Knock-Down the Expression of Brassinosteroid Receptor TaBRI1 Reduces Photosynthesis, Tolerance to High Light and High Temperature Stresses and Grain Yield in Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
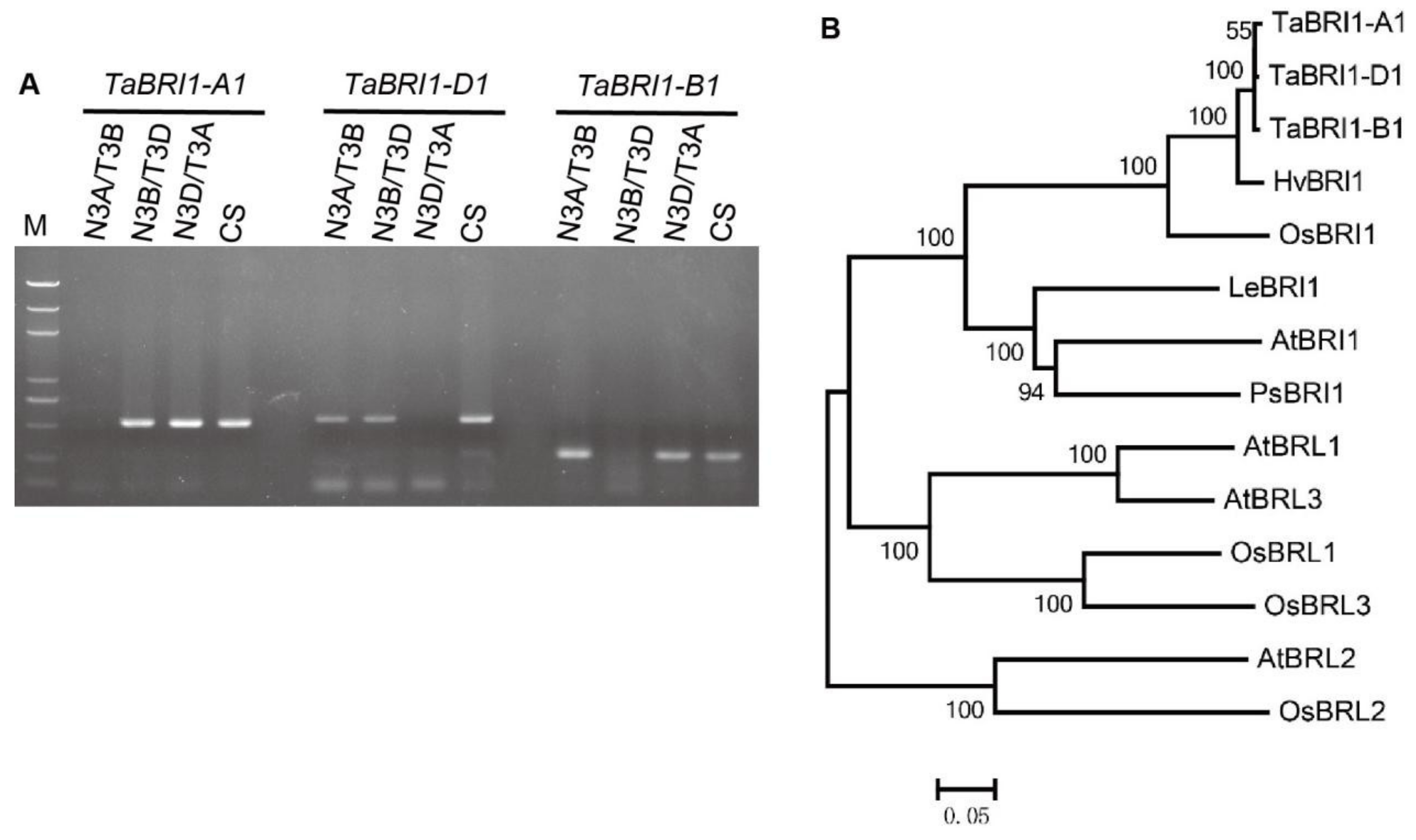
2.2. Gene Cloning and Phylogenetic Analysis
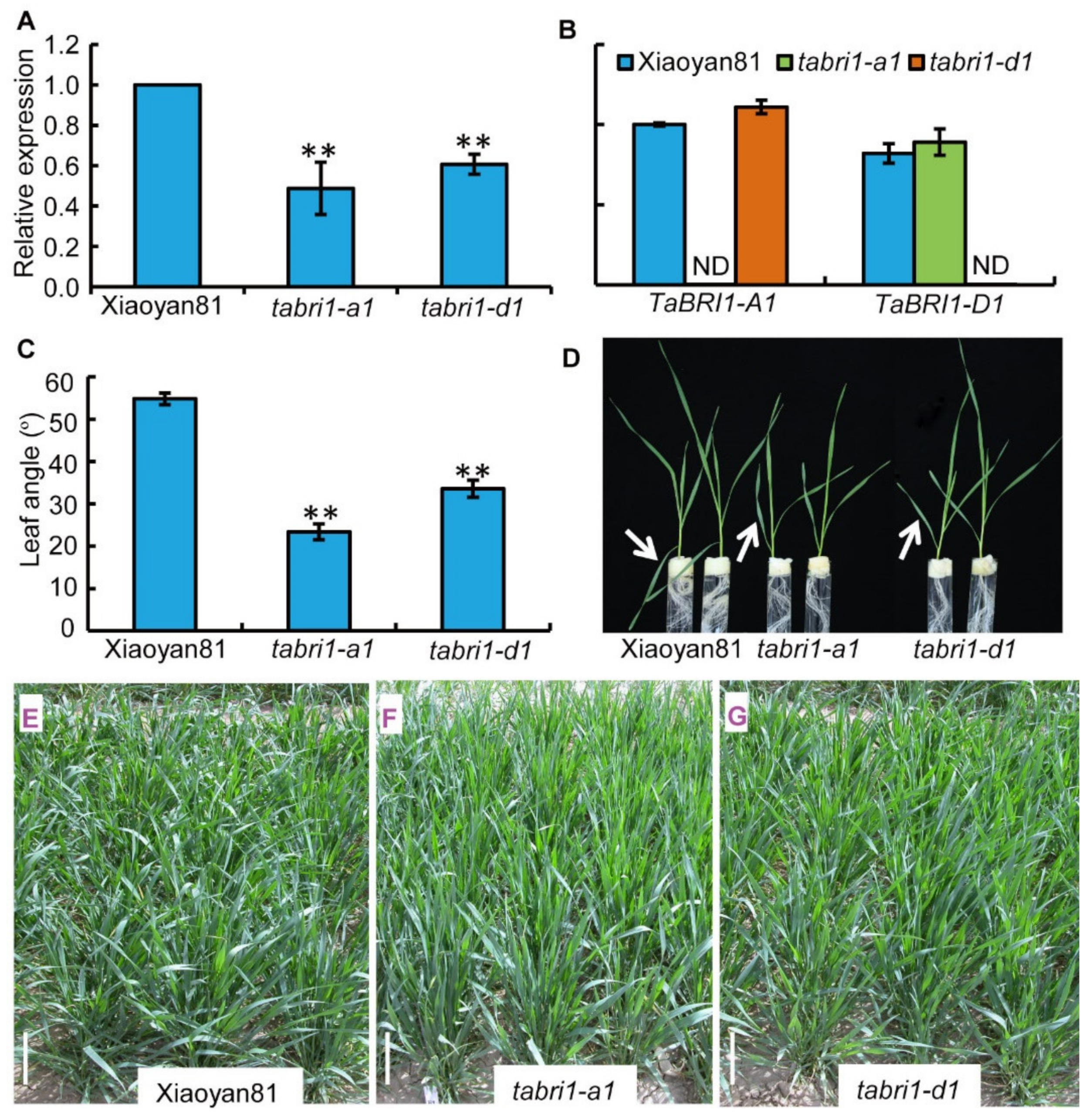
2.3. Screening for TaBRI1-A1, TaBRI1-D1 Deletion Mutants
2.4. Vector Construction and Transformation
2.5. Leaf Angle Measurements
2.6. Gas Exchange Measurements
2.7. Virus-Induced Gene Silencing (VIGS) Assay
2.8. Imaging Analysis of Chlorophyll Fluorescence
2.9. RNA Extraction and Quantitative RT-PCR
2.10. EBR Treatment
2.11. Statistical Analysis
3. Results
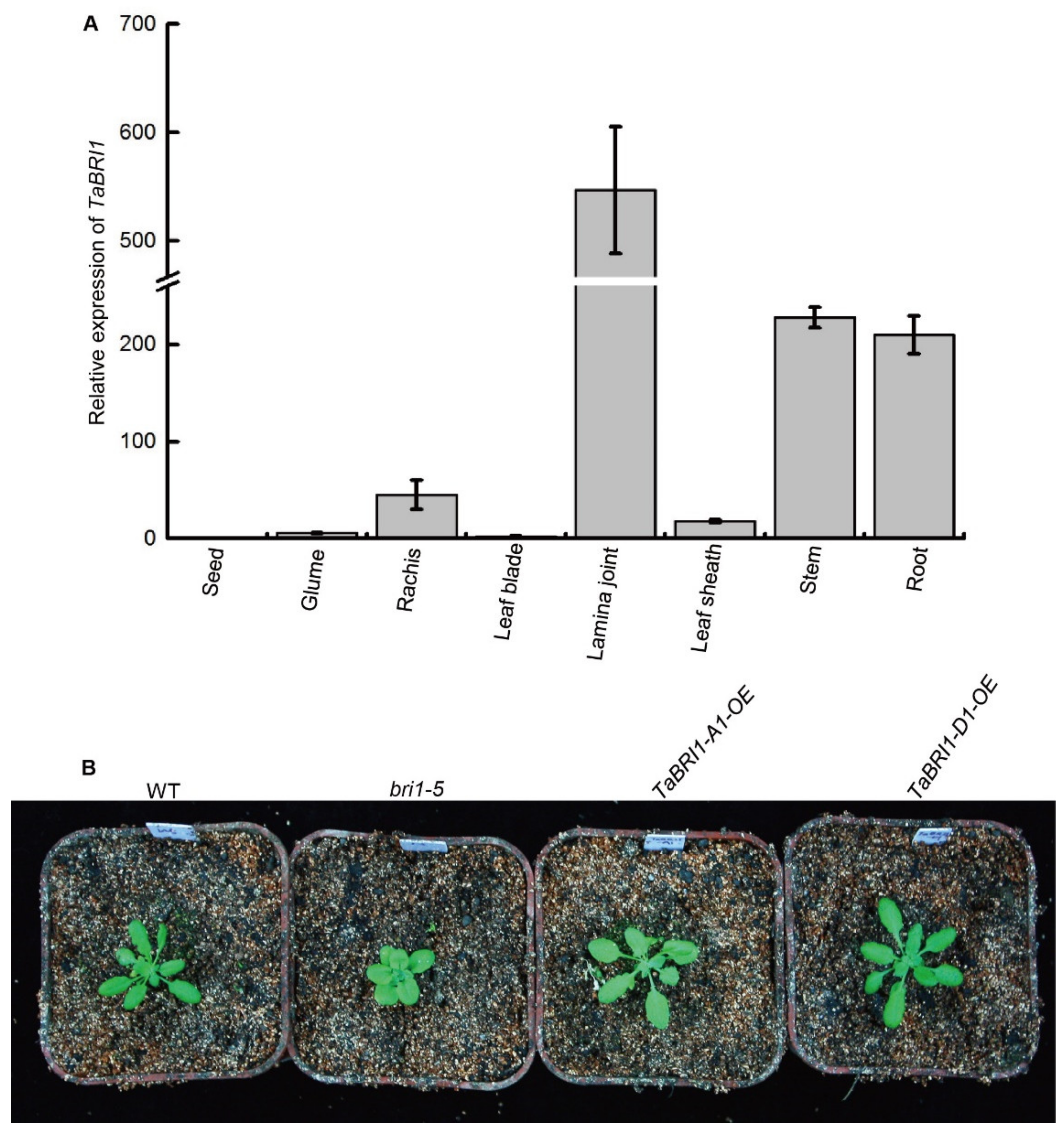
3.1. Cloning of Three TaBRI1 Homologous Genes and the Expression Pattern of TaBRI1
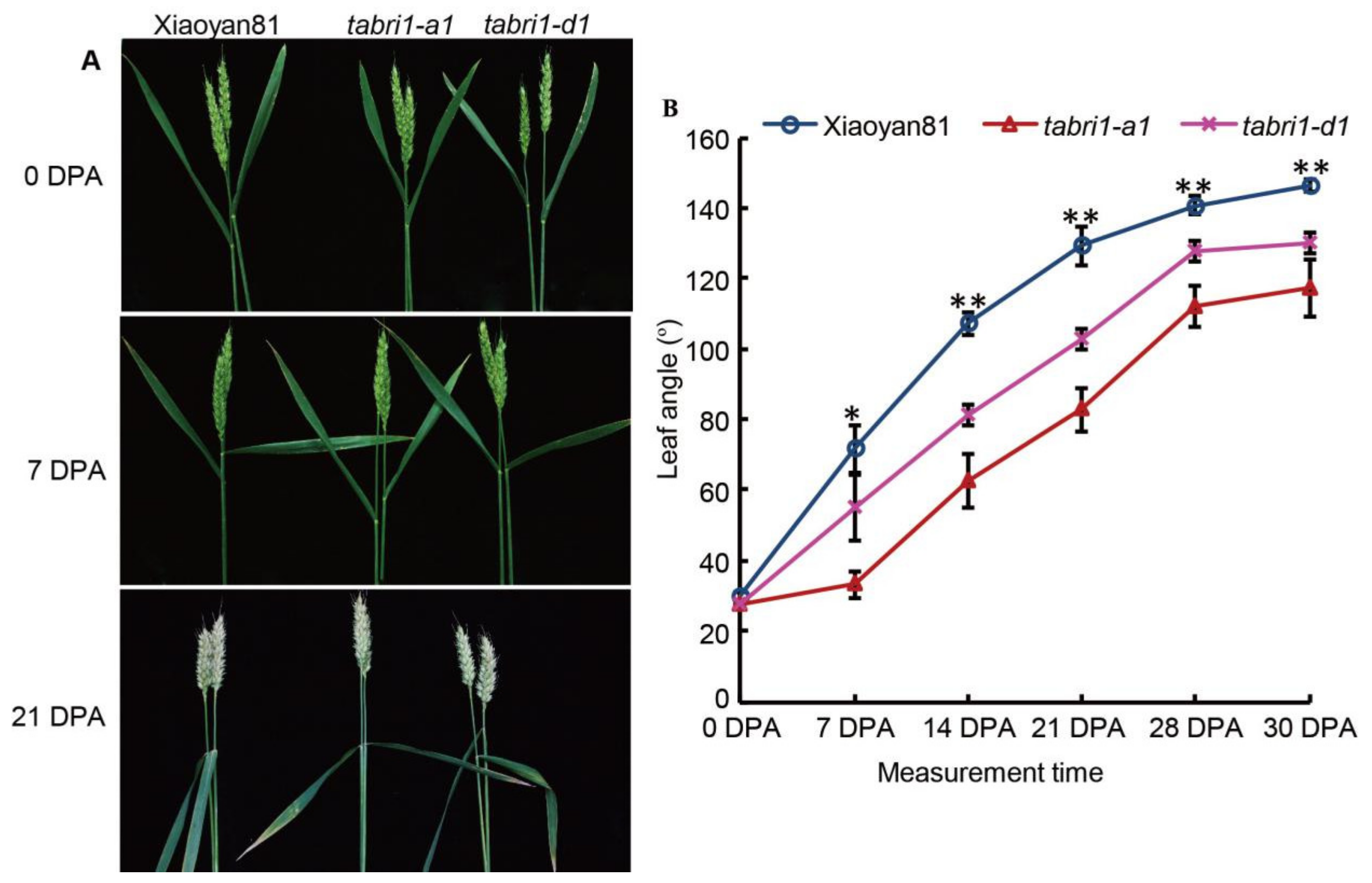
3.2. TaBRI1-A1 or TaBRI1-D1 Deletion Mutants with a Decreased TaBRI1 Expression Exhibited Relatively Erect Leaves
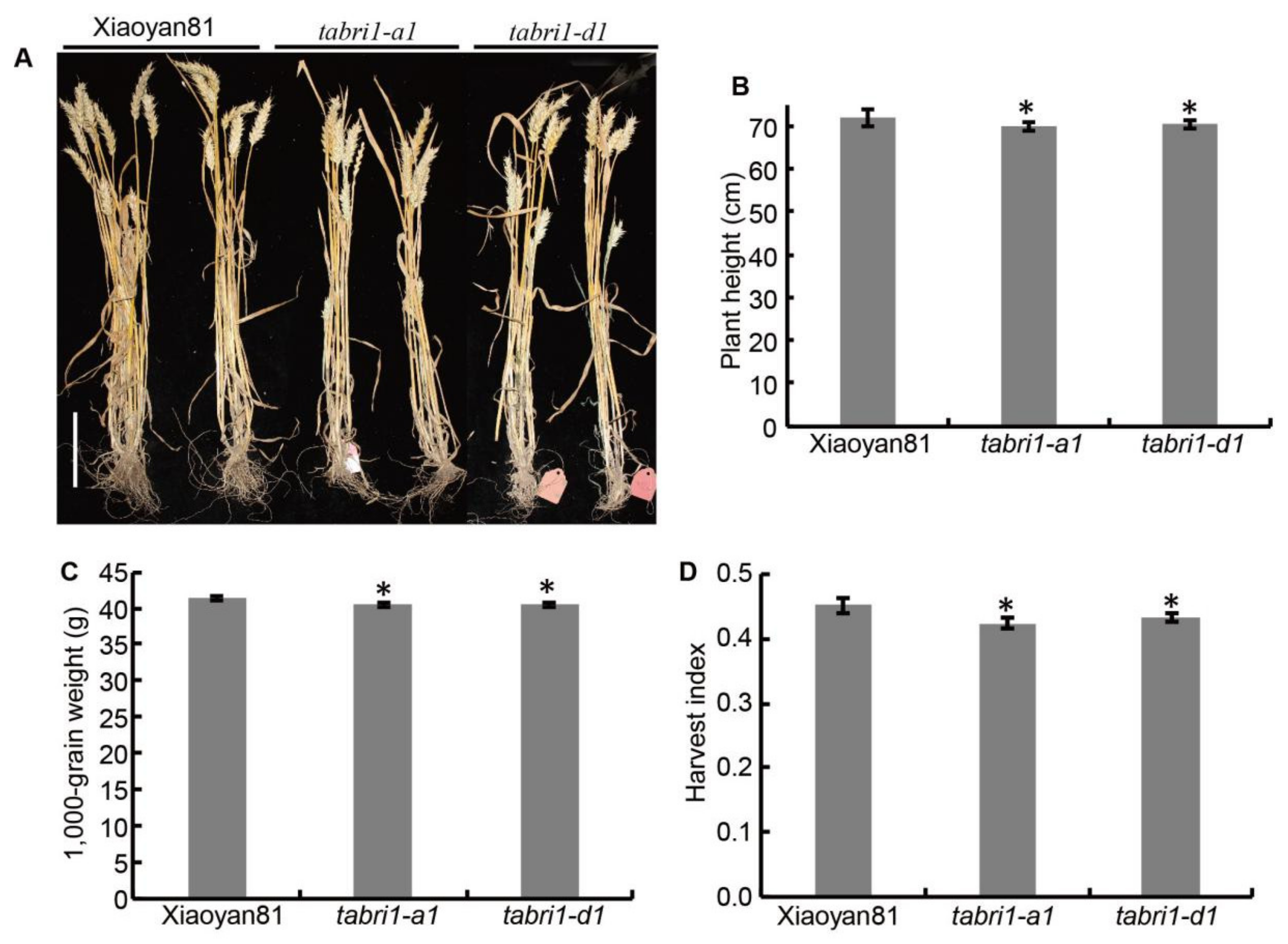
3.3. TaBRI1 Knock-Down Mutants Showed a Reduction in Grain Yield
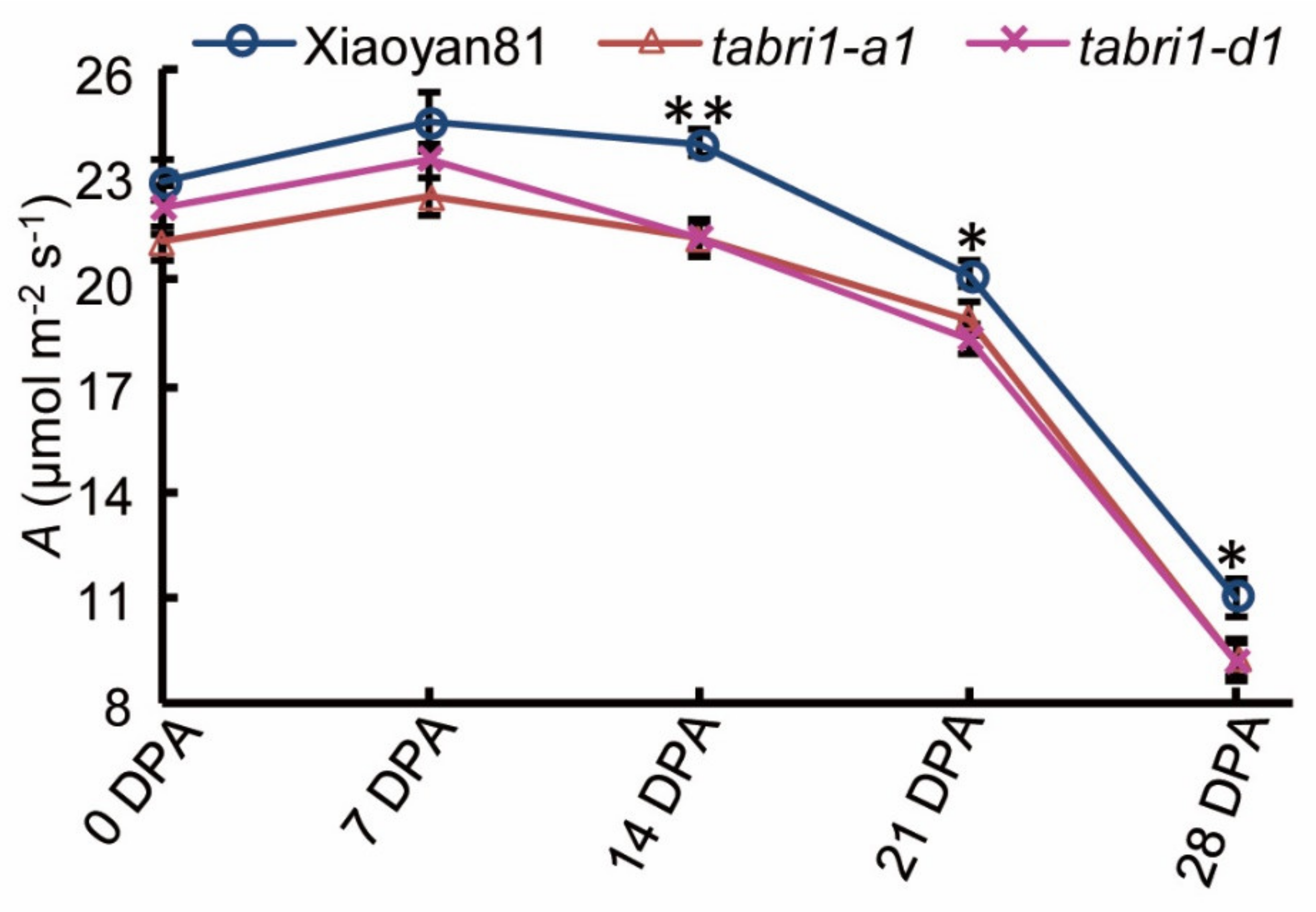
3.4. TaBRI1 Knock-Down Mutants Showed a Reduction in Photosynthetic Rate
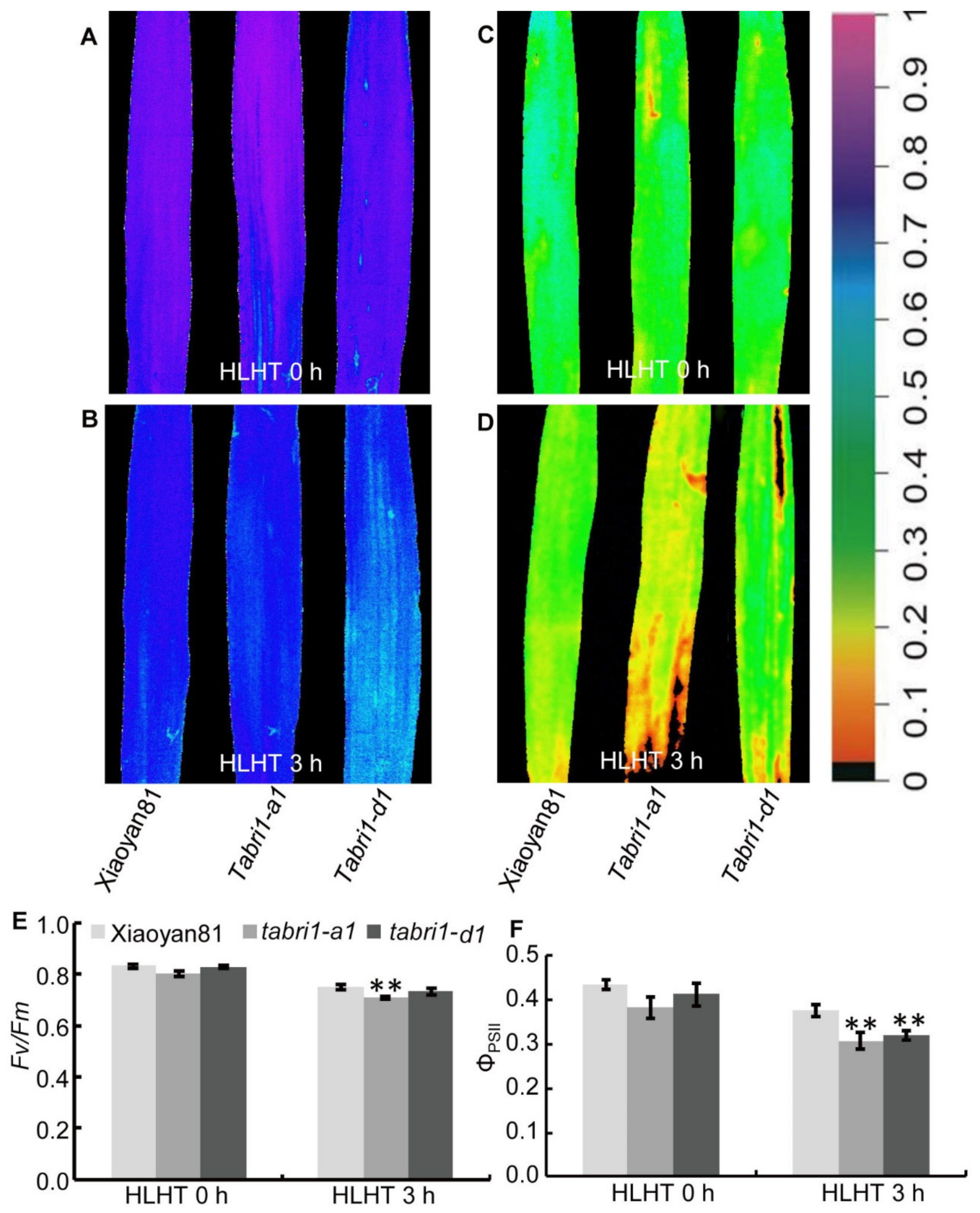
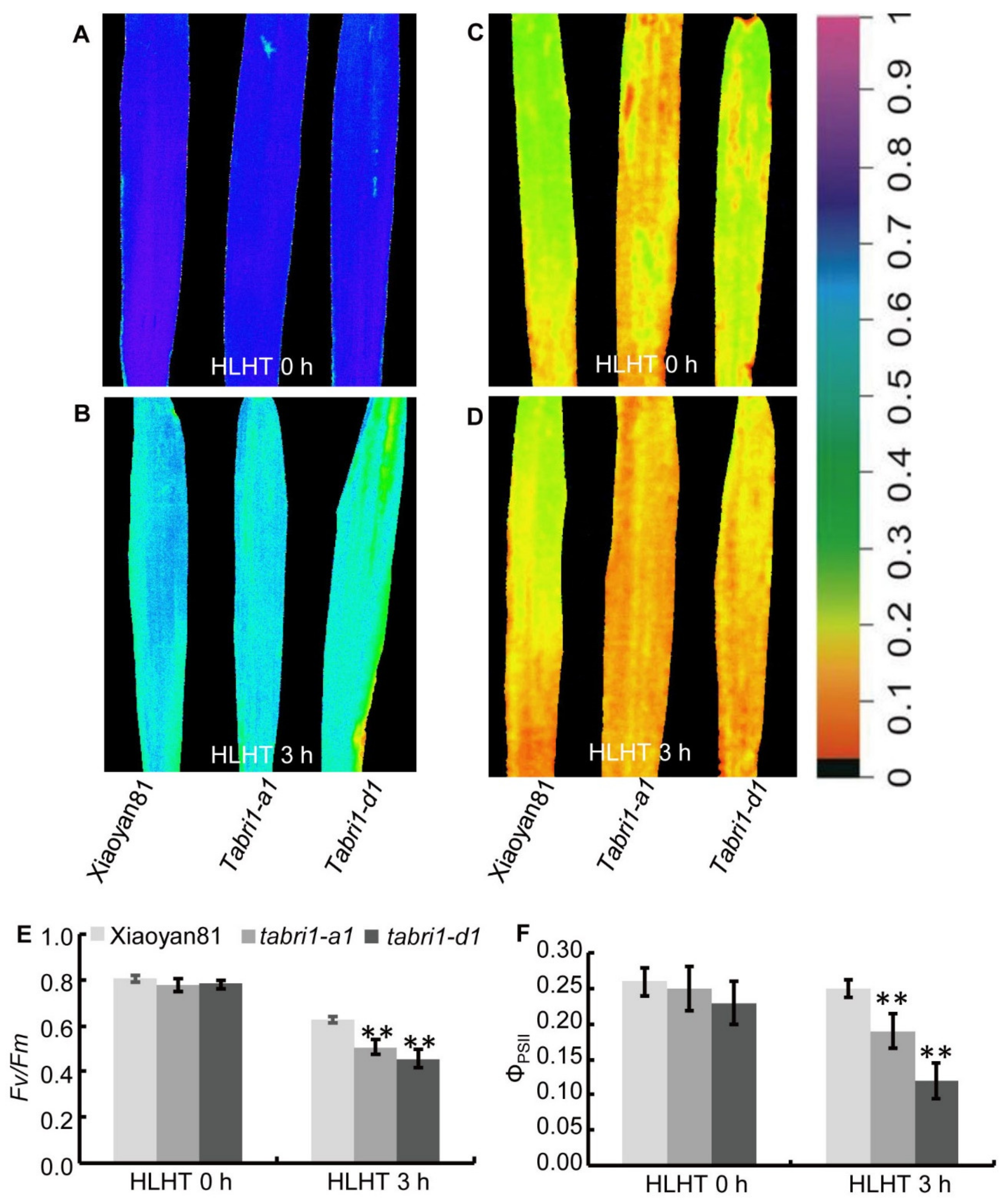
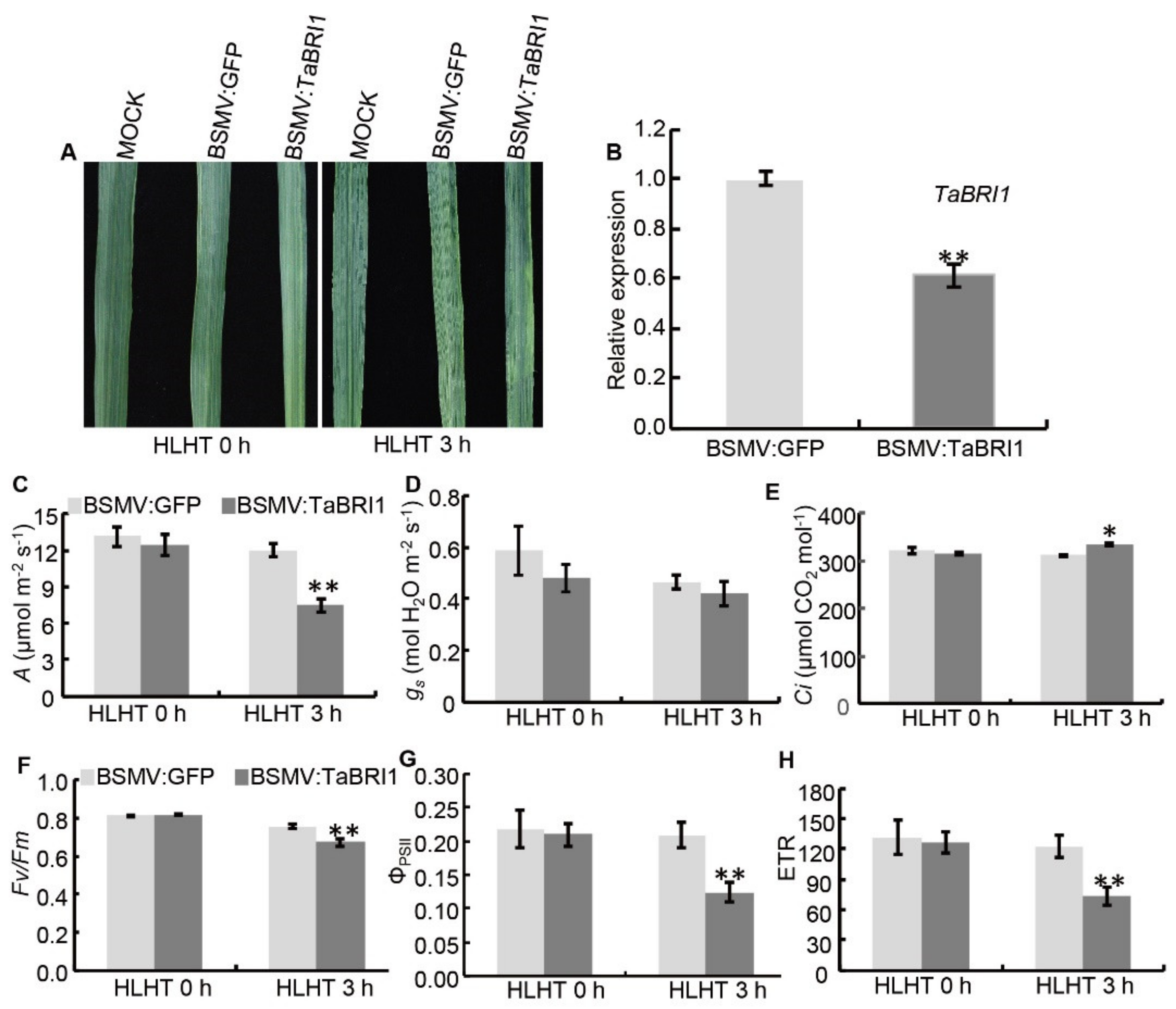
3.5. TaBRI1 Knock-Down Plants Generated by TaBRI1-A1, TaBRI1-D1 Deletion or Using Virus-Induced Gene Silencing Exhibited the Reduced Tolerance to High Light and High Temperature Stresses
3.6. TaBRI1 Knock-Down Mutants Is Less Sensitive to Exogenous BR Treatment than Wild Type Plants
4. Discussion
4.1. Knock-Down the Expression of TaBRI1 Reduced Leaf Angles and the 1000-Grain Weight
