Abstract
The ability of a seed to germinate and establish a plant at the right time of year is of vital importance from an ecological and economical point of view. Due to the fragility of these early growth stages, their swiftness and robustness will impact later developmental stages and crop yield. These traits are modulated by a continuous interaction between the genetic makeup of the plant and the environment from seed production to germination stages. In this review, we have summarized the established knowledge on the control of seed germination from a molecular and a genetic perspective. This serves as a “backbone” to integrate the latest developments in the field. These include the link of germination to events occurring in the mother plant influenced by the environment, the impact of changes in the chromatin landscape, the discovery of new players and new insights related to well-known master regulators. Finally, results from recent studies on hormone transport, signaling, and biophysical and mechanical tissue properties are underscoring the relevance of tissue-specific regulation and the interplay of signals in this crucial developmental process.
1. General Introduction
Seed production and germination are intimately connected and closely linked to the survival and dispersal of plant species. The main role of the seed is to protect the embryo and sense environmental information to couple germination with seasons compatible with the completion of the plant life cycle. Germination encompasses the events from imbibition to radicle protrusion through the seed coverings. In the field, the spatial pattern of seed dispersal depends on the habitat of the mother plant as well as on the fruit and seed morphology. In addition, the temporal distribution of germination mainly depends on the interaction between the environment and the plant’s genetic makeup, which conditions both dormancy and germination potential. For instance, it is known that seeds developed in plants exposed to low temperatures will have higher dormancy levels and the opposite when supplied with nitrate. Physiological dormancy, the most common type [1,2], provides seeds with valuable advantages. First, it maximizes dispersion, thus reducing competition for resources between the offspring and the mother plant. Second, it halts germination in the wrong season, even if short spells of favorable conditions occur. After reaching maturity, seeds undergo a process called after-ripening (AR) characterized by a gradual reduction in water content and dormancy level, whose speed depends on the relationship of seed moisture content and temperature during dry storage. At this point, non-dormant seeds can retrieve the dormancy program upon encountering inadequate conditions (secondary dormancy) or, if conditions are adequate, proceed to germination. In this case, the intake of water or imbibition by the non-dormant seed triggers different biochemical, metabolic and physiological processes, such as the resumption of respiratory activity, energy production, activation of repair mechanisms, protein biosynthesis from both stored and newly synthesized mRNAs and reserve mobilization. These events fuel the elongation of the embryonic axis and the weakening of the embryo surrounding tissues, leading to rupture of the seed coat (testa rupture), embryo radicle protrusion (germination sensu stricto) and seedling establishment.
To comprehend the processes taking place in the seed, it is necessary to have a deep understanding of the molecular and biochemical mechanisms that regulate them. This review intends to illustrate the key issues in a comprehensive and readable form, keeping a reasonable extension. Nevertheless, supplementary tables with a compilation of selected reviews that expand on specific aspects of seed biology (Table S1) and a list of complete gene names and their abbreviations (Table S2) have also been included. We have focused this review on Arabidopsis thaliana (Arabidopsis) although relevant findings in other plant species have been included. In the first part, we will describe the molecular players and networks controlling these processes, and their links to environmental and hormonal cues. In the second part we will review these processes from a genetic and physiological perspective.
2. Regulatory Layers Controlling Seed Germination
2.1. Hormone Metabolism and Signaling
Germination depends on the physiological state (dormancy) of the seed, which is partly caused by the interaction between the plant genotype and a wide spectrum of environmental factors, such as temperature, soil moisture, light and nutrient availability. This is mainly achieved through regulation of the metabolism and signaling of gibberellins (GAs) and abscisic acid (ABA), two phytohormones with antagonistic roles. Their spatio-temporal balance plays a pivotal role in seed biology by favoring dormancy over germination when the ABA/GA ratio is high, and the opposite when it is low [3]. In fact, the first dormancy- and germination-associated loci identified in Arabidopsis mutants included genes involved in GA and ABA biosynthesis, perception and signaling [3,4,5,6,7,8]. Bioactive GAs are formed in terminal reactions catalyzed by GA20ox and GA3ox oxidases. Deactivation of GAs by GA2ox oxidase and transcriptional feedback loops are also important features in GA homeostasis [9,10]. In particular, the GA3ox1 and GA3ox2 enzymes for biosynthesis, and GA2ox2 for catabolism, stand out for their key role in GA signaling during germination [11,12,13]. There are three main components involved in early perception and signaling by GAs: the GID1 (GA receptor) and the GID2 (F-box) proteins are positive signaling regulators [14,15,16], while DELLA proteins act as negative regulators [17,18]. The presence of GAs triggers the interaction of its receptor GID1 with DELLAs through their N-terminal domain (DELLA domain) and the formation of a ubiquitination complex via interaction with GID2. This interaction induces the proteasome-mediated degradation of DELLAs [19,20,21,22]. DELLA mutant versions lacking the DELLA domain are resistant to degradation and confer GA insensitivity [23,24]. DELLAs negatively regulate GA signaling through protein-protein interactions with several transcriptional regulators [19,20,25,26,27,28]. In Arabidopsis there are five DELLAs, of which RGL2 has a major role in regulating germination, since its loss-of-function mutants are able to restore germination of GA-deficient seeds [29,30]. Regarding the two Arabidopsis GID2 proteins, SLY1 seems to have the dominant role in germination, since SNE/SLY2 overexpression does not produce a decrease in RGL2 protein levels [31]. For the three GID1 genes found in Arabidopsis, a double mutant gid1ac had to be obtained to observe defects on germination [32], whereas GID1b has an ABA-independent role in AR [33].
