2017, 213, 874–885. [Google Scholar] [CrossRef]
- Davison, J.; Moora, M.; Öpik, M.; Adholeya, A.; Ainsaar, L.; Bâ, A.; Burla, S.; Diedhiou, A.; Hiiesalu, I.; Jairus, T. Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 2015, 349, 970–973. [Google Scholar] [CrossRef]
- Rasmussen, P.U.; Hugerth, L.W.; Blanchet, F.G.; Andersson, A.F.; Lindahl, B.D.; Tack, A.J.M. Multiscale patterns and drivers of arbuscular mycorrhizal fungal communities in the roots and root-associated soil of a wild perennial herb. New Phytol. 2018, 220, 1248–1261. [Google Scholar] [CrossRef]
- Torrecillas, E.; Torres, P.; Alguacil, M.M.; Querejeta, J.I.; Roldan, A. Influence of habitat and climate variables on arbuscular mycorrhizal fungus community distribution, as revealed by a case study of facultative plant epiphytism under semiarid conditions. Appl Env. Microbiol 2013, 79, 7203–7209. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, V.B.; Rua, M.A.; Antoninka, A.; Bever, J.D.; Cannon, J.; Craig, A.; Duchicela, J.; Frame, A.; Gardes, M.; Gehring, C.; et al. MycoDB, a global database of plant response to mycorrhizal fungi. Sci. Data 2016, 3, 160028. [Google Scholar] [CrossRef] [PubMed]
- Klironomos, J.N. Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 2003, 84, 2292–2301. [Google Scholar] [CrossRef]
- Neuenkamp, L.; Moora, M.; Opik, M.; Davison, J.; Gerz, M.; Mannisto, M.; Jairus, T.; Vasar, M.; Zobel, M. The role of plant mycorrhizal type and status in modulating the relationship between plant and arbuscular mycorrhizal fungal communities. New Phytol. 2018, 220, 1236–1247. [Google Scholar] [CrossRef]
- Lopez-Garcia, A.; Varela-Cervero, S.; Vasar, M.; Opik, M.; Barea, J.M.; Azcon-Aguilar, C. Plant traits determine the phylogenetic structure of arbuscular mycorrhizal fungal communities. Mol Ecol 2017, 26, 6948–6959. [Google Scholar] [CrossRef]
- Verbruggen, E.; Kuramae, E.E.; Hillekens, R.; de Hollander, M.; Kiers, E.T.; Roling, W.F.; Kowalchuk, G.A.; van der Heijden, M.G. Testing potential effects of maize expressing the Bacillus thuringiensis Cry1Ab endotoxin (Bt maize) on mycorrhizal fungal communities via DNA- and RNA-based pyrosequencing and molecular fingerprinting. Appl. Env. Microbiol. 2012, 78, 7384–7392. [Google Scholar] [CrossRef]
- Ribeiro, R.V.; Espinoza-Núñez, E.; Junior, J.P.; Mourão Filho, F.A.; Machado, E.C. Citrus rootstocks for improving the horticultural performance and physiological responses under constraining environments. In Improvement of Crops in the Era of Climatic Changes; Springer: Berlin, Germany, 2014; pp. 1–37. [Google Scholar]
- Bowman, K.D.; McCollum, G. Five new citrus rootstocks with improved tolerance to huanglongbing. HortScience 2015, 50, 1731–1734. [Google Scholar] [CrossRef]
- Marasco, R.; Rolli, E.; Fusi, M.; Michoud, G.; Daffonchio, D. Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 2018, 6, 3. [Google Scholar] [CrossRef]
- Ling, N.; Song, Y.; Raza, W.; Huang, Q.; Guo, S.; Shen, Q. The response of root-associated bacterial community to the grafting of watermelon. Plant. Soil 2015, 391, 253–264. [Google Scholar] [CrossRef]