4.2. Knock-Down the Expression of TaBRI1 Reduced Photosynthetic Rate and the Tolerance to High Light and High Temperature Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murchie, E.H.; Pinto, M.; Horton, P. Agriculture and the new challenges for photosynthesis research. New Phytol. 2009, 181, 532–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Bai, M.Y.; Chong, K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014, 33, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Savaldigoldstein, S. Growth control: Brassinosteroid activity gets context. J. Exp. Bot. 2015, 66, 1123. [Google Scholar] [CrossRef]
- Vert, G.; Nemhauser, J.L.; Geldner, N.; Hong, F.; Chory, J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 2005, 21, 177–201. [Google Scholar] [CrossRef]
- Tong, H.; Chu, C. Functional Specificities of Brassinosteroid and Potential Utilization for Crop Improvement. Trends Plant Sci. 2018, 23, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12, 1591–1605. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Wang, M.; Xu, Y.Y.; Luo, W.; Liu, Y.J.; Xu, Z.H.; Li, J.; Chong, K. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 2009, 7, 791–806. [Google Scholar] [CrossRef] [PubMed]
- Jaillais, Y.; Hothorn, M.; Belkhadir, Y.; Dabi, T.; Nimchuk, Z.L.; Meyerowitz, E.M.; Chory, J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Gene Dev. 2011, 25, 232–237. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR Receptor-like Protein Kinase, Interacts with BRI1 and Modulates Brassinosteroid Signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Tong, H.N.; Liu, L.C.; Jin, Y.; Du, L.; Yin, Y.H.; Qian, Q.; Zhu, L.H.; Chu, C.C. DWARF AND LOW-TILLERING Acts as a Direct Downstream Target of a GSK3/SHAGGY-Like Kinase to Mediate Brassinosteroid Responses in Rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar] [CrossRef]
