Distinct Responses to Light in Plants
Abstract
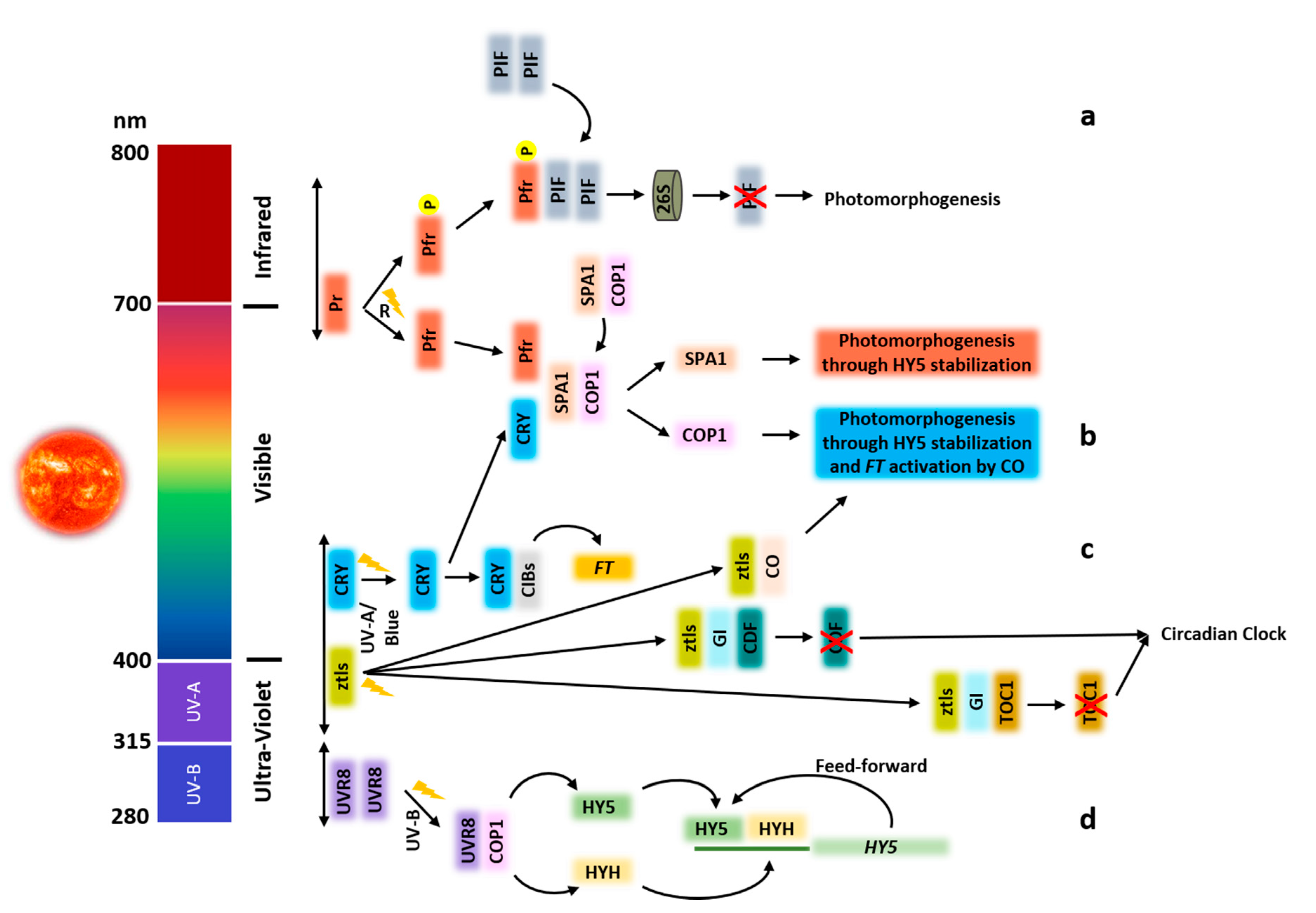
:1. Overview of Light Transduction Scheme
2. Introduction to Photoreceptors
2.1. Phytochromes
2.2. Cryptochromes
2.3. Phototropins
2.4. Zeitlupe Family
2.5. UVR8
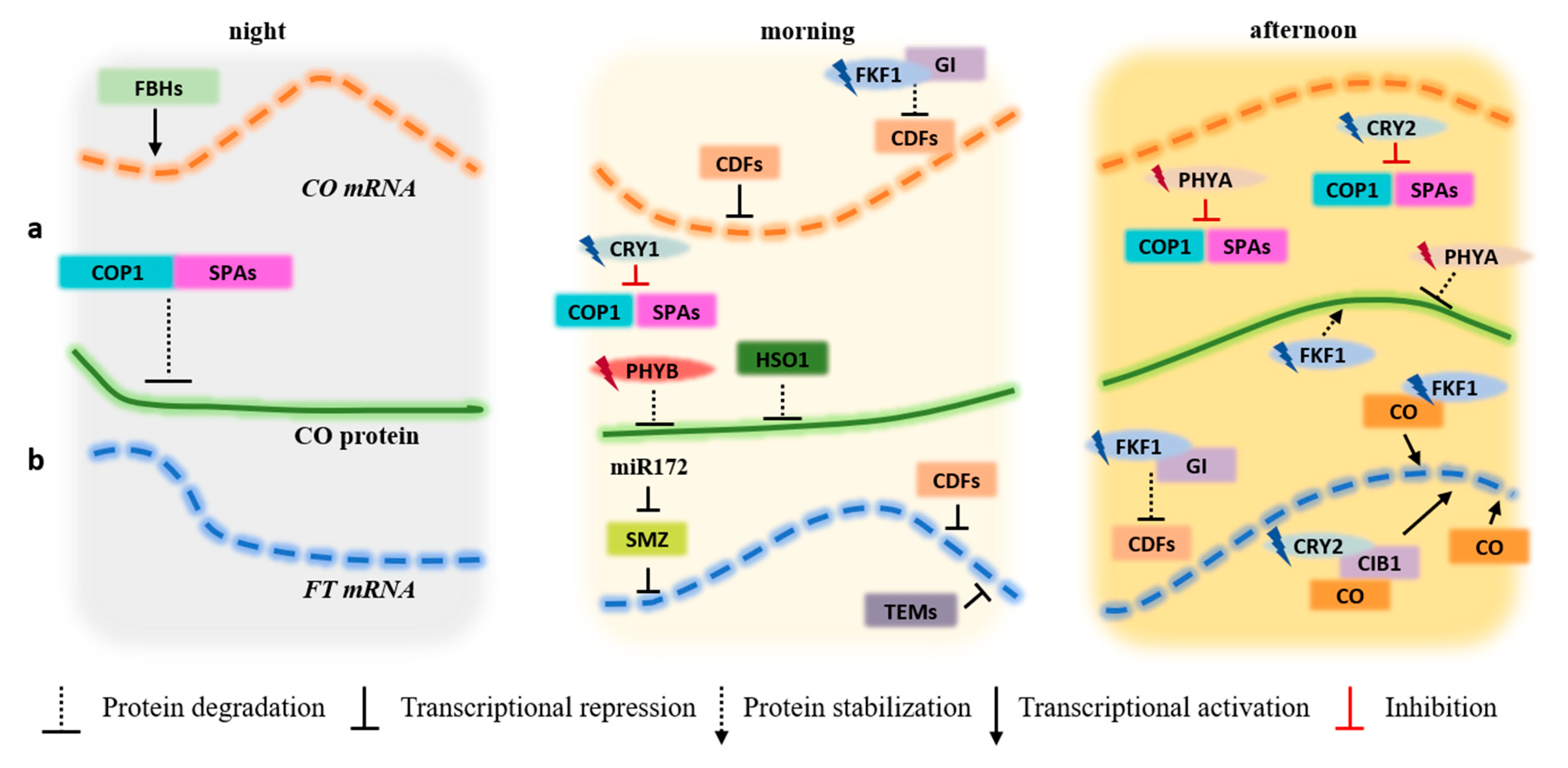
3. Plant Development under Light
3.1. Mechanisms Regulated by the Action of Light
3.2. Flowering Control by CO/FT Regulation
4. Conclusions
Funding
Conflicts of Interest
References
- Karpiński, S.; Szechyńska-Hebda, M.; Wituszyńska, W.; Burdiak, P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 2013, 36, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Terashima, I.; Hanba, Y.T.; Tazoe, Y.; Vyas, P.; Yano, S. Irradiance and phenotype: Comparative eco-development of sun and shade leaves in relation to photosynthetic CO2 diffusion. J. Exp. Bot. 2006, 57, 343–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kono, M.; Terashima, I. Long-term and short-term responses of the photosynthetic electron transport to fluctuating light. J. Photochem. Photobiol. B Biol. 2014, 137, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Light-regulated plant growth and development. In Current Topics in Developmental Biology; Academic Press Inc.: Cambridge, MA, USA, 2010; Volume 91, pp. 29–66. [Google Scholar]
- Higa, T.; Suetsugu, N.; Kong, S.G.; Wada, M. Actin-dependent plastid movement is required for motive force generation in directional nuclear movement in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 4327–4331. [Google Scholar] [CrossRef] [Green Version]
- Yamori, W. Photosynthetic response to fluctuating environments and photoprotective strategies under abiotic stress. J. Plant. Res. 2016, 129, 379–395. [Google Scholar] [CrossRef]
- Murchie, E.H.; Hubbart, S.; Peng, S.; Horton, P. Acclimation of photosynthesis to high irradiance in rice: Gene expression and interactions with leaf development. J. Exp. Bot. 2005, 56, 449–460. [Google Scholar] [CrossRef]
- Sinclair, T.R.; Muchow, R.C. Radiation Use Efficiency. Adv. Agron. 1999, 65, 215–265. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Pierik, R.; Ruban, A.; Wingler, A. The dynamic plant: Capture, transformation, and management of energy. Plant Physiol. 2018, 176, 961–966. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.P. Photosynthesis. Essays Biochem. 2016, 60, 255–273. [Google Scholar] [CrossRef]
- Song, Y.H.; Ito, S.; Imaizumi, T. Similarities in the circadian clock and photoperiodism in plants. Curr. Opin. Plant. Biol. 2010, 13, 594–603. [Google Scholar] [CrossRef] [Green Version]
- Harmer, S.L. The Circadian System in Higher Plants. Ann. Rev. Plant Biol. 2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farré, E.M.; Harmer, S.L.; Harmon, F.G.; Yanovsky, M.J.; Kay, S.A. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 2005, 15, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.L.; Newton, L.; Liu, M.J.; Shiu, S.H.; Farré, E.M. A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in arabidopsis. Plant Physiol. 