- Song, F.; Pan, Z.; Bai, F.; An, J.; Liu, J.; Guo, W.; Bisseling, T.; Deng, X.; Xiao, S. The Scion/Rootstock Genotypes and Habitats Affect Arbuscular Mycorrhizal Fungal Community in Citrus. Front. Microbiol. 2015, 6, 1372. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 2012, gks1219. [Google Scholar] [CrossRef]
- Öpik, M.; Vanatoa, A.; Vanatoa, E.; Moora, M.; Davison, J.; Kalwij, J.; Reier, Ü.; Zobel, M. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 2010, 188, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Nemec, S.; Menge, J.; Platt, R.; Johnson, E. Vesicular: Arbuscular mycorrhizal fungi associated with citrus in Florida and California and notes on their distribution and ecology. Mycologia 1981, 112–127. [Google Scholar]
- Wang, P.; Shu, B.; Wang, Y.; Zhang, D.; Liu, J.; Xia, R. Diversity of arbuscular mycorrhizal fungi in red tangerine (Citrus reticulata Blanco) rootstock rhizospheric soils from hillside citrus orchards. Pedobiologia 2013, 56, 161–167. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, J.J.; Shu, B.; Xia, R.X. Arbuscular mycorrhizal fungi associated with citrus orchards under different types of soil management, southern China. Plant. Soil Env. 2012, 58, 302–308. [Google Scholar] [CrossRef]
- Faggioli, V.S.; Cabello, M.N.; Grilli, G.; Vasar, M.; Covacevich, F.; Öpik, M. Root colonizing and soil borne communities of arbuscular mycorrhizal fungi differ among soybean fields with contrasting historical land use. Agric. Ecosyst. Environ. 2019, 269, 174–182. [Google Scholar] [CrossRef]
- Saks, Ü.; Davison, J.; Öpik, M.; Vasar, M.; Moora, M.; Zobel, M. Root-colonizing and soil-borne communities of arbuscular mycorrhizal fungi in a temperate forest understorey. Botany 2014, 92, 277–285. [Google Scholar] [CrossRef]
- Davison, J.; Moora, M.; Jairus, T.; Vasar, M.; Öpik, M.; Zobel, M. Hierarchical assembly rules in arbuscular mycorrhizal (AM) fungal communities. Soil Biol. Biochem. 2016, 97, 63–70. [Google Scholar] [CrossRef]
- Martínez-García, L.B.; Richardson, S.J.; Tylianakis, J.M.; Peltzer, D.A.; Dickie, I.A. Host identity is a dominant driver of mycorrhizal fungal community composition during ecosystem development. New Phytol. 2015, 205, 1565–1576. [Google Scholar] [CrossRef]
- Krüger, C.; Kohout, P.; Janoušková, M.; Püschel, D.; Frouz, J.; Rydlová, J. Plant communities rather than soil properties structure arbuscular mycorrhizal fungal communities along primary succession on a mine spoil. Front. Microbiol. 2017, 8, 719. [Google Scholar] [CrossRef]
- Landis, F.C.; Gargas, A.; Givnish, T.J. Relationships among arbuscular mycorrhizal fungi, vascular plants and environmental conditions in oak savannas. New Phytol. 2004, 164, 493–504. [Google Scholar] [CrossRef]
- An, J.; Sun, M.; van Velzen, R.; Ji, C.; Zheng, Z.; Limpens, E.; Bisseling, T.; Deng, X.; Xiao, S.; Pan, Z. Comparative transcriptome analysis of Poncirus trifoliata identifies a core set of genes involved in arbuscular mycorrhizal symbiosis. J. Exp. Bot 2018, 69, 5255–5264. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhao, S.; Xu, Q.-L.; Xiao, J.-X. Transcriptome responses of grafted Citrus sinensis plants to inoculation with the arbuscular mycorrhizal fungus Glomus versiforme. Trees 2016, 30, 1073–1082. [Google Scholar] [CrossRef]