- Tang, W.Q.; Yuan, M.; Wang, R.J.; Yang, Y.H.; Wang, C.M.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.P.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.J.; Huang, L.F.; Zhou, Y.H.; Mao, W.H.; Shi, K.; Wu, J.X.; Asami, T.; Chen, Z.X.; Yu, J.Q. Brassinosteroids promote photosynthesis and growth by enhancing activation of Rubisco and expression of photosynthetic genes in Cucumis sativus. Planta 2009, 230, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.C.; Zhai, Z.X.; Tian, X.L.; Duan, L.S.; Li, Z.H. Brassinolide alleviated the adverse effect of water deficits on photosynthesis and the antioxidant of soybean (Glycine max L.). Plant Growth Regul. 2008, 56, 257–264. [Google Scholar] [CrossRef]
- Shahbaz, M.; Ashraf, M.; Athar, H.U.R. Does exogenous application of 24-epibrassinolide ameliorate salt induced growth inhibition in wheat (Triticum aestivum L.)? Plant Growth Regul. 2008, 55, 51–64. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Yuan, H.L.; Ogweno, J.O.; Zhou, Y.H.; Xia, X.J.; Mao, W.H.; Shi, K.; Yu, J.Q. Brassinosteroid alleviates phenanthrene and pyrene phytotoxicity by increasing detoxification activity and photosynthesis in tomato. Chemosphere 2012, 86, 546–555. [Google Scholar] [CrossRef]
- Sharma, I.; Pati, P.K.; Bhardwaj, R. Effect of 24-epibrassinolide on oxidative stress markers induced by nickel-ion in Raphanus sativus L. Acta Physiol. Plant 2011, 33, 1723–1735. [Google Scholar] [CrossRef]
- Gururani, M.A.; Mohanta, T.K.; Bae, H. Current understanding of the interplay between phytohormones and photosynthesis under environmental stress. Int. J. Mol. Sci. 2015, 16, 19055–19085. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Murata, N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 2008, 13, 178–182. [Google Scholar] [CrossRef]