2016, 170, 528–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009, 323, 1481–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamichi, N.; Kita, M.; Ito, S.; Yamashino, T.; Mizuno, T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, Together Play Essential Roles Close to the Circadian Clock of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 686–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE ReGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Imaizumi, T.; Tran, H.G.; Swartz, T.E.; Briggs, W.R.; Kay, S.A. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 2003, 426, 302–306. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Niinuma, K.; Ito, S.; Yamashino, T.; Mizoguchi, T.; Mizuno, T. Arabidopsis Clock-Associated Pseudo-Response Regulators PRR9, PRR7 and PRR5 Coordinately and Positively Regulate Flowering Time Through the Canonical CONSTANS-dependent Photoperiodic Pathway. Plant Cell Physiol. 2007, 48. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Bueno, G.; Romero-Campero, F.J.; Lucas-Reina, E.; Romero, J.M.; Valverde, F. Evolution of photoperiod sensing in plants and algae. Curr. Opin. Plant. Biol. 2017, 37, 10–17. [Google Scholar] [CrossRef]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S.; Kim, S.K.; Lee, S.Y.; MacCoss, M.J.; Imaizumi, T. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of constans stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef] [Green Version]
- Fornara, F.; Panigrahi, K.C.S.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF Transcription Factors Act Redundantly to Reduce CONSTANS Expression and Are Essential for a Photoperiodic Flowering Response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Geng, R.; Gallenstein, R.A.; Somers, D.E. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 2013, 140, 4060–4069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanovsky, M.J.; Kay, S.A. Molecular basis of seasonal time measurement in Arabidopsis. Nature 2002, 419, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Payyavula, R.S.; Chen, L.; Zhang, J.; Zhang, C.; Turgeon, R. FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proc. Natl. Acad. Sci. USA 2018, 115, 2830–2835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaeger, K.E.; Wigge, P.A. FT Protein Acts as a Long-Range Signal in Arabidopsis. Curr. Biol. 2007, 17, 1050–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.-Y.; Adams, J.P.; Kim, H.; No, K.; Ma, C.; Strauss, S.H.; Drnevich, J.; Vandervelde, L.; Ellis, J.D.; Rice, B.M.; et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA 2011, 108, 10756–10761. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, R.T.; Sheng, X.; Brunner, A.M. Activity of the shoot apical and cambial meristems: Coordination and responses to environmental signals. In Advances in Botanical Research; Academic Press Inc.: Cambridge, MA, USA, 2019; Volume 89, pp. 185–199. ISBN 9780128154656. [Google Scholar]
- Maurya, J.P.; Bhalerao, R.P. Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: A molecular perspective. Ann. Bot. 2017, 120, 351–360. [Google Scholar] [CrossRef]
- Burgie, E.S.; Vierstra, R.D. Phytochromes: An atomic perspective on photoactivation and signaling. Plant Cell 2014, 26, 568–4583. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, S.E.; Rugnone, M.L.; Kay, S.A. Light Perception: A Matter of Time. Mol. Plant 2020, 13, 363–385. [Google Scholar] [CrossRef]