- Zou, Y.-N.; Wang, P.; Liu, C.-Y.; Ni, Q.-D.; Zhang, D.-J.; Wu, Q.-S. Mycorrhizal trifoliate orange has greater root adaptation of morphology and phytohormones in response to drought stress. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Wu, J.; Cao, J.; Su, M.; Feng, G.; Xu, Y.; Yi, H. Genome-wide comprehensive analysis of transcriptomes and small RNAs offers insights into the molecular mechanism of alkaline stress tolerance in a citrus rootstock. Hortic. Res. 2019, 6, 1–19. [Google Scholar] [CrossRef]
- Tan, F.-Q.; Tu, H.; Liang, W.-J.; Long, J.-M.; Wu, X.-M.; Zhang, H.-Y.; Guo, W.-W. Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biol. 2015, 15, 89. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, F.K.; Prudente, D.A.; Bueno, A.C.R.; Machado, E.C.; Ribeiro, R.V. Drought tolerance in citrus trees is enhanced by rootstock-dependent changes in root growth and carbohydrate availability. Environ. Exp. Bot. 2014, 101, 26–35. [Google Scholar] [CrossRef]
- Albrecht, U.; Fiehn, O.; Bowman, K.D. Metabolic variations in different citrus rootstock cultivars associated with different responses to Huanglongbing. Plant. Physiol. Biochem. 2016, 107, 33–44. [Google Scholar] [CrossRef]
- Benjamin, G.; Tietel, Z.; Porat, R. Effects of rootstock/scion combinations on the flavor of citrus fruit. J. Agric. Food Chem. 2013, 61, 11286–11294. [Google Scholar] [CrossRef]
- Broeckling, C.D.; Broz, A.K.; Bergelson, J.; Manter, D.K.; Vivanco, J.M. Root exudates regulate soil fungal community composition and diversty. Appl Env. Microb 2008, 74, 738–744. [Google Scholar] [CrossRef]
- Micallef, S.A.; Shiaris, M.P.; Colon-Carmona, A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J. Exp. Bot 2009, 60, 1729–1742. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Krusell, L.; Madsen, L.H.; Sato, S.; Aubert, G.; Genua, A.; Szczyglowski, K.; Duc, G.; Kaneko, T.; Tabata, S.; de Bruijn, F.; et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 2002, 420, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.; Hayashi, M.; Wu, G.J.; Kouchi, H.; Imaizumi-Anraku, H.; Murakami, Y.; Kawasaki, S.; Akao, S.; Ohmori, M.; Nagasawa, M.; et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature 2002, 420, 426–429. [Google Scholar] [CrossRef]
- Poudel, R.; Jumpponen, A.; Kennelly, M.M.; Rivard, C.L.; Gomez-Montano, L.; Garrett, K.A. Rootstocks shape the rhizobiome: Rhizosphere and endosphere bacterial communities in the grafted tomato system. Appl. Env. Microbiol. 2019, 85, e01765-18. [Google Scholar] [CrossRef]
- Liu, J.; Abdelfattah, A.; Norelli, J.; Burchard, E.; Schena, L.; Droby, S.; Wisniewski, M. Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome 2018, 6, 18. [Google Scholar] [CrossRef]
- Singh, G.; Mukerji, K.G. Root exudates as determinant of rhizospheric microbial biodiversity. In Microbial Activity in the Rhizoshere; Springer: Berlin, Germany, 2006; pp. 39–53. [Google Scholar]
- Hugoni, M.; Luis, P.; Guyonnet, J.; el Zahar Haichar, F. Plant host habitat and root exudates shape fungal diversity. Mycorrhiza 2018, 28, 451–463. [Google Scholar] [CrossRef]