The key enzymes involved in ABA biosynthesis are NCED dioxygenases, while CYP707A monooxygenases are central to ABA catabolism through 8’-hydroxylation [34,35]. In particular, the loss of function of two ABA biosynthetic enzymes, NCED6 and NCED9, results in dormancy reduction [36] while mutation of the CYP707A2 gene decreases germination potential [37,38,39]. As in the case of GAs, three main components are involved in early ABA perception and signaling: the ABA receptor (PYR/PYL/RCAR) proteins and the SnRK2 protein kinases, which are positive regulators of the pathway, and PP2Cs protein phosphatases as negative regulators. ABA-bound receptors are able to bind and inhibit PP2Cs which, in turn, allow phosphorylation and activation of SnRK2s [35,40,41,42]. Activated SnRK2s phosphorylate downstream targets, such as the ABI5 and other members of the AREB/ABFs bZIP transcription factors (TF) family [43], among others, to activate the ABA response in plants [35]. In the seed, responses to ABA and related physiological processes are mainly under the control of ABI5 together with two other non-bZIP TFs, ABI3 (B3 family TF) and ABI4 (ERF family TF) [35,42]. Among these regulators, ABI3 is the one acting upstream ABI5 and ABI4 and is essential for ABI5 expression in germination arrest [44], whereas ABI4 acts as a repressor of lipid breakdown in the embryo [45]. Both TFs are positive regulators of the expression of ABI5 during seed germination [44,46]. Furthermore, ABI5 activates its own expression by binding to its own promoter [47].
2.2. Hormone Dynamics and Transport
The spatio-temporal action of plant hormones is crucial for proper development and germination [48,49,50,51]. Compelling evidence of temporal and tissue-specific regulation of hormone metabolism and signaling in seeds have been obtained, and recent results are improving our view on hormone transport in this organ [11,45,52,53,54,55,56,57,58,59,60,61,62]. For instance, the release of ABA from the endosperm into the embryo controls its growth and maintains its dormancy in dry seeds, a role that requires RGL2 function [63]. It has also been found that temperature shifts alter the spatial distribution of GAs and ABA in dormant embryos, suggesting that crosstalk mediated by hormone transport occurs between cell types in the embryonic axis [61]. Four AtABCG transporters expressed specifically in seed tissues were found to act in concert to correctly deliver ABA to control seed germination: AtABCG25 and AtABCG31 export ABA from the endosperm to the embryo, whereas AtABCG30 and AtABCG40 import ABA into the embryo from the endosperm. Consequently, it has been proposed that radicle extension and subsequent embryonic growth are suppressed by the coordinated activity of multiple ABA transporters expressed in different seed tissues [64].
The AtSWEET13 and AtSWEET14 transporter proteins were found to mediate cellular GA uptake when expressed in yeast and oocytes [65]. The sweet13/sweet14 double mutant exhibits altered long distance transport of exogenously applied GAs and their wild type (WT) versions are required for proper development of seeds and seedlings. SWEET family proteins were initially identified as sugar transporters and specific members of the family are involved in seed filling [66,67,68]. However, their role as GA transporters during seed development may not be so relevant for germination. In fact, GAs stored in dry seeds are not, or not sufficiently, transmitted to the offspring to successfully complete germination under permissive conditions, since de novo synthesis of GAs is required at this stage [69]. An intriguing observation is that the seeds produced by sweet13/sweet14 plants were larger than WT seeds but less sensitive to inhibition of germination by ABA or paclobutrazol (PAC, a GA biosynthesis inhibitor). If the SWEET proteins promoted GA influx into seed tissues, their loss of function would be expected to reduce seed size and increase sensitivity to ABA and PAC-mediated inhibition of germination [65].
The NPF3 gene encodes a protein targeted to the plant cell membrane where it functions as a GA influx transporter [70,71]. NPF3 belongs to the NPF gene family previously reported to encode nitrate or peptide transporters, some of which are also able to transport hormones [72,73]. Whereas NPF3 is expressed during seed development, npf3 mutants do not show altered germination under standard conditions, maybe due to genetic redundancy. Yet, these mutants showed a reduced germination response to GAs under nitrogen-limiting conditions [70,71]. NPF3 is also an ABA transporter in vitro and its expression is upregulated by low nitrogen, light and ABA and downregulated by high GA levels [70,71]. Interestingly, enhanced expression of NPF3 has been associated with a greater propensity to break dormancy. This effect has been proposed to be related to altered ABA/GA balance due to enhanced capacity for GA intake [61]. These findings suggest a role for NPF3 as a negative regulator of dormancy subjected to GA negative feedback. Another member of this family previously known as a low-affinity nitrate transporter (NPF4.6/AIT1/NRT1.2) [74], also mediates ABA uptake in yeast and insect cells. Compared with WT plants, the npf4.6/ait1/nrt1.2 mutant was less sensitive to exogenously applied ABA during seed germination and/or post-germination growth [75]. The fact that NPF proteins may act in vivo as dual transporters of nitrate and hormones, along with the dependence of their mutants on nitrogen availability to show altered germination responses, suggests the existence of molecular crosstalk that adapts germination to the nutrient environment. NLP6 TF is a NIN-like (NLP) protein that was found to control gene expression in response to the nitrate signal in vegetative stages [76]. NLP6 as well as other NLP family proteins bind a specific cis-element (NRE) in the promoters of nitrate-responsive genes and activate their expression. Genes involved in nitrate assimilation (NIA, NIR1) and transport (NRT1.1, NRT2.1) as well as regulatory genes of both processes are targets of NLP6 [76,77]. When nitrate is fed to the mother plant, seeds have reduced levels of dormancy, partly because ABA levels are reduced [78]. Not surprisingly, the CYP707A2 gene is induced by nitrate and, when mutated, seeds are less sensitive to nitrate-induced dormancy release [79]. Recently, a direct link between nitrate and ABA metabolism was found by revealing the role of NLP8, a TF that reduces ABA levels in a nitrate-dependent manner by directly binding to the promoter of CYP707A2 [80].