- Anjum, S.A.; Wang, L.C.; Farooq, M.; Hussain, M.; Xue, L.L.; Zou, C.M. Brassinolide Application Improves the Drought Tolerance in Maize Through Modulation of Enzymatic Antioxidants and Leaf Gas Exchange. J. Agron. Crop Sci. 2011, 197, 177–185. [Google Scholar] [CrossRef]
- Friebe, A. Brassinosteroids in induced resistance and induction of tolerances to abiotic stress in plants. In Natural Products for Pest Management; American Chemical Society: Washington, DC, USA, 2006; Volume 927, pp. 233–242. [Google Scholar] [CrossRef]
- Sonjaroon, W.; Jutamanee, K.; Khamsuk, O.; Thussagunpanit, J.; Kaveeta, L.; Suksamrarn, A. Impact of brassinosteroid mimic on photosynthesis, carbohydrate content and rice seed set at reproductive stage under heat stress. Agric. Nat. Resour. 2018, 52, 234–240. [Google Scholar] [CrossRef]
- Sharma, I.; Kaur, N.; Pati, P.K. Brassinosteroids: A Promising Option in Deciphering Remedial Strategies for Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2017, 8, 2151. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Liu, L.J.; Zhao, Q.Z.; Zhang, Z.H.; Li, Q.L.; Lin, B.Y.; Wu, Y.R.; Tang, S.Y.; Xie, Q. Arabidopsis Ubiquitin Conjugase UBC32 Is an ERAD Component That Functions in Brassinosteroid-Mediated Salt Stress Tolerance. Plant Cell 2012, 24, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Li, M.; Chen, W.; Wang, Y.; Sun, C.; Yin, H.; He, K.; Li, J. Thermal-Enhanced bri1-301 Instability Reveals a Plasma Membrane Protein Quality Control System in Plants. Front. Plant Sci. 2018, 9, 1620. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Ueguchi-Tanaka, M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 2005, 17, 2243–2254. [Google Scholar] [CrossRef]
- Tanabe, S.; Ashikari, M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Yano, M.; Yoshimura, A.; Kitano, H.; Matsuoka, M.; Fujisawa, Y.; et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 2005, 17, 776–790. [Google Scholar] [CrossRef]
