First Report of Root Rot Caused by Phytophthora bilorbang on Olea europaea in Italy
Abstract
1. Introduction
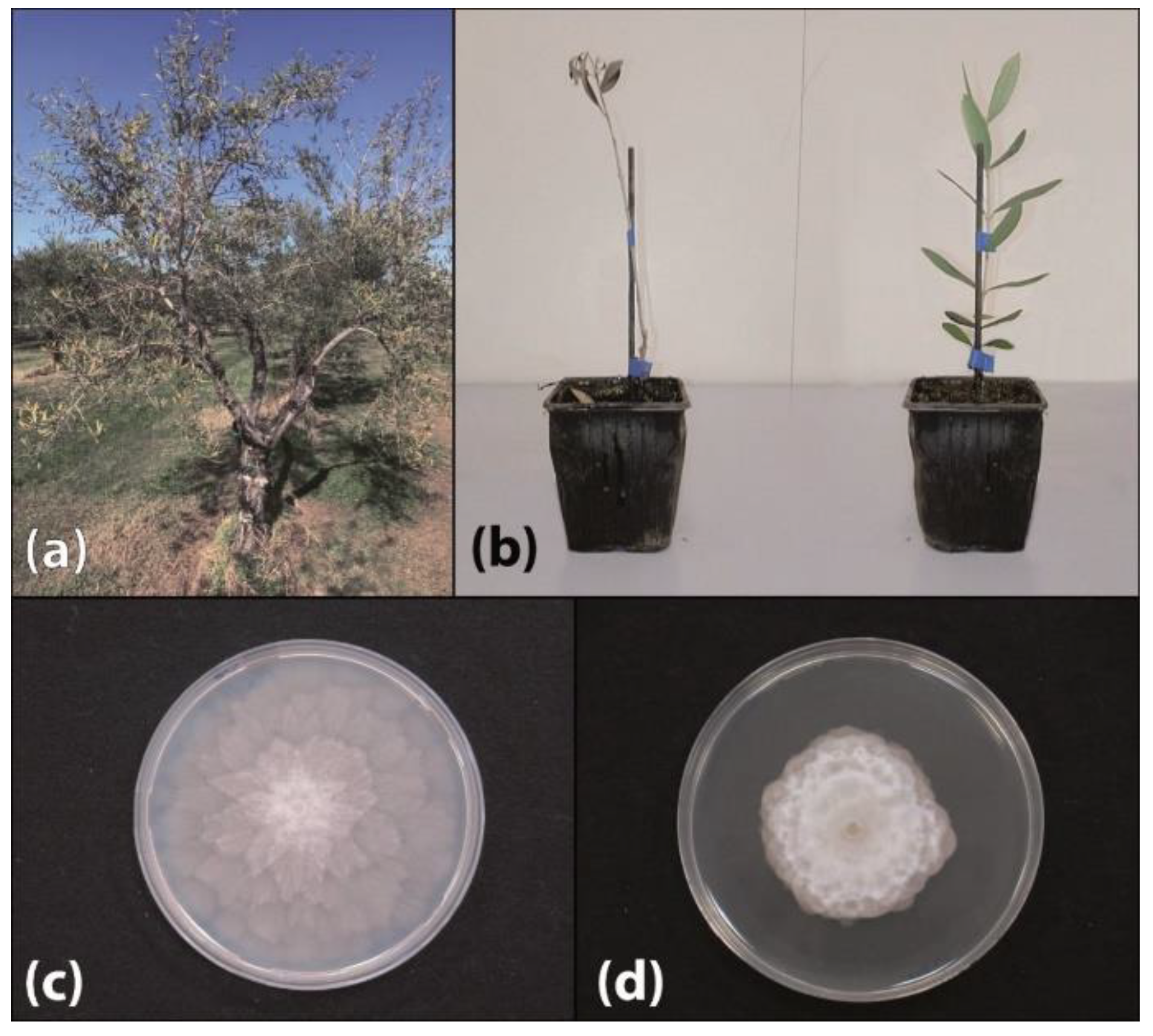
2. Results
2.1. Species Identification and Morphological Features of Isolates
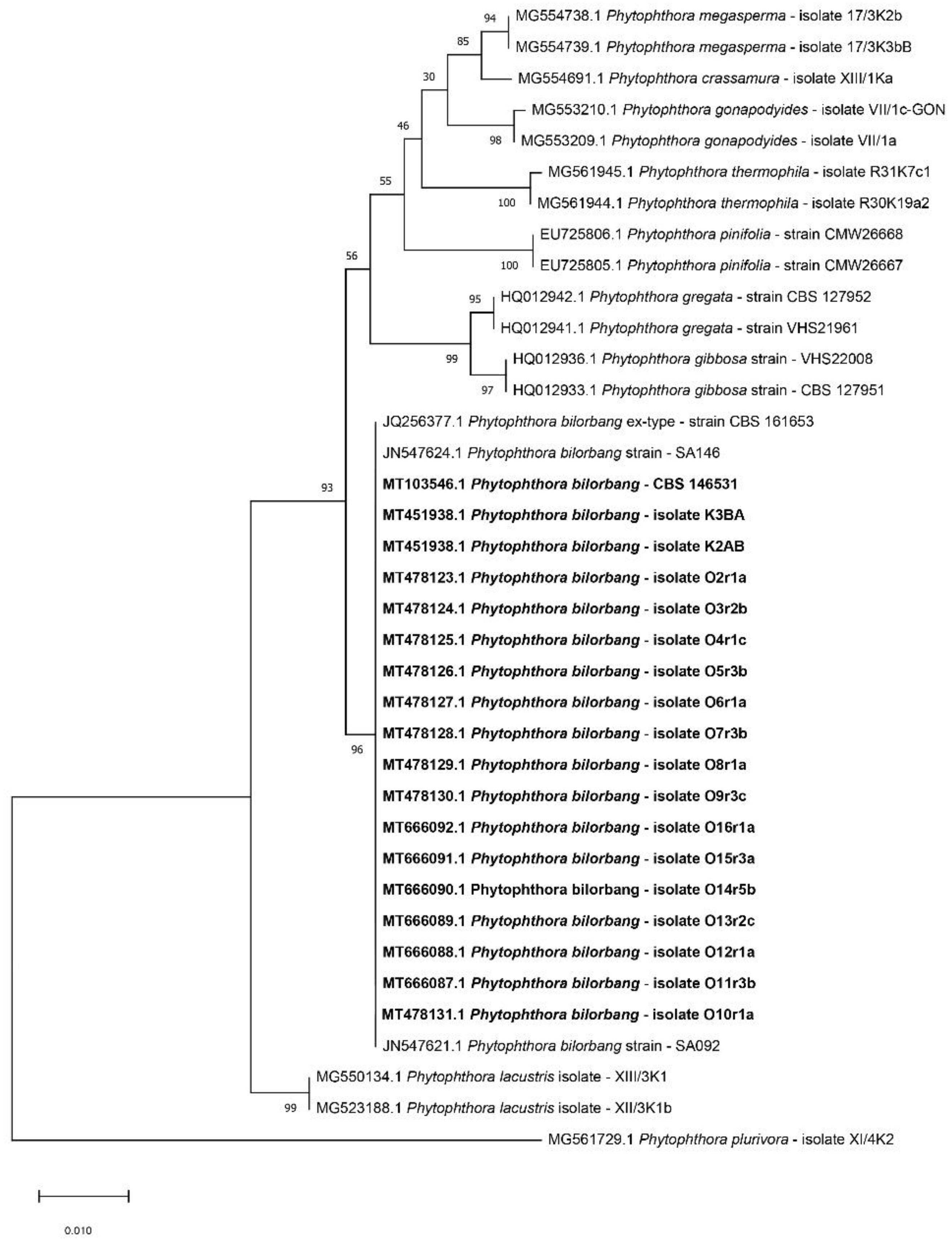
2.2. Molecular Identification
2.3. Pathogenicity
3. Discussion
4. Materials and Methods
4.1. Isolation and Morphological Identification of Isolates
4.2. Molecular Identification of Isolates
4.3. Pathogenicity Test
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartolini, G.; Petrucelli, R. Classification, Origin, Diffusion and History of the Olive; Food and Agriculture Organization of the United Nations: Rome, Italy, 2002; pp. 13–19. [Google Scholar]
- Krishnamurthy, K.V. Textbook of Biodiversity; Science Publishers: Enfield, NH, USA, 2003; pp. 39–51. [Google Scholar]
- Russo, C.; Cappelletti, G.M.; Nicoletti, G.M.; Di Noia, A.E.; Michalopoulos, G. Comparison of European olive production systems. Sustainability 2016, 8, 825. [Google Scholar] [CrossRef]
- ISTAT. Available online: http://www.istat.it (accessed on 23 June 2020).