In summary, it is now clear that seeds respond to the nutrient environment perception by linking transport of nitrogen to hormones. By using specific nitrogen-dependent regulators, seeds can also modify gene networks and hormonal balance to modulate dormancy and germination. Additional information on this regulation can be found at the end of the next section.
2.3. Environmental Influence of Transcriptional Regulation: Expanding the Regulatory Breadth of Known/Classic TFs
The antagonism between GAs and ABA fine tunes seed germination to environmental conditions. The regulation exerted by the ABI5 TF (Figure 1) is one of the central nodes of this antagonism [81]. Expression of ABI5 is stimulated by ABA, water stress and high salinity, a response that relies on three TFs that bind to and activate its promoter, namely HY5/HYH, RSM1 and AGL21. Loss-of-function mutants of these genes decrease sensitivity to ABA, salinity and water stress during seed germination [82,83,84]. Although HY5 and BBX21 TFs are positive regulators of photomorphogenesis, their interaction counteracts HY5 upregulation of the ABI5 promoter. This mechanism is also used by BBX21 to interfere with ABI5 upregulation of its own promoter [85]. Similarly, VQ18 and VQ26, which belong to a recently identified family of plant-specific transcriptional regulators [86], bind to ABI5 to interfere with its transcriptional activity [87]. Reduction of ABI5 function is also controlled through negative feedback by the ABI5-induced AHT1, a BTB/POZ-domain containing protein which is a potential receptor for the proteasome CRL3 complex [88].
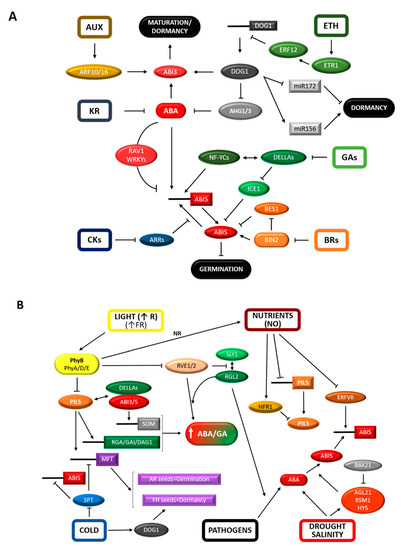
Figure 1.
Interplay between core components of molecular mechanisms controlling seed dormancy and germination with hormones and environmental signals. (A) Hormonal and molecular regulatory networks involved in dormancy and germination. DOG1 increases abscisic acid (ABA) sensitivity through sequestration of PP2Cs (AHG1/3) and genetically interacts with ABI3 to ensure ABA signaling during seed maturation and the establishment of dormancy. DOG1 expression is regulated by ethylene (ETH) signaling through the ETR1/ERF12 pathway and has an impact on dormancy release through the control of two antagonistic miRNAs. Other hormones such as auxin (AUX) by the ARF10/16 pathway and karrikins (KR) have a role in dormancy by altering ABA content or signaling. ABI5 plays a key role in ABA signaling to repress germination. ABI5 expression is upregulated under conditions unfavorable for germination by several TFs (ABA-related TFs or NF-YC3/4/9). Negative feedback by RAV1 and WRKY18/40/60 or conditions promoting germination counteract this upregulation. Also, other hormonal signaling pathways (gibberellins, GAs; brassinosteroids, BRs; cytokinins, CKs) interfere with ABI5-mediated transcription or stability through several regulatory proteins (DELLAs, ICE1, BES1, BIN2 or ARR4/5/6). (B) Effects of environmental factors on the regulation of seed dormancy and germination. PIL5 represses germination in the absence of light. It increases the ABA/GA balance partly through direct upregulation of SOM, DELLAs (RGA/GAI) and DAG1 transcription. ABI3/5 and DELLAs also participate in the upregulation of SOM gene transcription. Upon light perception, PIL5 activity is counteracted by different mechanisms mostly mediated by phytochromes (PhyB): increased degradation and reduced transcription in response to higher NO levels and reduced function through HFR1 competitive interaction. NO has also an effect on the stability of class VII ERFs, mediating their degradation and thus reducing ABI5 expression. In addition, activated Phys also reduce DELLA (RGL2) expression and increases its degradation by reducing the expression of circadian genes (RVE1/2). DOG1 integrates temperature cues to regulate dormancy release in fresh-harvested seeds. In dry seeds, SPT negatively regulates germination in the absence of low temperatures. SPT activates ABI5 and represses MFT expression. MFT induces dormancy in fresh seeds but promotes germination in AR seeds, and it is a convergence point between PIL5 and SPT regulation. Pathogen perception triggers DELLA-dependent and GA-independent ABI5 expression to block germination in anticipation of potential seedling damage. Drought and salinity stimulate ABA biosynthesis and induce ABI5 expression, a response mediated by HY5, RSM1 and AGL21 and counteracted by BBX21.
