Development of In Vivo Haploid Inducer Lines for Screening Haploid Immature Embryos in Maize
Abstract
:1. Introduction
2. Results
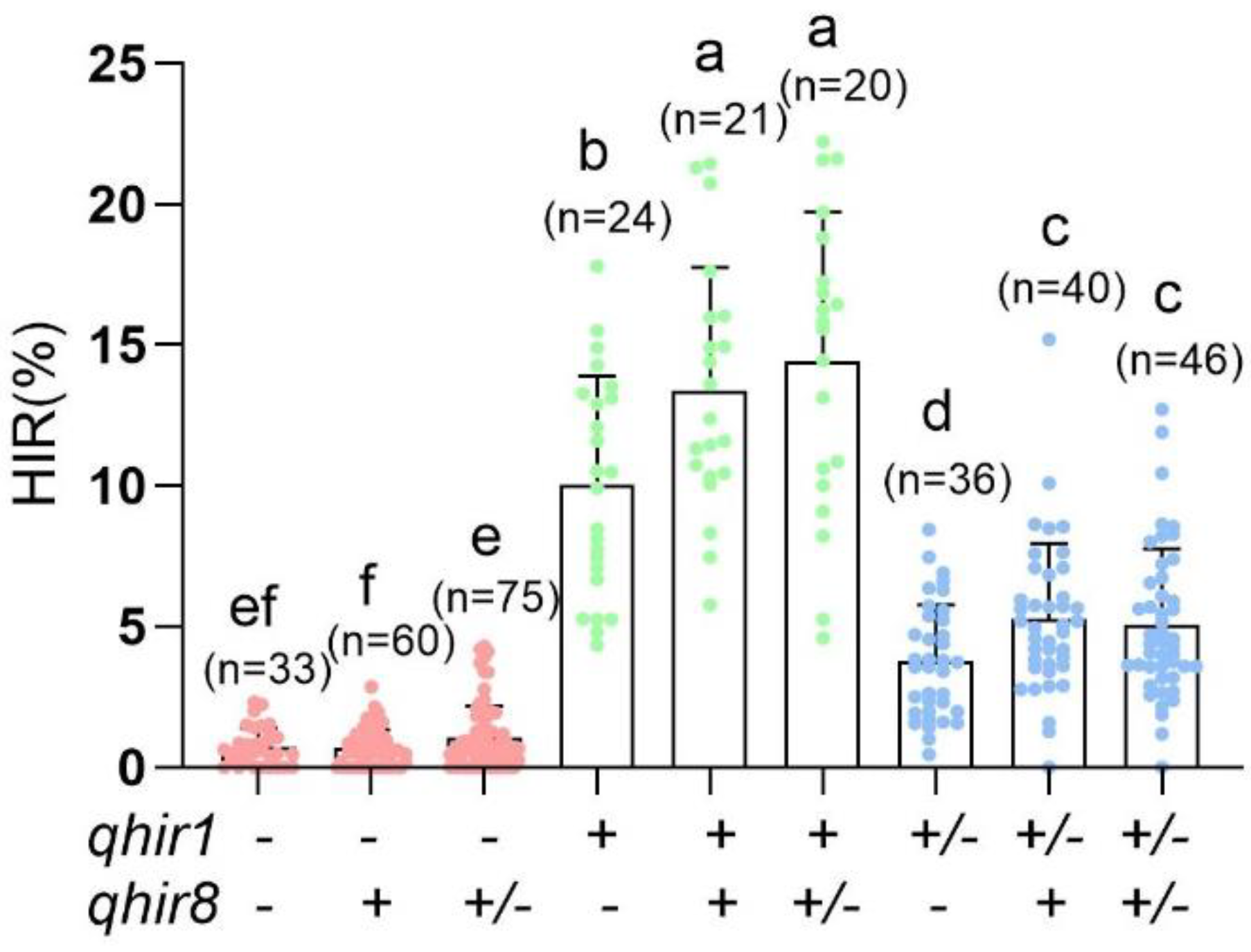
2.1. Validating the Effectiveness of MAS for qhir1 and qhir8
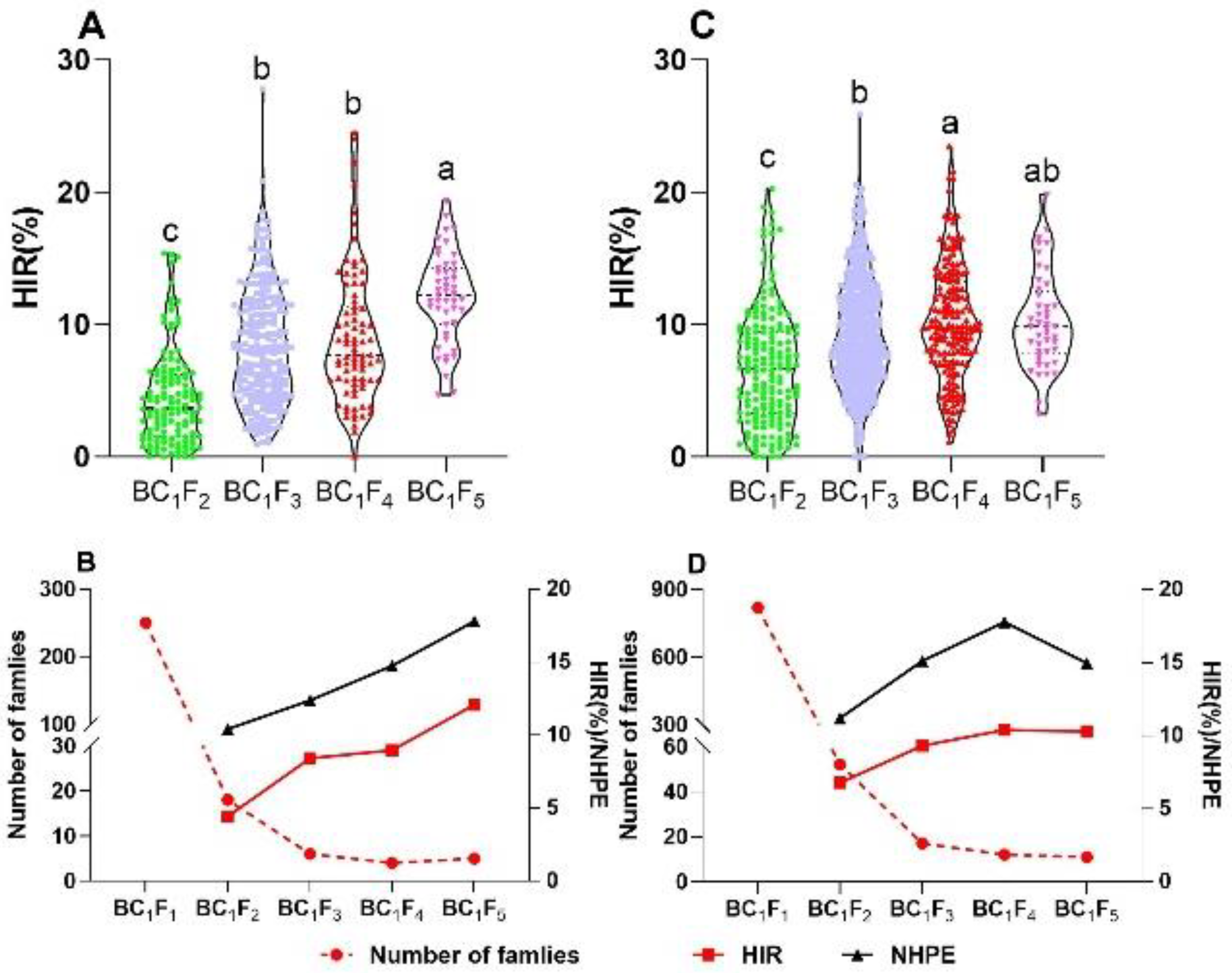
2.2. Selection for HIR and Number of Haploids per Ear (NHPE)
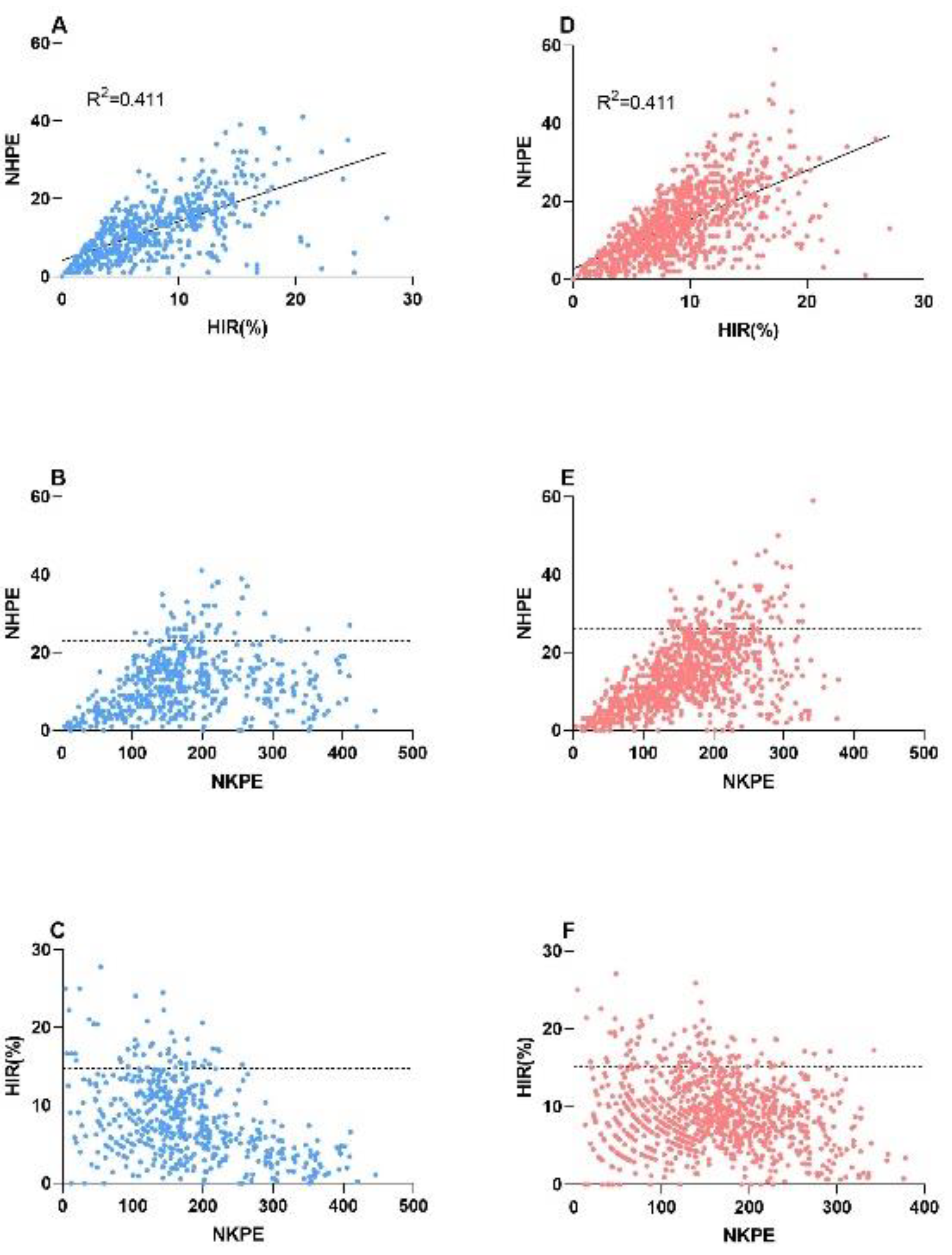
2.3. Relationship between the NHPE, HIR, and Number of Kernels per Ear (NKPE)
2.4. Evaluating the HIR among the Different Varieties
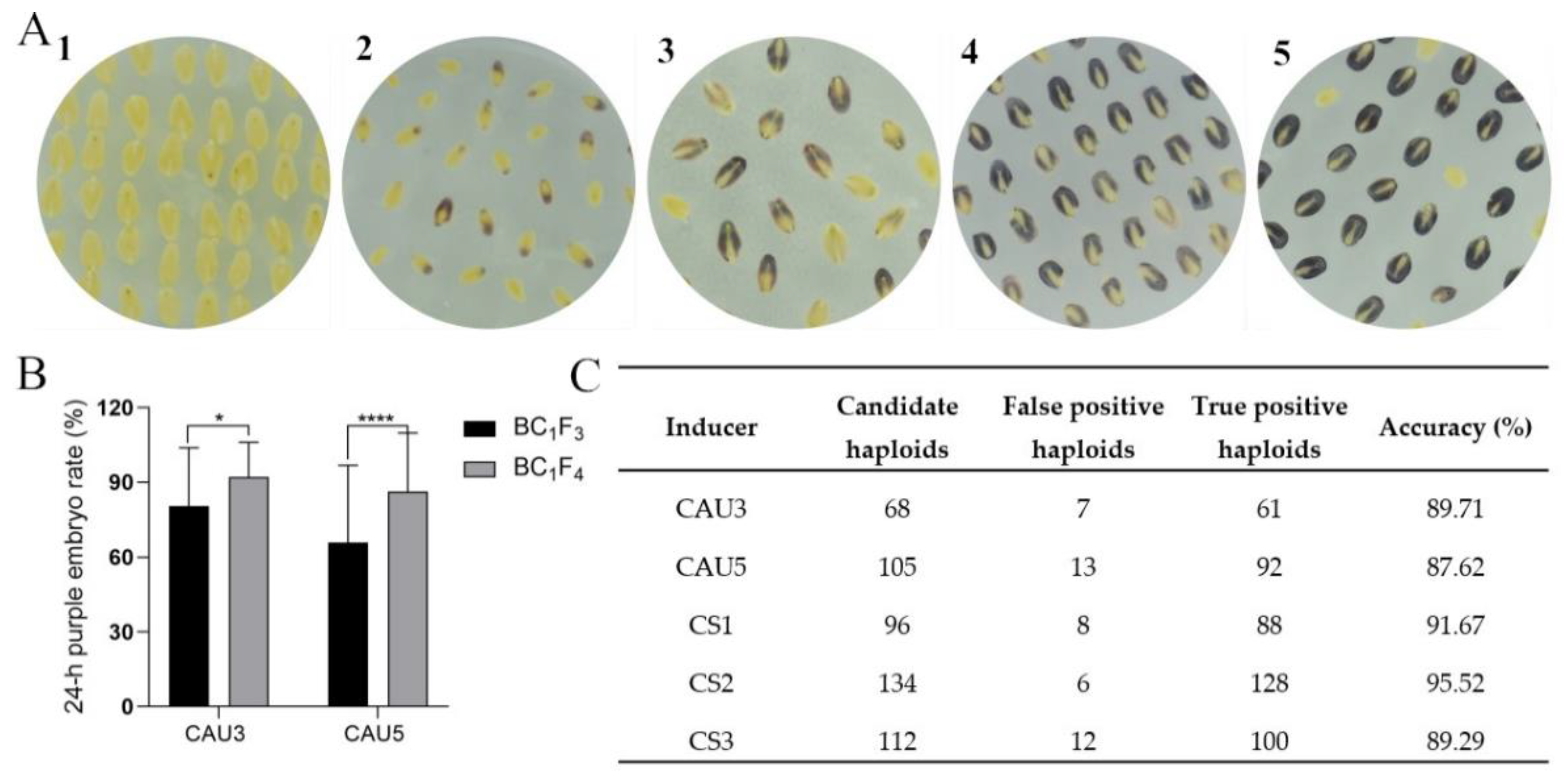
2.5. Evaluation of the Pigmentation in Crossing Embryos and Identification of the Haploid Embryos
2.6. Agronomic Traits
3. Discussion
3.1. MAS for Multiple Loci Related to Haploid Induction
3.2. Development of an Efficient Haploid Inducer Lines
4. Materials and Methods
4.1. Materials
4.2. Breeding Scheme
4.3. Verification of the MAS for qhir1 and qhir8 during the Selection for HIR
4.4. Haploid Identification Based on the Pigmentation of Crossing Embryos
4.5. Assessment of the Agronomic Performance
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chaikam, V.; Molenaar, W.; Melchinger, A.E.; Boddupalli, P.M. Doubled haploid technology for line development in maize: Technical advances and prospects. Theor. Appl. Genet. 2019, 132, 3227–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coe, J.E.H. A Line of Maize with High Haploid Frequency. Am. Nat. 1959, 93, 381–382. [Google Scholar] [CrossRef]
- Dong, X.; Xu, X.; Li, L.; Liu, C.; Tian, X.; Li, W.; Chen, S. Marker-assisted selection and evaluation of high oil in vivo haploid inducers in maize. Mol. Breed. 2014, 34, 1147–1158. [Google Scholar] [CrossRef]