- Jach, M.; Ceulemans, R. Effects of season, needle age and elevated atmospheric CO2 on photosynthesis in Scots pine (Pinus sylvestris). Tree Physiol. 2000, 20, 145–157. [Google Scholar] [CrossRef]
- Shiroya, T.; Lister, G.; Krotkov, G.; Nelson, C.; Slankis, V. Translocation of the products of photosynthesis to roots of pine seedlings. Can. J. Bot. 1962, 40, 1125–1135. [Google Scholar] [CrossRef]
- Hazard, C.; Gosling, P.; Van Der Gast, C.J.; Mitchell, D.T.; Doohan, F.M.; Bending, G.D. The role of local environment and geographical distance in determining community composition of arbuscular mycorrhizal fungi at the landscape scale. ISME J. 2013, 7, 498–508. [Google Scholar] [CrossRef]
- Vieira, L.C.; Silva, D.K.A.d.; Escobar, I.E.C.; Silva, J.M.d.; Moura, I.A.d.; Oehl, F.; Silva, G.A.d. Changes in an Arbuscular Mycorrhizal Fungi Community Along an Environmental Gradient. Plants 2020, 9, 52. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Zhang, Z.; Sun, Z.; Chen, Y.; Jiang, J.; Shen, Z. The influence of environmental factors on communities of arbuscular mycorrhizal fungi associated with Chenopodium ambrosioides revealed by MiSeq sequencing investigation. Sci. Rep. 2017, 7, 45134. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-J.; Guo, W.-W.; Yi, H.-L.; Pang, X.-M.; Deng, X. An efficient protocol for genomic DNA extraction from Citrus species. Plant. Mol. Biol. Report. 2003, 21, 177–178. [Google Scholar] [CrossRef]
- Lumini, E.; Orgiazzi, A.; Borriello, R.; Bonfante, P.; Bianciotto, V. Disclosing arbuscular mycorrhizal fungal biodiversity in soil through a land-use gradient using a pyrosequencing approach. Environ. Microbiol. 2010, 12, 2165–2179. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Feng, Y.; Zhang, H.; Chen, R.; Wang, J.; Zhang, J.; Chu, H. Long-term balanced fertilization decreases arbuscular mycorrhizal fungal diversity in an arable soil in North China revealed by 454 pyrosequencing. Environ. Sci. Technol. 2012, 46, 5764–5771. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Milne, I.; Wright, F.; Rowe, G.; Marshall, D.F.; Husmeier, D.; McGuire, G. TOPALi: Software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics 2004, 20, 1806–1807. [Google Scholar] [CrossRef] [PubMed]
- Holzer, C.; Precht, M. Multiple comparison procedures for normally distributed ANOVA models in SAS, SPSS, BMDP, and MINITAB. Computation Stat 1992, 13, 351–358. [Google Scholar] [CrossRef]
| Phylum | OTUs | Tags |
|---|---|---|
| Glomeromycota | 87 (65.91%) | 613,408 (99.81%) |
| Basidiomycota | 13 (9.85%) | 794 (0.13%) |
| Ascomycota | 8 (6.06%) | 41 (0.01%) |
| Chytridiomycota | 1 (0.76%) | 13 (0.00%) |
| unclassified | 23 (17.42%) | 334 (0.05%) |
| AMF Family | AMF Species | OTU | Tags |
|---|---|---|---|
| Glomeraceae (39 VT, corresponding to 474,991 clean reads, accounting for 99.81% of the total (475,917) clean reads against the MAARJAM database) | Glomus.Yamato08.A1_VTX00100 | 2 | 134,970 |
| Glomus.acnaGlo2_VTX00155 | 3 | 71,418 | |
| Glomus.Yamato09.A2_VTX00248 | 1 | 66,821 | |
| Glomus.viscosum_VTX00063 | 8 | 37,975 | |