Additional proteins participate in the negative regulation of ABA signaling in germinating seeds: (1) the RAV1 TF represses ABI5 expression when is phosphorylated by SnRK2 kinases, in sharp contrast with the positive effect that these kinases have on the ABA pathway through phosphorylation of some early signaling components [89]; (2) a loss-of-function mutant of an MDN1 domain-containing protein (SAG) shows higher levels of ABI5 and ABI3 mRNAs in the presence of ABA [90]; (3) three WRKY-domain TFs (WRKY18, WRKY40, and WRKY60) repress ABI4 and ABI5 expression by direct binding to their gene promoters [91,92]; (4) ABI5 mRNA levels increase in the AtMyb7 TF loss-of-function mutant [93].
A clear example of ABA and GA signaling integration during seed germination is provided by the interaction between the RGL2 DELLA protein and three NF-YC TF homologs (NF-YC3, NF-YC4 and NF-YC9). This module directly upregulates ABI5 gene expression through specific binding to CCAAT elements in the ABI5 promoter [94]. Another example is the ICE1 TF, previously known for its positive role in cold-induced responses [95]. It has been reported that ICE1 binds to ABI5 and interferes with its transcriptional activity. In addition, DELLAs interact with ICE1 to repress its effect on ABI5 [96].
The central role of the ABA/GA balance in seed biology is integrated with other hormones, such as brassinosteroids (BRs) and cytokinins (CKs). The binding of ABI5 to the BIN2 kinase, a key repressor of BR signaling, promotes its phosphorylation and stability [97]. On the other hand, the positive regulator of BR signaling BES1 TF interacts with ABI5 to interfere with its transcriptional activity [98]. These findings are in line with the role proposed for BRs in reducing ABA sensitivity during germination [99]. The A-type ARR TF proteins are primary targets of cytokinin signaling and negative feedback regulators of the pathway. Specific members of this family (ARR4, ARR5 and ARR6) were found to interact with ABI5 and reciprocally downregulate their transcription [100]. After germination sensu stricto, cytokinin promotes the proteasomal degradation of ABI5 and regulates cotyledon greening, mainly via ARR12 [101]. Pathogens also influence germination. A biotic compound released by Pseudomonas aeruginosa is perceived by the seed and triggers DELLA-dependent and GA-independent ABI5 expression to block germination in anticipation of potential seedling damage [102]. In summary, current knowledge suggests that ABI5 is an important hub protein, regulated at different levels, where several hormonal and environmental signals converge. The roles of additional hormones are reviewed elsewhere in the text.
Light is one of the most important environmental cues and molecular links have been found between seed responses to adverse light conditions and ABA-mediated repression of seed germination. In AR seeds and under unfavorable conditions, ABI5, DELLAs and ABI3 form a complex that directly activates the transcription of the SOM TF. This factor then negatively regulates seed germination by increasing the ABA/GA balance [103,104,105]. ABI3 also interacts with PIL5/PIF1 TF to collaboratively activate SOM expression by binding to its promoter [104]. PIL5 represents a link between hormonal signaling and light-regulated germination [106]. Phytochromes (Phy) are a class of plant photoreceptors that enter the nucleus upon light activation, recruiting PIL5 away from its DNA-binding sites and triggering fast PIL5 degradation, and hence seed germination [107,108,109]. PIL5 degradation under light conditions is mediated by the COP1–SPA1–CUL4 E3 ubiquitin ligase (CUL4 complex) [110], where SPA1 is necessary for PIL5 phosphorylation and subsequent ubiquitination by the CUL4 complex [111]. In addition to CUL4 complex-mediated degradation, PIL5 is polyubiquitinated and subsequently degraded by the KELCH F-box protein CTG10 in association with an E3 ubiquitin ligase (SCF-complex) [112,113]. When light influx is low, Phy-mediated signaling is not enough to remove PIL5 [108]. Under these conditions, the HFR1 TF is able to effectively sequester the remaining PIL5, so its transcriptional activity is suppressed to ensure rapid germination [114]. In darkness, Phys are inactive and the COP10–DET1–DDB1–CUL4 E3 ligase complex targets and degrades HFR1 by using DET1 and COP10 as substrate receptors. Moreover, DET1 and COP10 directly interact with PIL5 to prevent its 26S proteasome-mediated degradation and favor its stability [115]. PIL5 then directly binds to and activates the GAI and RGA promoters [116]. It also represses GA biosynthesis (GA3ox1 and GA3ox2) and activates GA catabolism (GA2ox) partly through promoter binding and activation of SOM and DAG1 TFs transcription [103,117,118,119]. PIL5 typically binds to G-box elements in target promoters [116,120], but it can target additional binding sites depending on its interaction with other TFs [121]. Another aspect of the germination response to light is that far-red light inactivates PhyB mainly in the endosperm, initially preventing germination through PIL5 stabilization. Simultaneous activation of PhyA in the embryo leads to a slow destabilization of PIL5, accompanied by a weakening of ABA-dependent responses and eventually to germination in the absence of testa rupture [122,123,124]. PhyA-mediated germination has been interpreted as the last opportunity for seeds to develop a seedling despite the presence of unfavorable light conditions (e.g., far-red-enriched canopy light). The expression of several genes involved in this response has been found to be independent of PIL5, suggesting that PhyA action is regulated by additional TFs [125,126]. Interestingly, the PIF8/UNE10 TF inhibits phyA-induced seed germination without affecting phyB-mediated responses, suggesting a role as an attenuator of the photomorphogenic development under long term far-red conditions [127]. Besides PIL5 and PIF8, PIF6/PIL2 TF also plays a role in seeds. It is expressed strongly during seed development and its loss increases primary dormancy [128]. Less studied phytochromes such as PhyD and PhyE, stimulate germination under high far-red light fluence, probably by promoting PhyA action [129,130]. PhyD also has a role in relieving secondary dormancy in seeds exposed to high temperature through PIL5 removal [131]. Oppositely, PhyC negatively regulates seed responses to light, a function that depends on other phytochromes and the formation of heterodimers between them [130,132,133]. However, these phytochromes play minor roles compared with PhyB [130].