- Röber, F.; Gordila, G.A.; Geiger, H.H. In vivo haploid induction in maize—Performance of new inducers and significance of doubled haploid lines in hybrid breeding. Maydica 2005, 50, 275–283. [Google Scholar]
- Prigge, V.; Xu, X.; Li, L.; Babu, R.; Chen, S.; Atlin, G.N.; Melchinger, A.E. New Insights into the Genetics of in Vivo Induction of Maternal Haploids, the Backbone of Doubled Haploid Technology in Maize. Genetics 2011, 190, 781–793. [Google Scholar] [CrossRef] [Green Version]
- Rotarenco, D.G.; Fuia, S. New inducers of maternal haploids in maize. Maize Genet. Coop. Newsl. 2010, 84, 21–22. [Google Scholar]
- Chaikam, V.; Nair, S.K.; Martinez, L.; Lopez, L.A.; Utz, H.F.; Melchinger, A.E.; Prasanna, B.M. Marker-Assisted Breeding of Improved Maternal Haploid Inducers in Maize for the Tropical/Subtropical Regions. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Wu, P.; Trampe, B.; Tian, X.; Lübberstedt, T.; Chen, S. Novel technologies in doubled haploid line development. Plant Biotechnol. J. 2017, 15, 1361–1370. [Google Scholar] [CrossRef] [Green Version]
- Andorf, C.; Beavis, W.D.; Hufford, M.; Smith, S.; Suza, W.P.; Wang, K.; Woodhouse, M.; Yu, J.; Lübberstedt, T. Technological advances in maize breeding: Past, present and future. Theor. Appl. Genet. 2019, 132, 817–849. [Google Scholar] [CrossRef] [Green Version]