- Cacciola, S.O.; Faedda, R.; Pane, A.; Scarito, G. Root and crown rot of olive caused by Phytophthora spp. In Olive Diseases and Disorders; Schena, L., Agosteo, G.E., Cacciola, S.O., Eds.; Transworld Research Network: Trivandrum, Kerala, India, 2011; pp. 305–327. [Google Scholar]
- Riddell, C.E.; Frederickson-Matika, D.; Armstrong, A.C.; Elliot, M.; Forster, J.; Hedley, P.E.; Morris, J.; Thorpe, P.; Cooke, D.E.L.; Pritchard, L.; et al. Metabarcoding reveals a high diversity of woody host-associated Phytophthora spp. in soils at public gardens and amenity woodlands in Britain. Peer J. 2019, 7, e6931. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Horta Jung, M.; Cacciola, S.O.; Cech, T.; Bakonyi, J.; Seress, D.; Mosca, S.; Schena, L.; Seddaiu, S.; Pane, A.; et al. Multiple new cryptic pathogenic Phytophthora species from Fagaceae forests in Austria, Italy and Portugal. IMA Fungus 2017, 8, 219–244. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tyler, B.; Hong, C. An expanded phylogeny for the genus Phytophthora. IMA Fungus 2017, 8, 355–384. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hong, C. Differential usefulness of nine commonly used genetic markers for identifying Phytphthora species. Front. Microbiol. 2018, 9, 2334. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Horta Jung, M.; Scanu, B.; Seress, D.; Kovács, G.M.; Maia, C.; Pérez-Sierra, A.; Chang, T.-T.; Chandelier, A.; Heungens, K.; et al. Six new Phytophthora species from ITS Clade 7a including two sexually functional heterothallic hybrid species detected in natural ecosystems in Taiwan. Pers. Mol. Phylogeny Evol. Fungi 2017, 38, 100–135. [Google Scholar] [CrossRef]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora Diseases Worldwide; APS Press—The American Phytopathological Society: St. Paul, MN, USA, 1996; pp. 84–95, 96–144, 456–463. [Google Scholar]
- Cacciola, S.O.; Agosteo, G.E.; Pane, A. First report of Phytophthora palmivora as a pathogen of olive in Italy. Plant Dis. 2000, 84, 1153. [Google Scholar] [CrossRef]
- Cacciola, S.O.; Agosteo, G.E.; Magnano di San Lio, G. Collar and root rot of olive trees caused by Phytophthora megasperma in Sicily. Plant Dis. 2001, 85, 96. [Google Scholar] [CrossRef]
- Sánchez, M.E.; Muñoz-Garcia, M.; Brasier, C.M. Identity and pathogenicity of two Phytophthora taxa associated with a new root disease of olive trees. Plant Dis. 2001, 85, 411–416. [Google Scholar] [CrossRef]
- Brasier, C.M.; Sánchez-Hernandez, E.; Kirk, S.A. Phytophthora inundata sp. nov., a part heterothallic pathogen of trees and shrubs in wet or flooded soils. Mycol. Res. 2003, 107, 477–484. [Google Scholar] [CrossRef]
- Lucero, G.; Vettraino, A.M.; Pizzuolo, P.; Di Stefano, C.; Vannini, A. First report of Phytophthora palmivora on olive trees in Argentina. Plant Pathol. 2007, 56, 728. [Google Scholar] [CrossRef]
- Taheri, M.; Mirabolfathy, M.; Rahnama, K. Phytophthora root and crown rot of several field and orchard crops in Gorgan area, Iran. J. Plant Pathol. 2012, 48, 85–88. [Google Scholar]
- Kurbetli, I.; Sulu, G.; TaŞtekİn, E.; Polat, I. First report of Phytophthora inundata causing olive tree decline in Turkey. Can. J. Plant Pathol. 2016, 38, 254–257. [Google Scholar] [CrossRef]