- Saijo, Y.; Sullivan, J.A.; Wang, H.; Yang, J.; Shen, Y.; Rubio, V.; Ma, L.; Hoecker, U.; Deng, X.W. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003, 17, 2642–2647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Lory, N.; Stauber, J.; Hoecker, U. Photoreceptor Specificity in the Light-Induced and COP1-Mediated Rapid Degradation of the Repressor of Photomorphogenesis SPA2 in Arabidopsis. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef] [Green Version]
- Podolec, R.; Ulm, R. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 2018, 45, 18–25. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012, 17, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.-G.; Okajima, K. Diverse photoreceptors and light responses in plants. J. Plant Res. 2016, 129, 111–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgie, E.S.; Bussell, A.N.; Walker, J.M.; Dubiel, K.; Vierstra, R.D. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. USA 2014, 111, 10179–10184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christie, J.M.; Blackwood, L.; Petersen, J.; Sullivan, S. Plant Flavoprotein Photoreceptors. Plant Cell Physiol. 2015, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paik, I.; Huq, E. Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 2019, 92, 114–121. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 2010, 13, 571–577. [Google Scholar] [CrossRef]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvão, V.C.; Fankhauser, C. Sensing the light environment in plants: Photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 2015, 34, 46–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Ouyang, X.; Deng, X.W. Beyond repression of photomorphogenesis: Role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 2014, 21, 96–103. [Google Scholar] [CrossRef]
- Medzihradszky, M.; Bindics, J.; Ádám, É.; Viczián, A.; Klement, É.; Lorrain, S.; Gyula, P.; Mérai, Z.; Fankhauser, C.; Medzihradszky, K.F.; et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 2013, 25, 535–544. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Galvão, R.M.; Li, M.; Burger, B.; Bugea, J.; Bolado, J.; Chory, J. Arabidopsis HEMERA/pTAC12 Initiates photomorphogenesis by phytochromes. Cell 2010, 141, 1230–1240. [Google Scholar] [CrossRef] [Green Version]
- Kaiserli, E.; Páldi, K.; O’Donnell, L.; Batalov, O.; Pedmale, U.V.; Nusinow, D.A.; Kay, S.A.; Chory, J. Integration of Light and Photoperiodic Signaling in Transcriptional Nuclear Foci. Dev. Cell 2015, 35, 311–321. [Google Scholar] [CrossRef] [Green Version]
- Van Buskirk, E.K.; Decker, P.V.; Chen, M. Photobodies in light signaling. Plant Physiol. 2012, 158, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, R.; Nakamura, M.; Mochizuki, N.; Kay, S.A.; Nagatani, A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 1999, 145, 437–445. [Google Scholar] [CrossRef]
- Lin, C.; Ahmad, M.; Cashmore, A.R. Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 1996, 10, 893–902. [Google Scholar] [CrossRef]