- Barret, P.; Brinkmann, M.; Beckert, M. A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor. Appl. Genet. 2008, 117, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Xu, X.; Miao, J.; Li, L.; Zhang, D.; Mi, X.; Liu, C.; Tian, X.; Melchinger, A.E.; Chen, S. Fine mapping of qhir1 influencing in vivo haploid induction in maize. Theor. Appl. Genet. 2013, 126, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, W.; Zhong, Y.; Dong, X.; Hu, H.; Tian, X.; Wang, L.; Chen, B.; Chen, C.; Melchinger, A.E.; et al. Fine mapping of qhir8 affecting in vivo haploid induction in maize. Theor. Appl. Genet. 2015, 128, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Schrag, T.A.; Peis, R.; Unterseer, S.; Schipprack, W.; Chen, S.; Lai, J.; Yan, J.; Prasanna, B.M.; Nair, S.; et al. The Genetic Basis of Haploid Induction in Maize Identified with a Novel Genome-Wide Association Method. Genetics 2016, 202, 1267–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, X.; Meng, D.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.; Chen, B.; Li, W.; Li, L.; et al. A 4-bp Insertion at ZmPLA1 Encoding a Putative Phospholipase A Generates Haploid Induction in Maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef] [Green Version]
- Gilles, L.M.; Khaled, A.; Laffaire, J.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Bergès, H.; Beydon, G.; Bayle, V.; Barret, P.; et al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, N.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Nanda, D.K.; Chase, S.S. An Embryo Marker for Detecting Monoploids of Maize (Zea Mays L). Crop Sci. 1966, 6, 213–219. [Google Scholar] [CrossRef]