| Glomus.Glo7_VTX00214 | 3 | 30,951 | |
| Glomus.NF13_VTX00419 | 2 | 21,670 | |
| Glomus.Torrecillas12b.Glo.G13_VTX00409 | 3 | 19,765 | |
| Glomus.sp._VTX00304 | 1 | 16,515 | |
| Glomus.Wirsel.OTU16_VTX00156 | 1 | 16,510 | |
| Glomus.Glo-C_VTX00323 | 2 | 10,763 | |
| Glomus.sp._VTX00412 | 1 | 8217 | |
| Glomus.Yamato09.A1_VTX00092 | 2 | 6499 | |
| Glomus.Glo8_VTX00175 | 1 | 3781 | |
| Glomus.MO-G5_VTX00219 | 1 | 3736 | |
| Glomus.MO-G8_VTX00130 | 1 | 3311 | |
| Glomus_sp._Glo56 | 1 | 3092 | |
| Glomus.MO-G44_VTX00410 | 2 | 3038 | |
| Glomus.Alguacil09b.Glo.G3_VTX00389 | 1 | 2478 | |
| Glomus.MO-G23_VTX00222 | 1 | 2337 | |
| Glomus.sp._VTX00301 | 2 | 2292 | |
| Glomus.PF9_VTX00154 | 1 | 1567 | |
| Glomus_sp._C/3-10 | 2 | 1305 | |
| Glomus.ORVIN.GLO3E_VTX00309 | 1 | 1290 | |
| Glomus.MO-G20_VTX00143 | 1 | 1279 | |
| Glomus.Alguacil09b.Glo.G3_VTX00312 | 1 | 802 | |
| Glomus.NF05_VTX00322 | 1 | 657 | |
| Glomus.sp._VTX00420 | 1 | 548 | |
| Glomus.MO-G22_VTX00125 | 1 | 404 | |
| Glomus.Alguacil12a.Glo.G7_VTX00363 | 1 | 300 | |
| Glomus.NF23_VTX00326 | 2 | 260 | |
| Glomus.Wirsel.OTU13_VTX00140 | 1 | 159 | |
| Glomus.Glo2_VTX00280 | 1 | 96 | |
| Glomus.Wirsel.OTU6_VTX00202 | 1 | 75 | |
| Glomus.ORVIN.GLO3D_VTX00310 | 1 | 68 | |
| Glomus.Wirsel.OTU14_VTX00137 | 2 | 17 | |
| Glomus.mosseae_VTX00067 | 1 | 9 | |
| Glomus.MO-G16_VTX00072 | 1 | 6 | |
| Glomus.sp._VTX00365 | 1 | 5 | |
| Glomus.Whitfield.type.17_VTX00195 | 1 | 5 | |
| Claroideoglomeraceae (3 VT, 88 reads, 0.02%) | Claroideoglomus.Torrecillas12b.Glo.G5_VTX00402 | 2 | 37 |
| Claroideoglomus.lamellosum_VTX00193 | 1 | 37 | |
| Claroideoglomus.ORVIN.GLO4_VTX00278 | 2 | 14 | |
| Paraglomeraceae (3 VT, 832 reads, 0.17%) | Paraglomus.Alguacil12a.Para.1_VTX00348 | 12 | 787 |
| Paraglomus.laccatum_VTX00281 | 1 | 37 | |
| Paraglomus.Alguacil12b.ACA1_VTX00352 | 1 | 8 | |
| Gigasporaceae (1 VT, 6 reads, 0.00%) | Gigaspora.decipiens_VTX00039 | 1 | 6 |
| Sample | Sobs | Ace | Shannon | Simpson |
|---|---|---|---|---|
| Citrange/Citrange | 55.00 ± 2.65 ab | 60.74 ± 4.56 ab | 1.79 ± 0.2 b | 0.34 ± 0.07 a |
| Orange/Citrange | 48.33 ± 4.26 b | 54.83 ± 3.46 b | 1.80 ± 0.52 b | 0.35 ± 0.16 a |
| Poncirus/Poncirus | 55.33 ± 2.91 ab | 59.37 ± 3.93 ab | 2.43 ± 0.18 ab | 0.15 ± 0.04 ab |
| Pummelo/Poncirus | 56.33 ± 6.36 ab | 58.56 ± 5.99 ab | 2.27 ± 0.17 ab | 0.16 ± 0.02 ab |
| Mandarin/Poncirus | 58.00 ± 3.79 ab | 64.47 ± 5.04 ab | 2.28 ± 0.14 ab | 0.15 ± 0.03 ab |
| Orange/Poncirus | 63.00 ± 4.04 a | 77.40 ± 12.21 a | 2.64 ± 0.14 a | 0.11 ± 0.02 b |
| AMF Species | Citrange/ Citrange | Orange/ Citrange | Poncirus/ Poncirus | Pummelo/ Poncirus | Mandarin/ Poncirus | Orange/ Poncirus |
|---|---|---|---|---|---|---|
| Glomus.MO-G44_VTX00410 | 0 ± 0 b | 0 ± 0 b | 0.05 ± 0.05 b | 0.38 ± 0.38 b | 0.26 ± 0.18 b | 2.27 ± 1.29 a |
| Glomus.MO-G8_VTX00130 | 0.01 ± 0 b | 0 ± 0 b | 0.07 ± 0.05 b | 0.38 ± 0.2 ab | 0.47 ± 0.27 ab | 2.29 ± 1.65 a |
| Glomus.sp._VTX00301 | 0.03 ± 0.01 b | 0.01 ± 0.01 b | 0.1 ± 0.02 b | 0.74 ± 0.26 ab | 1.33 ± 0.61 a | 0.04 ± 0.02 b |
| Glomus.NF05_VTX00322 | 0.03 ± 0.03 b | 0 ± 0 b | 0.04 ± 0.03 b | 0.08 ± 0.08 b | 0 ± 0 b | 0.49 ± 0.25 a |