- Tanaka, A.; Nakagawa, H.; Tomita, C.; Shimatani, Z.; Ohtake, M.; Nomura, T.; Jiang, C.J.; Dubouzet, J.G.; Kikuchi, S.; Sekimoto, H.; et al. BRASSINOSTEROID UPREGULATED1, Encoding a Helix-Loop-Helix Protein, Is a Novel Gene Involved in Brassinosteroid Signaling and Controls Bending of the Lamina Joint in Rice. Plant Physiol. 2009, 151, 669–680. [Google Scholar] [CrossRef]
- Choe, S.; Fujioka, S.; Noguchi, T.; Takatsuto, S.; Yoshida, S.; Feldmann, K.A. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001, 26, 573–582. [Google Scholar] [CrossRef]
- Wu, C.Y.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.L.; Wan, J.M.; Zhai, H.Q.; Takatsuto, S.; et al. Brassinosteroids regulate grain filling in rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S.; et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Morinaka, Y.; Sakamoto, T.; Inukai, Y.; Agetsuma, M.; Kitano, H.; Ashikari, M.; Matsuoka, M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 2006, 141, 924–931. [Google Scholar] [CrossRef]
- Chono, M.; Honda, I.; Zeniya, H.; Yoneyama, K.; Saisho, D.; Takeda, K.; Takatsuto, S.; Hoshino, T.; Watanabe, Y. A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 2003, 133, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Koka, C.V.; Cerny, R.E.; Gardner, R.G.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Clouse, S.D. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 2000, 122, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Bishop, G.J.; Kaneta, T.; Reid, J.B.; Chory, J.; Yokota, T. The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J. 2003, 36, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Goddard, R.; Peraldi, A.; Ridout, C.; Nicholson, P. Enhanced disease resistance caused by BRI1 mutation is conserved between Brachypodium distachyon and barley (Hordeum vulgare). Mol. Plant Microbe Interact. 2014, 27, 1095–1106. [Google Scholar] [CrossRef]