- Chaikam, V.; Nair, S.K.; Babu, R.; Martinez, L.; Tejomurtula, J.; Boddupalli, P.M. Analysis of effectiveness of R1-nj anthocyanin marker for in vivo haploid identification in maize and molecular markers for predicting the inhibition of R-nj expression. Theor. Appl. Genet. 2015, 128. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Chandler, V.L.; Robbins, T.P.; Chen, J.; Turks, D. Two regulatory genes of the maize anthocyanin pathway are homologous isolation of B utilizing R genomic sequences. Plant Cell 1989, 1, 1175–1183. [Google Scholar]
- Goff, S.A.; Klein, T.M.; Roth, B.A.; Fromm, M.E.; Cone, K.C.; Radicella, J.P.; Chandler, V.L. Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 1990, 9, 2517–2522. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Li, L.; Liu, C.; Liu, C.; Geng, S.; Li, X.; Huang, C.; Mao, L.; Chen, S.; Xie, C. Genome Editing and Double-Fluorescence Proteins Enable Robust Maternal Haploid Induction and Identification in Maize. Mol. Plant 2018, 11, 1214–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, J.E.; Maddock, S.E.; Wu, X.E.; Zhao, Z.; Willimas, M.E.; Hussain, T.; Gordon-kamm, W.J. Doubling of Chromosomes in Haploid Embryos. U.S. Patent Application No 11/532,921, 4 September 2008. [Google Scholar]
- Vicient, P.F.A.C.M. Plant Embryogenesis; Humana Press: Totowa, NJ, USA, 2008; pp. 17–23. [Google Scholar]
- Chen, S.; Song, T. Identification Haploid with High Oil Xenia Effect in Maize. Acta Agron. Sin. 2003, 29, 587–590. [Google Scholar]
- Melchinger, A.E.; Schipprack, W.; Mi, X.; Mirdita, V. Oil Content is Superior to Oil Mass for Identification of Haploid Seeds in Maize Produced with High-Oil Inducers. Crop. Sci. 2015, 55, 188–195. [Google Scholar] [CrossRef]