- González, M.; Pérez-Sierra, A.; Serrano, M.S.; Sanchez, M.E. Two Phytophthora species causing decline of wild olive (Olea europaea subsp. europaea var. sylvestris). Plant Pathol. 2017, 66, 941–948. [Google Scholar]
- González, M.; Pérez-Sierra, A.; Sánchez, M.E. Phytophthora oleae, a new root pathogen of wild olives. Plant Pathol. 2019, 68, 901–907. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Bregant, C.; Montecchio, L.; Favaron, F.; Sella, L. First report of Phytophthora acerina, P. pini, and P. plurivora causing root rot and sudden death of olive trees in Italy. Plant Dis. 2020, 104, 996. [Google Scholar] [CrossRef]
- Lo Giudice, V.; Raudino, F.; Magnano di San Lio, R.; Cacciola, S.O.; Faedda, R.; Pane, A. First report of a decline and wilt of young olive trees caused by simultaneous infections of Verticillium dahliae and Phytophthora palmivora in Sicily. Plant Dis. 2010, 94, 1372. [Google Scholar] [CrossRef]
- Riolo, M.; Evoli, M.; Schena, L.; Aloi, F.; Santilli, E.; Ruano-Rosa, D.; Agosteo, G.E.; Pane, A.; La Spada, F.; Cacciola, S.O. Distribution of Phytophthora oleae in southern Italy. In Abstracts of Presentations at the XXV Congress of the Italian Phytopathological Society (SIPaV). J. Plant Pathol. 2019, 101, 811–847. [Google Scholar]
- Riolo, M.; Schena, L.; Aloi, F.; Santilli, E.; Ruano-Rosa, D.; Agosteo, G.E.; Pane, A.; La Spada, F.; Cacciola, S.O. Phytophthora oleae a widespread species in soil of olive (Olea europaea) orchards in Southern Italy. In Proceedings of the Phytophthora in Forests and Natural Ecosystems Proceeding 9th Meeting—IUFRO—Working Party 7.02.09, La Maddalena, Sardinia, Italy, 17–26 October 2019; p. 35. [Google Scholar]
- Cacciola, S.O.; Scarito, G.; Salamone, A.; Fodale, A.S.; Mulé, R.; Pirajno, G.; Sammarco, G. Phytophthora species associated to root rot of olive in Sicily. In Proceedings of the IOBC/WPRS, Working Group “Integrated Protection of Olive Crops”, Florence, Italy, 26–28 October 2005; Kalaitzaki, A., Ed.; Bull.OILB/SROP. International Organization for Biological and Integrated Control of Noxious Animals and Plants (OIBC/OILB), West Palaearctic Regional Section (WPRS/SROP): Dijon, France, 2007; Volume 30, p. 251. [Google Scholar]
- Nigro, F.; Ippolito, A. Occurrence of new rots of olive drupes in Apulia. Acta Hort. 2002, 586, 777–780. [Google Scholar] [CrossRef]
- Vettraino, A.M.; Lucero, G.; Pizzuolo, P.; Franceschini, S.; Vannini, A. First report of root rot and twigs wilting of olive trees in Argentina caused by Phytophthora nicotianae. Plant Dis. 2009, 93, 765. [Google Scholar] [CrossRef]
- Ruano-Rosa, D.; Schena, L.; Agosteo, G.E.; Magnano di San Lio, G.; Cacciola, S.O. Phytophthora oleae sp. nov. causing fruit rot of olive in southern Italy. Plant Pathol. 2018, 67, 1362–1373. [Google Scholar] [CrossRef]
- Aghighi, S.; Hardy, G.E.S.J.; Scott, J.K.; Burgess, T.I. Phytophthora bilorbang sp. nov., a new species associated with the decline of Rubus anglocandicans (European blackberry) in Western Australia. Eur. J. Plant Pathol. 2012, 133, 841–855. [Google Scholar] [CrossRef]