Besides light, other environmental factors such as the presence of nitrate or low temperatures stimulate the biosynthesis of GAs and promote germination of mature seeds [11,78]. In the absence of low temperatures the light-stable SPT TF suppresses the expression of GA3ox and represses germination. SPT is degraded upon cold treatment thus removing the repression of germination [134]. In the absence of light and cold stimuli PIL5 and SPT block germination in a complementary manner [135]. In addition, SPT binds the ABI5 and MFT (phosphatidylethanolamine-binding protein) promoters, activating ABI5 and repressing MFT expression, respectively [136]. Interestingly, while MFT induces dormancy in freshly-harvested seeds, it promotes germination in AR seeds through negative feedback on the ABA signaling pathway [136,137]. An interaction between light and cold signaling has also been observed in the control of germination. It has been proposed that MFT functions as a convergence point between light and cold regulation since its expression is promoted by PIL5 under far red light and, in the absence of cold stimulus, repressed by SPT under red light. This is in agreement with the observation that PIL5 downregulates SPT expression, an additional checkpoint to block germination in the dark [135].
Nitrate is the major nitrogen source for most plant species and plants sense the nutritional environment through nitric oxide (NO) synthesis from nitrate. NO has been indeed proposed as the key signaling element mediating nitrogen responses which promotes dormancy breaking and germination [138,139]. PhyB activation has been linked to the stimulation of nitrate reductase (NR) activity and NO accumulation [140]. In turn, NO signals downregulate the transcription of PIL5 and stabilize HFR1 protein to intensify the HFR1-PIL5 interaction, which counteracts the inhibitory effect of PIL5 on its target genes [140]. NO also reduces ABA sensitivity at least by promoting CYP707A2 synthesis, probably through enhanced transcription mediated by NLP8 [80], thus leading to ABA degradation [141]. Molecular evidence of the crosstalk between NO and ABA signaling was provided by showing that ABI5 expression is reduced through the NO-mediated activation of the N-end rule pathway targeting class VII ERFs TFs for degradation [142]. Another study found that NO controls ABI5 protein stability through S-nitrosylation, which triggers ABI5 ubiquitination by the KEG E3 ligase and degradation by the 26S proteasome [143]. Interestingly, ABA antagonizes this effect by promoting KEG degradation [144]. In addition, NO produces S-nitrosylation and inactivation of SnRK2 kinases required for phosphorylation and activation of ABI5 [145].
2.4. Germination Control by the Epigenome
Several studies have revealed that epigenomic mechanisms are able to modulate the expression of genes related to dormancy, maturation and germination. Specific chromatin modifiers and remodelers have been shown to promote seed dormancy or germination by enhancing and/or repressing the expression of specific gene subsets (Table 1).

Table 1.
Modifiers and remodelers altering the chromatin status of genes promoting dormancy and germination.
A number of chromatin modifications which alter transcription initiation and elongation positively regulate the expression of dormancy-related genes. The HUB1/RDO4 E3 ubiquitin ligase gene, like its homolog HUB2, is required for H2B histone monoubiquitination and expression of dormancy-related genes [146]. The H2B monoubiquitination is a chromatin modification associated with promoting transcription initiation and early elongation events [147]. RDO2 encodes a transcription elongation factor (TFIIS) and, similarly to hub1, the rdo2 single mutant has reduced dormancy and share with it about 30% of downregulated genes [148,149]. The reduction of DOG1 levels, a central regulator of dormancy (discussed below), partly explains the phenotype of the rdo2 mutant [150]. Arabidopsis ATXR7 is a H3K4 methyltransferase that, when mutated, produce similar effects on dormancy as those described for hub1 and rdo2 mutants [149]. Interestingly, the human counterparts of these genes interact with the RNA Polymerase II-Associated PAF1C factors and mutations in the Arabidopsis PAF1C-associated genes also produce early flowering, thereby linking two important developmental transitions, flowering time and seed dormancy [149]. The PIL5 promoter is also a target for histone modification, since the EFS methyltransferase increases the level of H3K36me2 and H3K36me3 to promote recruitment of RNA polymerase II and thus enhance PIL5 transcription [151]. Acetylation is another chromatin modification associated with active gene expression. Mechanisms involving deacetylation of genes with a negative effect on dormancy have been observed. In particular, members of the SNL deacetylation complex regulate key genes involved in ABA, ethylene and auxin pathways. The expression of SNL1 and SNL2 increases gradually during embryo development and seed maturation, causing a decrease in the acetylation level (H3K9/K18 and H3K14) of ABA hydrolytic genes (CYP707A1 and CYP707A2) and some ethylene-related genes (ACO1 and ACO4). This favors higher ABA levels and blocks the ethylene pathway [152]. During imbibition of AR seeds, the expression of SNL1 and SNL2 declines, causing an increase in the acetylation levels of auxin pathway genes (e.g., the auxin importer AUX1). Subsequent activation of AUX1 transcription leads to increased auxin levels and signaling, followed by enhanced cell division that promotes seed germination [153]. The plant homeodomain (PHD) motif-containing EBS is involved in the control of flowering time by binding to H3K4me2/3 and recruiting histone deacetylases (HDACs) to H3 [154]. It was shown that the ebs mutant also showed a reduction in seed dormancy independent of DOG1 [155]. In addition, PIL5 recruits HDA15 deacetylase to decrease the H3 acetylation levels of its target gene promoters to repress germination in the dark [156]. Furthermore, the LUH protein, a member of the Groucho family of transcriptional corepressors (known to recruit either HDAC or mediator complexes [157]), acts as PIL5 corepressor in the dark [158]. Another deacetylase, HDA9, negatively influences germination and promotes dormancy. HDA9 is involved in the transcriptional repression of genes related to the transition from seed to seedling during seed development, probably through H3K9 deacetylation [159].