- Li, L.; Xu, X.; Jin, W.; Chen, S. Morphological and molecular evidences for DNA introgression in haploid induction via a high oil inducer CAUHOI in maize. Planta 2009, 230, 367–376. [Google Scholar] [CrossRef]
- Prigge, V.; Schipprack, W.; Mahuku, G.; Atlin, G.N.; Melchinger, A.E. Development of in vivo haploid inducers for tropical maize breeding programs. Euphytica 2012, 185, 481–490. [Google Scholar] [CrossRef]
- Chase, S.S. Monoploids and monoploid-derivatives of maize (Zea mays L.). Bot. Rev. 1969, 35, 117–168. [Google Scholar] [CrossRef]
- Geiger, H.H.; Gordillo, G.A.; Koch, S. Genetic Correlations among Haploids, Doubled Haploids, and Testcrosses in Maize. Crop. Sci. 2013, 53, 2313–2320. [Google Scholar] [CrossRef]
- Yu, W.; Birchler, J.A. A Green Fluorescent Protein-Engineered Haploid Inducer Line Facilitates Haploid Mutant Screens and Doubled Haploid Breeding in Maize. Mol. Breed. 2016, 36, 1–12. [Google Scholar] [CrossRef]
- Chen, S.; Liu, C.; Zhong, Y. Maize haploid inducer primer pair used for assisting maize breeding, and preparing PCR kit in identifying maize plant’s haploid inducing trait comprises DNA molecule containing specific nucleobases. Patent CN104846104-A, CN104846104-B, 19 August 2015. (In Chinese). [Google Scholar]
- Chen, S.; Xu, X.; Li, L.; Dong, X. New Specific Primer, Useful for Efficiently and Rapidly Auxiliary identifying Corn Haploid Induction Line. Patent CN103088109-A, CN103088109-B, 8 May 2013. (In Chinese). [Google Scholar]
| Inducer | Tester | Total Number | NHPE | NKPE | EmAR (%) | HIR (%) |
|---|---|---|---|---|---|---|
| CAU3 | ZD958 | 475 | 10.00 ± 3.11 | 95.00 ± 26.89 | 3.14 ± 1.42 | 11.22 ± 1.79 |