- Engelbrecht, B.M.J.; Tyree, M.T.; Kursar, T.A. Visual assessment of wilting as a measure of leaf water potential and seedling drought survival. J. Trop. Ecol. 2007, 23, 497–500. [Google Scholar] [CrossRef]
- Aghighi, S.; Burgess, T.I.; Scott, J.K.; Calver, M.; Hardy, G.E.S.J. Isolation and pathogenicity of Phytophthora species from declining Rubus anglocandicans. Plant Pathol. 2016, 65, 451–461. [Google Scholar] [CrossRef]
- Brasier, C.; Grunwald, N.; Hansen, E.; Reeser, P.; Sims, L.; Sutton, W. What is a species?—You be the judge. In Proceedings of the Phytophthora in Forests and Natural Ecosystems Proceeeding 7th Meeting—IUFRO—Working Party 7.02.09, Esquel, Argentina, 10–14 November 2014; pp. 109–111. [Google Scholar]
- Kroon, L.P.N.M.; Brouwer, H.; De Cock, A.W.A.M.; Govers, F. The genus Phytophthora anno 2012. Phytopathology 2012, 102, 348–364. [Google Scholar] [CrossRef]
- Mora-Sala, B.; Gramaje, D.; Abad-Campos, P.; Berbegal, M. Diversity of Phytophthora species associated with Quercus ilex L. in three Spanish regions evaluated by NGS. Forests 2019, 10, 979. [Google Scholar] [CrossRef]
- Brasier, C.M.; Cooke, D.E.L.; Duncan, J.M.; Hansen, E.M. Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides-P. megasperma ITS Clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile. Mycol. Res. 2003, 107, 277–290. [Google Scholar] [CrossRef]
- Jung, T.; Stukely, M.J.C.; Hardy, G.E.S.J.; White, D.; Paap, T.; Dunstan, W.A.; Burgess, T.I. Multiple new Phytophthora species from ITS Clade 6 associated with natural ecosystems in Australia: Evolutionary and ecological implications. Persoonia 2011, 26, 13–39. [Google Scholar] [CrossRef]
- Yang, X.; Copes, W.E.; Hong, C. Phytophthora mississippiae sp. nov., a new species recovered from irrigation reservoirs at a plant nursery in Mississippi. J. Plant Pathol. Microb. 2013, 4. [Google Scholar] [CrossRef]
- Nechwatal, J.; Bakonyi, J.; Cacciola, S.O.; Cooke, D.E.L.; Jung, T.; Nagy, Z.Á.; Vannini, A.; Vettraino, A.M.; Brasier, C.M. The morphology, behaviour and molecular phylogeny of Phytophthora taxon Salixsoil and its redesignation as Phytophthora lacustris sp. nov. Plant Pathol. 2013, 62, 355–369. [Google Scholar] [CrossRef]
- Sims, L.L.; Sutton, W.; Reeser, P.; Hansen, E.M. The Phytophthora species assemblage and diversity in riparian alder ecosystems of western Oregon, USA. Mycologia 2015, 107, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Scanu, B.; Linaldeddu, B.T.; Deidda, A.; Jung, T. Diversity of Phytophthora species from declining Mediterranean maquis vegetation, including two new species, Phytophthora crassamura and P. ornamentata sp. nov. PLoS ONE 2015, 10, e0143234. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; La Spada, F.; Pane, A.; Aloi, F.; Evoli, M.; Horta Jung, M.; Scanu, B.; Faedda, R.; Rizza, C.; Puglisi, I.; et al. Diversity and distribution of Phytophthora species in protected natural areas in Sicily. Forests 2019, 10, 259. [Google Scholar] [CrossRef]
- Oliva, J.; Redondo, M.A.; Stenlid, J. Functional ecology of forest disease. Annu. Rev. Phytopathol. 2020, 58, 8.1–8.19. [Google Scholar] [CrossRef]