- Yang, Y.; Li, S.; Zhang, K.; Dong, Z.; Li, Y.; An, X.; Chen, J.; Chen, Q.; Jiao, Z.; Liu, X.; et al. Efficient isolation of ion beam-induced mutants for homoeologous loci in common wheat and comparison of the contributions of Glu-1 loci to gluten functionality. Theor. Appl. Genet. 2014, 127, 359–372. [Google Scholar] [CrossRef]
- Ren, Y.Z.; He, X.; Liu, D.C.; Li, J.J.; Zhao, X.Q.; Li, B.; Tong, Y.P.; Zhang, A.M.; Li, Z.S. Major quantitative trait loci for seminal root morphology of wheat seedlings. Mol. Breed. 2012, 30, 139–148. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Petty, I.T.; Hunter, B.G.; Wei, N.; Jackson, A.O. Infectious barley stripe mosaic virus RNA transcribed in vitro from full-length genomic cDNA clones. Virology 1989, 171, 342–349. [Google Scholar] [CrossRef]
- Lu, C.; Qiu, N.; Wang, B.; Zhang, J. Salinity treatment shows no effects on photosystem II photochemistry, but increases the resistance of photosystem II to heat stress in halophyte Suaeda salsa. J. Exp. Bot. 2003, 54, 851–860. [Google Scholar] [CrossRef]
- Cui, K.H.; Peng, S.B.; Xing, Y.Z.; Yu, S.B.; Xu, C.G.; Zhang, Q. Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor. Appl. Genet. 2003, 106, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.W.; Luo, L.J.; Ying, C.S.; Wang, Y.P.; Yu, X.Q.; Guo, L.B.; Paterson, A.H.; Li, Z.K. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two testcross populations. Theor. Appl. Genet. 2003, 107, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhou, G.; Yu, H.; Yu, S. Fine mapping a major QTL for flag leaf size and yield-related traits in rice. Theor. Appl. Genet. 2011, 123, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Gladun, I.V.; Karpov, E.A. Distribution of assimilates from the flag leaf of rice during the reproductive period of development. Russ. J. Plant Physiol. 1993, 40, 215–218. [Google Scholar]