The HDA9 function is opposite to that of its homologous genes HDA6 and HDA19, which have been reported to repress embryonic properties upon seed imbibition, probably via H3K9 deacetylation [160]. This is another indication that active chromatin modifications are required to promote germination. A similar effect is produced by Arabidopsis PHD-domain H3K4me3-binding AL proteins. AL6 and AL7 are able to interact and build complexes with PRC1 polycomb proteins around H3K4me3 marks, leading to a switch from the H3K4me3-associated active to the H3K27me3-associated repressive transcription state of genes associated to seed development (e.g., ABI3, DOG1, CRU3, CHO1) during seed germination [161]. Additional repressors of seed maturation genes have been found in vegetative organs and germinating seeds [162]. These include the polycomb EMF2-PRC2 complex combined with the SDG8 methyltransferase, which are required to maintain the H3K27me3 repressive mark in seedlings [163]; two ZRF proteins that contribute to PCR1-mediated repression by binding to monoubiquitinated H2As and H3K27me3 [164]; the SUVH5 methyltransferase mediating repressive dimethylation of H3K9 [165]; LDL1/2 demethylases, which potentially remove activating histone modifications (H3K4me2/3) from seed dormancy genes [166,167]. Finally, deacetylation of H2B by the HD2B deacetylase is associated with reduced dormancy and increased GA levels in imbibed seeds. HD2B expression is upregulated by cold or AR in accessions of Arabidopsis with low dormancy (Columbia-0; Col-0), and correlates with a reduction in the expression levels of GA inactivating genes (GA2ox2) as well as increased expression of GA biosynthetic genes (GA3ox1/2). This upregulation of HD2B expression is significantly suppressed in Arabidopsis accessions showing high dormancy (Cape Verde Islands; Cvi-0) [168]. Several TFs have also been found to recruit some of these modifiers to negatively regulate specific dormancy-associated chromatin locations. The BES1 TF is able to form a transcriptional repressor complex with the TPL corepressor and HDA19 at the ABI3 locus [169]; the SCL15 TF recruits HDA19 at a subset of embryonic-specific loci in vegetative tissues [170]; the HSI2 TF recruits HDA6 to repress seed maturation genes upon post-germination [171]. Some of these chromatin changes may also affect TF loci. The repressive role of the H3K27me3 mark on specific negative regulators of germination such as DOG1, DAG1 or SOM, seems to be particularly important for developmental phase transitions, especially from the embryonic to seedling stage [172,173]. During germination, the SANT domain-containing protein PWR, previously reported to act in a complex with HDA9 in leaves [174], suppresses ABI3-dependent SOM transcription by accelerating histone H3 deacetylation levels and H2A.Z deposition at the SOM locus. Seed imbibition under high temperature stress blocks PWR transcription and triggers secondary dormancy [175]. H3 deacetylation of SOM is also a target in carbon monoxide (CO) signaling. Light and PhyB-mediated germination increase transcription of HY1 oxygenase for CO production (a molecular signal with a positive role in stress-mediated germination) by inducing antioxidant metabolism as well as the degradation of storage reserves [176,177,178]. CO signaling recruits HDA6 to the promoter of SOM to decrease its expression by H3 deacetylation [178].
Derepression of gene expression is also required during germination. Two histone arginine demethylases (JMJ20/JMJ22) have been shown to be positive regulators of light-induced germination through the removal of repressive H4R3me2s at GA3ox1/GA3ox2 resulting in increased GA levels. This regulation is mediated by light, as JMJ20/JMJ22 are directly repressed by SOM when PhyB is inactive [179].
Chromatin structure can also be changed by ATP-dependent remodeling complexes [180]. Several of their components have been found to repress dormancy genes or embryonic traits in post-germinative growth, such as BRM (SWI2/SNF2 subgroup ATPase [181]) and PKL (CHD3 class [182]). In seeds, BRM promotes germination and directly associates with the promoters of two positive regulators of GA signaling, GA3ox1 and SCL3 TF [183]. PKL is required for about 80% of the gene expression changes triggered by GAs [184]. On the other hand, the repression of the CHR12/23 genes (SWI2/SNF2 subgroup ATPases) is required for full germination since their overexpression represses germination by elevating the levels of maturation-related genes [185].
DNA methylation is another epigenetic modification usually associated with transcriptional repression. Extensive gain of CHH methylation during seed development and drastic loss of CHH methylation during germination have been observed. These findings hint at dynamic DNA methylation reprogramming events as probable mechanisms regulating both developmental stages [186]. Such notion was corroborated in another study detecting large-scale CHH demethylation levels towards the end of germination. However, it cannot be ruled out that these events are the result of passive demethylation, as they coincide with the onset of DNA replication and hence could not be strongly associated with gene expression changes [187].