| CAU5 | ZD958 | 1843 | 17.00 ± 2.70 | 131.64 ± 12.82 | 10.96 ± 0.50 | 11.89 ± 0.77 |
| CS1 | ZD958 | 3341 | 20.71 ± 1.45 ** | 196.53 ± 7.56 **** | 6.72 ± 0.67 * | 10.62 ± 0.68 |
| CS2 | ZD958 | 4059 | 33.93 ± 3.42 ** | 289.93 ± 16.44 **** | 6.31 ± 0.59 * | 11.54 ± 0.87 |
| CS3 | ZD958 | 3107 | 33.18 ± 3.39 ** | 282.45 ± 17.56 **** | 5.47 ± 0.91 ** | 11.71 ± 1.01 |
| CS1 | DK653 | 1358 | 24.57 ± 2.64 | 194.00 ± 19.95 | 5.35 ± 0.76 | 12.62 ± 0.61 |
| JK968 | 993 | 19.33 ± 3.73 | 165.50 ± 22.29 | 7.67 ± 1.29 | 11.56 ± 1.16 | |
| ND372 | 1244 | 16.89 ± 2.34 | 138.22 ± 10.20 | 9.34 ± 1.87 | 11.97 ± 1.07 | |
| ND678 | 1324 | 24.71 ± 3.04 | 189.14 ± 11.84 | 8.77 ± 1.11 | 13.25 ± 1.57 | |
| XY335 | 439 | 13.00 ± 4.93 | 146.33 ± 39.08 | 5.56 ± 0.84 | 8.29 ± 1.35 | |
| CS2 | DK653 | 3167 | 29.93 ± 3.49 | 211.13 ± 17.20 | 5.68 ± 0.72 | 13.72 ± 0.78 |
| JK968 | 1612 | 26.63 ± 5.37 | 201.50 ± 27.60 | 7.54 ± 1.04 | 12.59 ± 1.46 | |
| ND372 | 2648 | 27.42 ± 2.01 | 220.67 ± 10.67 | 9.61 ± 0.61 | 12.49 ± 0.84 | |
| ND678 | 2741 | 30.00 ± 3.18 | 195.79 ± 16.20 | 6.86 ± 0.69 | 15.54 ± 1.00 | |
| XY335 | 3685 | 39.18 ± 3.25 | 335.00 ± 20.03 | 5.46 ± 1.01 | 11.70 ± 0.68 | |
| CS3 | DK653 | 2933 | 25.86 ± 2.78 | 209.50 ± 14.41 | 4.35 ± 0.59 | 12.27 ± 0.94 |
| JK968 | 3674 | 31.90 ± 2.37 | 367.40 ± 13.62 | 2.88 ± 0.47 | 8.66 ± 0.54 | |
| ND372 | 3762 | 23.77 ± 2.37 | 289.38 ± 16.35 | 5.11 ± 0.64 | 8.14 ± 0.58 | |
| ND678 | 2135 | 18.21 ± 2.26 | 152.50 ± 16.15 | 4.24 ± 0.91 | 11.82 ± 0.73 | |
| XY335 | 3340 | 34.42± 3.62 | 278.33 ± 16.31 | 5.87 ± 1.03 | 12.28 ± 0.99 |
| Inducer | Days to Anthesis | Plant Height (cm) | Ear Height (cm) | Tassel Branches Number | Tassel Size Score | Number of Kernels per Ear |