- Long, S.P.; Humphries, S.; Falkowski, P.G. Photoinhibition of Photosynthesis in Nature. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1994, 45, 633–655. [Google Scholar] [CrossRef]
- Sinclair, T.R.; Sheehy, J.E. Erect Leaves and Photosynthesis in Rice. Science 1999, 283, 1455. [Google Scholar] [CrossRef]
- Wu, X.; Tang, D.; Li, M.; Wang, K.; Cheng, Z. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013, 161, 317–329. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Fujioka, S.; Shimada, Y.; Takatsuto, S.; Agetsuma, M.; Yoshida, S.; Watanabe, Y.; et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002, 32, 495–508. [Google Scholar] [CrossRef]
- Wu, C.Y.; Kwok, S.; Harris, S.; Trieu, A.; Radhakrishnan, P.; Salazar, A.; Zhang, K.; Wang, J.L.; Wan, J.M.; Fujioka, S.; et al. Enhancement of the Brassinosteroid Biosynthesis Pathway Improves Grain Yield in Rice. In Biotechnology and Sustainable Agriculture 2006 and Beyond; Springer: Dordrecht, The Netherlands, 2007; pp. 319–322. [Google Scholar]
- Makino, A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2011, 155, 125–129. [Google Scholar] [CrossRef]
- Long, S.P.; Zhu, X.G.; Naidu, S.L.; Ort, D.R. Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 2006, 29, 315–330. [Google Scholar] [CrossRef]
- Oh, M.H.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation of the BRI1 receptor kinase occurs via a post-translational modification and is activated by the juxtamembrane domain. Front. Plant Sci. 2012, 3, 175. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, B.H.; Nam, K.H. Reduced expression of the genes encoding chloroplast-localized proteins in a cold-resistant bri1 (brassinosteroid-insensitive 1) mutant. Plant Signal. Behav. 2010, 5, 458–463. [Google Scholar] [CrossRef]