| Paraglomus.Alguacil12a.Para.1_ VTX00348 | 0.04 ± 0.01 b | 0.06 ± 0.01 b | 0.05 ± 0.02 b | 0.1 ± 0.07 b | 0.4 ± 0.23 a | 0.13 ± 0.04 ab |
| Glomus.sp._VTX00304 | 0.18 ± 0.06 b | 0.08 ± 0.08 b | 0.63 ± 0.13 ab | 6.71 ± 0.48 a | 8.05 ± 4.37 a | 0.51 ± 0.29 ab |
| Glomus.Wirsel.OTU16_VTX00156 | 0.21 ± 0.09 b | 0.08 ± 0.08 b | 0.65 ± 0.16 b | 5.4 ± 1.62 ab | 9.29 ± 4.52 a | 0.5 ± 0.26 b |
| Glomus.ORVIN.GLO3E_VTX00309 | 0.86 ± 0.33 a | 0 ± 0 b | 0 ± 0 b | 0 ± 0 b | 0 ± 0 b | 0.37 ± 0.37 ab |
| Glomus.Torrecillas12b.Glo.G13_ VTX00409 | 0.91 ± 0.33 b | 0.7 ± 0.26 b | 0.64 ± 0.35 b | 8.15 ± 5.2 a | 5.83 ± 0.66 ab | 3.08 ± 0.14 ab |
| Glomus.NF13_VTX00419 | 1.55 ± 1.29 ab | 6.74 ± 3.42 ab | 2.47 ± 0.3 ab | 2.19 ± 0.79 ab | 0.83 ± 0.2 b | 7.79 ± 4 a |
| Glomus.Glo-C_VTX00323 | 1.59 ± 0.59 b | 0.74 ± 0.40 b | 8.03 ± 3.78 a | 0.19 ± 0.18 b | 0.02 ± 0.01 b | 0.22 ± 0.19 b |
| Glomus.Glo7_VTX00214 | 3.04 ± 1.14 b | 3.65 ± 2.81 b | 15.18 ± 7.15 a | 2.68 ± 2.23 b | 0.73 ± 0.59 b | 5.34 ± 3.88 ab |
| Glomus.Yamato09.A2_VTX00248 | 3.52 ± 2.41 b | 2.75 ± 1.38 b | 20.17 ± 1.71 a | 11.79 ± 6.66 a | 7.13 ± 5.3 ab | 20.2 ± 5.78 a |
| Glomus.acnaGlo2_VTX00155 | 3.58 ± 0.63 b | 6.61 ± 4.42 b | 6.71 ± 1.49 b | 27.42 ± 4.17 a | 12.83 ± 5.49 b | 12.64 ± 3.68 b |
| Glomus.Yamato08.A1_VTX00100 | 54.49 ± 8.96 a | 50.67 ± 17.75 a | 19.04 ± 10.93 b | 0.22 ± 0.17 b | 0.05 ± 0.01 b | 5.42 ± 4.54 b |
| Sample Name | Scion | Scion Genotype | Rootstock | Rootstock Genotype |
|---|---|---|---|---|
| Poncirus/Poncirus | Poncirus | Poncirus trifoliata | Poncirus | Poncirus trifoliata |
| Citrange/Citrange | Citrange | Citrus sinensis×Poncirus trifoliata | Citrange | Citrus sinensis×Poncirus trifoliata |
| Mandarin/Poncirus | Mandarin | Citrus reticulate | Poncirus | Poncirus trifoliata |
| Pummelo/Poncirus | Pummelo | Citrus grandis | Poncirus | Poncirus trifoliata |
| Orange/Poncirus | Newhall sweet orange | Citrus sinensis | Poncirus | Poncirus trifoliata |
| Orange/Citrange | Newhall sweet orange | Citrus sinensis | Citrange | Citrus sinensis×Poncirus trifoliata |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, F.; Bai, F.; Wang, J.; Wu, L.; Jiang, Y.; Pan, Z. Influence of Citrus Scion/Rootstock Genotypes on Arbuscular Mycorrhizal Community Composition under Controlled Environment Condition. Plants 2020, 9, 901. https://doi.org/10.3390/plants9070901
Song F, Bai F, Wang J, Wu L, Jiang Y, Pan Z. Influence of Citrus Scion/Rootstock Genotypes on Arbuscular Mycorrhizal Community Composition under Controlled Environment Condition. Plants. 2020; 9(7):901. https://doi.org/10.3390/plants9070901
Chicago/Turabian StyleSong, Fang, Fuxi Bai, Juanjuan Wang, Liming Wu, Yingchun Jiang, and Zhiyong Pan. 2020. "Influence of Citrus Scion/Rootstock Genotypes on Arbuscular Mycorrhizal Community Composition under Controlled Environment Condition" Plants 9, no. 7: 901. https://doi.org/10.3390/plants9070901
APA StyleSong, F., Bai, F., Wang, J., Wu, L., Jiang, Y., & Pan, Z. (2020). Influence of Citrus Scion/Rootstock Genotypes on Arbuscular Mycorrhizal Community Composition under Controlled Environment Condition. Plants, 9(7), 901. https://doi.org/10.3390/plants9070901