- Jung, T.; Orlikowski, L.; Henricot, B.; Abad-Campos, P.; Aday, A.G.; Aguin Casal, O.; Bakonyi, J.; Cacciola, S.O.; Cech, T.; Chavarriaga, D.; et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Pathol. 2016, 46, 134–163. [Google Scholar] [CrossRef]
- Scanu, B.; Linaldeddu, B.T.; Peréz Sierra, A.; Deidda, A.; Franceschini, A. Phytophthora ilicis as a leaf and stem pathogen of Ilex aquifolium in Mediterranean islands. Phytopathol. Mediterr. 2014, 53, 480–490. [Google Scholar]
- Jung, T.; Blaschke, H.; Neumann, P. Isolation, identification and pathogenicity of Phytophthora species from declining oak stands. Eur. J. For. Pathol. 1996, 26, 253–272. [Google Scholar] [CrossRef]
- Jung, T.; Cooke, D.E.L.; Blaschke, H.; Duncan, J.M.; Oßwald, W. Phytophthora quercina sp. nov., causing root rot of European oaks. Mycol. Res. 1999, 103, 785–798. [Google Scholar] [CrossRef]
- Cooke, D.E.L.; Drenth, A.; Duncan, J.M.; Wagels, G.; Brasier, C.M. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 2000, 30, 17–32. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: San Diego, CA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- FinchTV v.1.4.0. Available online: https://digitalworldbiology.com/FinchTV (accessed on 18 May 2020).
- BLAST Searches. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 18 May 2020).
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 18 May 2020).
- Cacciola, S.A.; Faedda, R.; Sinatra, F.; Agosteo, G.E.; Schena, L.; Frisullo, S.; Magnano di San Lio, G. Olive anthracnose. J. Plant Pathol. 2012, 94, 29–44. [Google Scholar]
- Talhinhas, P.; Loureiro, A.; Oliveira, H. Olive anthracnose: A yield- and oil quality-degrading disease caused by several species of Colletotrichum that differ in virulence, host preference and geographical distribution. Mol. Plant Pathol. 2018, 19, 1797–1807. [Google Scholar] [CrossRef] [PubMed]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.B.; Campos, M.D.; Gomes, L.; Nobre, T.; Rei, F.; Felix, M. Effect of long-term fungicide applications on virulence and diversity of Colletotrichum spp. associated to olive anthracnose. Plants 2019, 8, 311. [Google Scholar] [CrossRef] [PubMed]
- Cacciola, S.O.; Gullino, M.L. Emerging and re-emerging fungus and oomycete soil-borne plant diseases in Italy. Phytopathol. Mediterr. 2019, 58, 451–472. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santilli, E.; Riolo, M.; La Spada, F.; Pane, A.; Cacciola, S.O. First Report of Root Rot Caused by Phytophthora bilorbang on Olea europaea in Italy. Plants 2020, 9, 826. https://doi.org/10.3390/plants9070826
Santilli E, Riolo M, La Spada F, Pane A, Cacciola SO. First Report of Root Rot Caused by Phytophthora bilorbang on Olea europaea in Italy. Plants. 2020; 9(7):826. https://doi.org/10.3390/plants9070826
Chicago/Turabian StyleSantilli, Elena, Mario Riolo, Federico La Spada, Antonella Pane, and Santa Olga Cacciola. 2020. "First Report of Root Rot Caused by Phytophthora bilorbang on Olea europaea in Italy" Plants 9, no. 7: 826. https://doi.org/10.3390/plants9070826
APA StyleSantilli, E., Riolo, M., La Spada, F., Pane, A., & Cacciola, S. O. (2020). First Report of Root Rot Caused by Phytophthora bilorbang on Olea europaea in Italy. Plants, 9(7), 826. https://doi.org/10.3390/plants9070826