- Ogren, E.; Rosenqvist, E. On the significance of photoinhibition of photosynthesis in the field and its generality among species. Photosynth. Res. 1992, 33, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Breja, P.; Khurana, J.P.; Khurana, P. Wheat Brassinosteroid-Insensitive1 (TaBRI1) Interacts with Members of TaSERK Gene Family and Cause Early Flowering and Seed Yield Enhancement in Arabidopsis. PLoS ONE 2016, 11, e0153273. [Google Scholar] [CrossRef]
- Yu, J.Q.; Huang, L.F.; Hu, W.H.; Zhou, Y.H.; Mao, W.H.; Ye, S.F.; Nogues, S. A role for brassinosteroids in the regulation of photosynthesis in Cucumis sativus. J. Exp. Bot. 2004, 55, 1135–1143. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, J.; Zhu, W.; Tong, Y. Knock-Down the Expression of Brassinosteroid Receptor TaBRI1 Reduces Photosynthesis, Tolerance to High Light and High Temperature Stresses and Grain Yield in Wheat. Plants 2020, 9, 840. https://doi.org/10.3390/plants9070840
Fang J, Zhu W, Tong Y. Knock-Down the Expression of Brassinosteroid Receptor TaBRI1 Reduces Photosynthesis, Tolerance to High Light and High Temperature Stresses and Grain Yield in Wheat. Plants. 2020; 9(7):840. https://doi.org/10.3390/plants9070840
Chicago/Turabian StyleFang, Jingjing, Weiqi Zhu, and Yiping Tong. 2020. "Knock-Down the Expression of Brassinosteroid Receptor TaBRI1 Reduces Photosynthesis, Tolerance to High Light and High Temperature Stresses and Grain Yield in Wheat" Plants 9, no. 7: 840. https://doi.org/10.3390/plants9070840
APA StyleFang, J., Zhu, W., & Tong, Y. (2020). Knock-Down the Expression of Brassinosteroid Receptor TaBRI1 Reduces Photosynthesis, Tolerance to High Light and High Temperature Stresses and Grain Yield in Wheat. Plants, 9(7), 840. https://doi.org/10.3390/plants9070840