- Yu, X.; Klejnot, J.; Zhao, X.; Shalitin, D.; Maymon, M.; Yang, H.; Lee, J.; Liu, X.; Lopez, J.; Lina, C. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 2007, 19, 3146–3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.-O.; van der Horst, G.T.J.; Batschauer, A.; Ahmad, M. The Cryptochromes: Blue Light Photoreceptors in Plants and Animals. Annu. Rev. Plant. Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Liu, H.; Liu, B.; Liu, X.; Lin, C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in arabidopsis. Curr. Biol. 2011, 21, 841–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Zuo, Z.; Liu, H.; Liu, X.; Lin, C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011, 25, 1029–1034. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef] [Green Version]
- Lian, H.L.; He, S.B.; Zhang, Y.C.; Zhu, D.M.; Zhang, J.Y.; Jia, K.P.; Sun, S.X.; Li, L.; Yang, H.Q. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011, 25, 1023–1028. [Google Scholar] [CrossRef] [Green Version]
- Demarsy, E.; Fankhauser, C. Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 2009, 12, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Suetsugu, N.; Wada, M. Evolution of Three LOV Blue Light Receptor Families in Green Plants and Photosynthetic Stramenopiles: Phototropin, ZTL/FKF1/LKP2 and Aureochrome. Plant Cell Physiol. 2013, 54, 8–23. [Google Scholar] [CrossRef] [Green Version]
- Inoue, S.-I.; Kinoshita, T.; Matsumoto, M.; Nakayama, K.I.; Doi, M.; Shimazaki, K.-I. Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 5626–5631. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Oses-Prieto, J.A.; Kutschera, U.; Tseng, T.S.; Hao, L.; Burlingame, A.L.; Wang, Z.Y.; Briggs, W.R. Blue light-induced proteomic changes in etiolated Arabidopsis seedlings. J. Proteome Res. 2014, 13, 2524–2533. [Google Scholar] [CrossRef]
- Motchoulski, A.; Liscum, E. Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 1999, 286, 961–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esmon, C.A.; Tinsley, A.G.; Ljung, K.; Sandberg, G.; Hearne, L.B.; Liscum, E. A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc. Natl. Acad. Sci. USA 2006, 103, 236–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Li, J.; Qu, L.; Hager, J.; Chen, Z.; Zhao, H.; Deng, X.W. Light Control of Arabidopsis Development Entails Coordinated Regulation of Genome Expression and Cellular Pathways. Plant Cell 2001, 13, 2589–2607. [Google Scholar] [CrossRef] [PubMed]
- Inada, S.; Ohgishi, M.; Mayama, T.; Okada, K.; Sakai, T. RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 2004, 16, 887–896. [Google Scholar] [CrossRef] [Green Version]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.H.; Tobin, E.M.; et al. F-Box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 2010, 22, 606–622. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.C.; Lasswell, J.; Rogg, L.E.; Cohen, M.A.; Bartel, B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 2000, 101, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Fujiwara, S.; Somers, D.E. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010, 29, 1903–1915. [Google Scholar] [CrossRef] [Green Version]