2.5. Germination Control by Small RNAs and Post-Transcriptional Regulation
Small RNAs are known to regulate gene expression in developing and germinating seeds. Several gene mutations related to small RNA biogenesis display severe defects in embryogenesis and seed development. Likewise, mutants of miRNA coding genes have altered levels of regulatory genes controlling seed dormancy and germination [188,189]. Transcriptional profiling of miRNAs during seed production of two Arabidopsis accessions with contrasting dormancy levels has revealed that the more dormant accession contains higher levels of miRNAs. Although computational analyses identified specific TFs involved in hormone signaling as putative miRNA targets, these predictions remain to be validated [190]. Additional studies on the role of miRNAs upon seed imbibition have found several miRNAs belonging to different families which are up- and down-regulated during this process [191,192,193]. Various miRNAs have also been described to influence seed germination under various abiotic stresses [189] and specific links have been proposed between miRNAs and TFs mediating hormone signaling during seed germination [194,195,196]. In addition, miR166 has been shown to contribute to the repression of maturation and dormancy genes in vegetative tissues [197]. On the other hand, many maternally expressed siRNAs transcribed by the NRPD1 polymerase (the largest subunit of RNA polymerase IV) during seed development have recently been found to regulate temporal and spatial expression of endosperm genes [198,199,200,201]. For instance, the regulation of AGL40/91 TF mRNA levels by several siRNAs is responsible for changes in seed size [201]. One type of siRNAs, the trans-acting siRNAs (ta-siRNA), have also been implicated in plant development. Ta-siRNAs are generated from the Trans-Acting SiRNA locus (TAS) gene resulting in non-coding transcripts through specific miRNA guided cleavage [189,202]. miR390 is required for the processing of a functional ta-siRNA-ARF that targets and negatively regulates auxin response TFs (ARF2, ARF3, and ARF4) in early stages of seed germination, indicating a crosstalk of ta-siRNAs and miRNAs in this process [193].
A less studied molecular event controlling germination is mRNA stability. It is well known that dry seeds accumulate extant RNAs whose abundance change during AR towards a “germination-friendly” transcriptome [203,204,205,206]. Taking into account that very low or no transcription is supported by quiescent seeds, it makes sense that the up and down-regulation observed for sets of AR regulated genes correlates with their mRNA decay rates [207,208,209]. Moreover, several groups have obtained evidence supporting a role of active mRNA degradation in the control of dormancy/germination responses. Thus, a 3’-5’ exonuclease (RRP41L), a subunit of the core exosome in Arabidopsis, is responsible for cytoplasmic degradation of specific mRNAs related to ABA signaling. Therefore, rrp41L loss-of-function seeds and seedlings showed ABA-hypersensitive phenotypes [210]. Evidence suggesting the involvement of 5’-3’ RNA decay in the control of dormancy and germination has also been found. It was observed that two 5’-3’ RNA decay mutants (xrn4 and vcs8) have altered and opposite dormancy and germination phenotypes. Moreover, transcript abundance of specific ABA/GA metabolism and signaling genes was modified accordingly to their phenotypes, suggesting that they could be direct targets of those exoribonucleases [208]. In another study, several loss-of-function mutants of the Arabidopsis 5′-3′ mRNA decay machinery were found to have enhanced ABA sensitivity. While the DCP5 (decapping) component of this machinery affected seed germination, the LSM1 (decapping activation) and XRN4 (exonucleolytic degradation) components impacted early stages of vegetative growth. The DCP5 and LSM1 components were found to have a negative effect on mRNA and protein levels of specific PYL/PYR ABA receptors, but only the lsm1 mutants showed higher levels of the SnRK2.6 kinase mRNA and protein [211]. It is worth mentioning that not all the components of the 5′-3′ mRNA decay machinery showed the same responses at the phenotypic and molecular level, and that discrepancies were observed for the xrn4 mutant in two studies [208,211]. Therefore, the degree of functional/genetic redundancy under different growth conditions thus remains an open question. Thus, the existence of additional mechanisms controlling mRNA turnover in seeds, like the targeted oxidation reported in sunflower [212], cannot be ruled out.
Several studies have found that differential recruitment of mRNAs by ribosomes adds an extra layer of regulation in both dormant and nondormant seeds [213,214]. More dynamic changes in polysomal occupancy were observed for nondormant seeds upon imbibition than for dormant seeds. GC content and the number of upstream open reading frames (uORFs) were identified as transcript features with a possible role in this selective translation. Polysomal RNAs associated with germination fell essentially into the cell wall, hormone metabolism, and redox pathway categories, while mRNAs related to stress responses and hormone metabolism pathways were associated with polysomal RNAs connected to dormancy. These results along with the absence of correlations between transcriptome and translatome led to propose that the transition from dormancy to germination is regulated mainly at the translational level [214]. In agreement with these results, large changes in polysome occupancy were found to occur upon seed imbibition, being mainly associated to the transition from dry to hydrated seed (6 h after imbibition; hai) and from 26 hai to a germinated seed (48 hai) [215]. However, the same authors found that polysomal mRNAs do not change between imbibed dormant and AR seeds treated with a transcriptional inhibitor. Since both conditions block germination, this result suggests a relevant role of transcriptional regulation and transcript abundance in controlling the germination onset [216]. These apparently contradictory results underscore that more information is needed to assess the specific roles of transcription and translation in the control of germination. In dry seeds, ribosomes are mainly present in the monosome form and certain transcript features (i.e., uORFs, length, stability) differed significantly between the polysome populations associated to specific germination stages [215]. It has been shown that most mRNAs in dry seeds are stored as monosomes forming complexes with mRNA-binding proteins, stress granules (SG), and P-body proteins. About 17% of those mRNAs are translationally up-regulated during seed germination and transcribed during seed maturation [217]. Moreover, mRNAs are not likely to be translated when found as monosomes, or are translated at low levels, since levels of translation usually correlate with ribosome density [218]. All these pieces of evidence suggest a possible scenario in which environmental conditions may modulate regulated packing of mRNAs in dry seeds to impact on their translational fate and rate upon imbibition.