|---|---|---|---|---|---|---|
| CAU3 | 66.40 ± 0.60 | 174.80 ± 1.43a | 50.60 ± 1.55 | 5.80 ± 0.25 | 4.00 ± 0.00 | 49.00 ± 2.34 |
| CAU5 | 59.80 ± 0.72 | 149.00 ± 1.13 | 32.60 ± 0.82 | 9.80 ± 0.86 | 3.20 ± 0.13 | 37.20 ± 3.16 |
| CS1 | 67.80 ± 0.37 | 196.80 ± 6.26 * | 94.40 ± 3.08 **** | 9.20 ± 0.80 ** | 3.20 ± 0.20 ** | 86.00 ± 8.64 ** |
| CS2 | 66.60 ± 0.75 | 198.00 ± 3.15 *** | 82.60 ± 1.21 **** | 9.20 ± 0.37 *** | 3.00 ± 0.00 ** | 103.00 ± 7.46 *** |
| CS3 | 62.00 ± 0.32 | 162.80 ± 2.24 *** | 45.80 ± 2.01 *** | 28.20 ± 1.11 **** | 1.20 ± 0.20 *** | 110.60 ± 3.44 **** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Xiao, Z.; Zhang, J.; Li, W.; Li, J.; Liu, C.; Chen, S. Development of In Vivo Haploid Inducer Lines for Screening Haploid Immature Embryos in Maize. Plants 2020, 9, 739. https://doi.org/10.3390/plants9060739
Chen C, Xiao Z, Zhang J, Li W, Li J, Liu C, Chen S. Development of In Vivo Haploid Inducer Lines for Screening Haploid Immature Embryos in Maize. Plants. 2020; 9(6):739. https://doi.org/10.3390/plants9060739
Chicago/Turabian StyleChen, Chen, Zijian Xiao, Junwen Zhang, Wei Li, Jinlong Li, Chenxu Liu, and Shaojiang Chen. 2020. "Development of In Vivo Haploid Inducer Lines for Screening Haploid Immature Embryos in Maize" Plants 9, no. 6: 739. https://doi.org/10.3390/plants9060739
APA StyleChen, C., Xiao, Z., Zhang, J., Li, W., Li, J., Liu, C., & Chen, S. (2020). Development of In Vivo Haploid Inducer Lines for Screening Haploid Immature Embryos in Maize. Plants, 9(6), 739. https://doi.org/10.3390/plants9060739