- Andrade, M.A.; González-Guzmán, M.; Serrano, R.; Rodríguez, P.L. A combination of the F-box motif and kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol. Biol. 2001, 46, 603–614. [Google Scholar] [CrossRef]
- Imaizumi, T.; Schultz, T.; Harmon, F.; Ho, L.A.; Kay, A.S. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schäfer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef] [Green Version]
- Tossi, V.; Lamattina, L.; Jenkins, G.I.; Cassia, R.O. Ultraviolet-B-induced stomatal closure in arabidopsis is regulated by the UV RESISTANCE LOCUS8 photoreceptor in a nitric oxide-dependent mechanism. Plant Physiol. 2014, 164, 2220–2230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fehér, B.; Kozma-Bognár, L.; Kevei, É.; Hajdu, A.; Binkert, M.; Davis, S.J.; Schäfer, E.; Ulm, R.; Nagy, F. Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 2011, 67, 37–48. [Google Scholar] [CrossRef]
- Wu, D.; Hu, Q.; Yan, Z.; Chen, W.; Yan, C.; Huang, X.; Zhang, J.; Yang, P.; Deng, H.; Wang, J.; et al. Structural basis of ultraviolet-B perception by UVR8. Nature 2012, 484, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Cloix, C.; Kaiserli, E.; Heilmann, M.; Baxter, K.J.; Brown, B.A.; O’Hara, A.; Smith, B.O.; Christie, J.M.; Jenkins, G.I. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc. Natl. Acad. Sci. USA 2012, 109, 16366–16370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binkert, M.; Kozma-Bognár, L.; Terecskei, K.; De Veylder, L.; Nagy, F.; Ulm, R. UV-B-Responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 2014, 26, 4200–4213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heijde, M.; Ulm, R. UV-B photoreceptor-mediated signalling in plants. Trends Plant. Sci. 2012, 17, 230–237. [Google Scholar] [CrossRef]
- Ulm, R.; Nagy, F. Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant. Biol. 2005, 8, 477–482. [Google Scholar] [CrossRef]
- Ballaré, C.L. Light Regulation of Plant Defense. Annu. Rev. Plant. Biol. 2014, 65, 335–363. [Google Scholar] [CrossRef]
- Hideg, É.; Jansen, M.A.K.; Strid, Å. UV-B exposure, ROS, and stress: Inseparable companions or loosely linked associates? Trends Plant Sci. 2013, 18, 107–115. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef]
- Ni, W.; Xu, S.L.; Tepperman, J.M.; Stanley, D.J.; Maltby, D.A.; Gross, J.D.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 2014, 344, 1160–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Ni, W.; Yu, R.; Deng, X.W.; Chen, H.; Wei, N. Light-Dependent Degradation of PIF3 by SCFEBF1/2 Promotes a Photomorphogenic Response in Arabidopsis. Curr. Biol. 2017, 27, 2420–2430. [Google Scholar] [CrossRef] [Green Version]
- Ni, W.; Xu, S.L.; González-Grandío, E.; Chalkley, R.J.; Huhmer, A.F.R.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 2017, 8, 15236. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Kim, J.; Park, E.; Kim, J.I.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majee, M.; Kumar, S.; Kathare, P.K.; Wu, S.; Gingerich, D.; Nayak, N.R.; Salaita, L.; Dinkins, R.; Martin, K.; Goodin, M.; et al. KELCH F-BOX protein positively influences Arabidopsis seed germination by targeting PHYTOCHROME-INTERACTING FACTOR1. Proc. Natl. Acad. Sci. USA 2018, 115, E4120–E4129. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Barros-Galvão, T.; Cole, A.F.; Gilday, A.D.; He, Z.; Li, Y.; Harvey, D.; Larson, T.R.; Graham, I.A. MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, 8442–8447. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.S.; Sun, T.P.; Kamiya, Y.; Choi, G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.K.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 2008, 20, 1260–1277. [Google Scholar] [CrossRef] [Green Version]
- Parcy, F. Flowering: A time for integration. Int. J. Dev. Biol. 2005, 49, 585–593. [Google Scholar] [CrossRef] [Green Version]
- Shim, J.S.; Imaizumi, T. Circadian clock and photoperiodic response in arabidopsis: From seasonal flowering to redox homeostasis. Biochemistry 2015, 54, 157–170. [Google Scholar] [CrossRef] [Green Version]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomoto, Y.; Kubozono, S.; Yamashino, T.; Nakamichi, N.; Mizuno, T. Circadian Clock- and PIF4-Controlled Plant Growth: A Coincidence Mechanism Directly Integrates a Hormone Signaling Network into the Photoperiodic Control of Plant Architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1950–1964. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian clock and photoperiodic flowering in arabidopsis: CONSTANS is a Hub for Signal integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, S.; Marchal, V.; Panigrahi, K.C.S.; Wenkel, S.; Soppe, W.; Deng, X.W.; Valverde, F.; Coupland, G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008, 27, 1277–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazaro, A.; Valverde, F.; Piñ, M.; Jarillo, J.A. The Arabidopsis E3 Ubiquitin Ligase HOS1 Negatively Regulates CONSTANS Abundance in the Photoperiodic Control of Flowering W. Plant Cell 2012, 24, 982–999. [Google Scholar] [CrossRef] [Green Version]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor Regulation of CONSTANS Protein in Photoperiodic Flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, Q.; Liu, Y.; Zhao, X.; Imaizumi, T.; Somers, D.E.; Tobin, E.M.; Lin, C. Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl. Acad. Sci. USA 2013, 110, 17582–17587. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A Repressor Complex Governs the Integration of Flowering Signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Le, C.; Wang, Y.; Li, Z.; Jiang, D.; Wang, Y.; He, Y. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 2013, 4, 1–10. [Google Scholar] [CrossRef]
- Castillejo, C.; Pelaz, S. The Balance between CONSTANS and TEMPRANILLO Activities Determines FT Expression to Trigger Flowering. Curr. Biol. 2008, 18, 1338–1343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathieu, J.; Yant, L.J.; Mürdter, F.; Küttner, F.; Schmid, M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teixeira, R.T. Distinct Responses to Light in Plants. Plants 2020, 9, 894. https://doi.org/10.3390/plants9070894
Teixeira RT. Distinct Responses to Light in Plants. Plants. 2020; 9(7):894. https://doi.org/10.3390/plants9070894
Chicago/Turabian StyleTeixeira, Rita Teresa. 2020. "Distinct Responses to Light in Plants" Plants 9, no. 7: 894. https://doi.org/10.3390/plants9070894