Lastly, regarding post-translational modifications, changes in the phosphorylation levels of proteins during seed germination have been reported in rice, mainly during the first 12 hai. The first 12 hai are critical not only for posttranslational processes but also for transcription and metabolic changes, because the decision making for germination occurs during this period in rice [219]. Moreover, alterations in the phosphorylation/dephosphorylation patterns affect germination [219,220,221]. Besides the changes in phosphorylation status of early components of the ABA signaling pathway previously described, additional phosphorylation events have an impact on germination. FyPP1 and FyPP3 PP6 phosphatases act antagonistically with SnRK2 kinases, dephosphorylating and destabilizing ABI5 [222]. The Raf10 and Raf11 MAP3Ks are positive regulators of dormancy and ABI3 and ABI5 expression [223]. Specifically, Raf10 phosphorylates subclass III SnRK2s, which in turn phosphorylate ABI5, ABF2 and ABI3 TFs to enhance their activity [224]. TAP46 is a PP2A phosphatase-associated protein that binds and stabilizes the active phosphorylated form of ABI5, preventing its PP2A-mediated dephosphorylation [225]. Phosphorylation also affects GA signaling. Under salt stress, the GARU E3 ubiquitin ligase suppresses germination by ubiquitination of the GID1 GA receptor. The GID1-GARU interaction is counteracted by the phosphorylation of GARU by the TAGK2 Tyr-kinase [226]. DELLA stability is also associated with phosphorylation, since TOPP4 PP1 phosphatase directly binds and dephosphorylates the RGA and GAI DELLA proteins, promoting their GA-dependent destabilization [227]. The MYB44 TF activity on germination is also dependent of its phosphorylation by MPK3 and MPK6 kinases [228], conversely to RAV1 TF, which is deactivated upon phosphorylation by SNRK2 kinases [89]. Protein ubiquitination is another important protein modification known to negatively affect the stability of proteins with a crucial role in germination, such as DELLAs [14,229], ABI3 [230] and ABI5 [231,232]. Changes in ubiquitination were detected in more than 1000 proteins during rice seed germination and most changes occurred at 12 hai, as observed previously for phosphorylation [233]. Sumoylation has also been demonstrated to play a role in this process by providing stability to ABI5 and MYB30 TFs. Sumoylation protects ABI5 from degradation but makes it inactive [234], suggesting a protective role by maintaining a degradation-resistant inactive pool of ABI5 in the absence of ABA [235]. MYB30 is a negative regulator of ABA responses that seems to provide a balance for the positive regulation exerted by ABI5. Interestingly, both regulators are sumoylated at specific amino acid residues by the same SUMO E3 ligase (SIZ1) [234,236]. ABI5 sumoylation site is in the same domain (K391) as the lysine residue required for KEG-E3-ligase-dependent turnover (K344) [235]. This suggests that ABI5 sumoylation or ubiquitination depends on a direct physical competition of enzymatic activities. Other posttranslational modifications in key regulators of plant growth and development may be important in germination. It is the case of DELLA O-fucosylation and O-GlucosylNAcetylation. RGA DELLA protein interactions with BZR1, PIF3 and PIF4 TFs is promoted by mono-O-fucosylation mediated by the O-fucosyltransferase SPY [237]. On the contrary, RGA interactions with BZR1, PIF3, PIF4 and JAZ1 TFs are inhibited by O-GlucosylNAcetylation mediated by the SEC O-GlcNAc transferase [238].
3. Genetic Control from Dormancy to Germination Stages
3.1. Dormancy
The interaction between the maternal environment and the genetic makeup of the mother plant will determine primary seed dormancy levels during seed maturation. One of the regulatory routes for the establishment and maintenance of physiological dormancy involves the ABA/GA hormonal balance [10,239,240]. Most mutations altering the metabolism, perception and early signaling of these hormones show effects on dormancy levels [239,241,242]. Additional regulators of this hormonal crosstalk have been described. It is the case of ABI4 TF, which increases ABA/GA balance in freshly-harvested seeds and post-germinative stages through direct binding to promoters of some of their metabolic genes [243,244]. The CHO1 TF gene also contributes to ABI4-mediated regulation [245,246]. MYB96 TF also increases ABA/GA balance and seed dormancy by directly activating NCED2 and NCED6 and indirectly repressing GA3ox1 and GA20ox1 expression [247]. DOF6 TF promotes dormancy by enhancing ABA-related gene expression [248] and by activating GATA12 TF expression upon complex formation with the RGL2 protein [249]. Other proteins regulating dormancy are involved in feedback hormone control. The AtSdr4L gene encodes a protein of unknown function which is required for the negative feedback control of GA biosynthetic genes, with an impact on dormancy and germination [250]. The WRKY41 TF directly upregulates ABI3 expression, a function requiring ABA but subjected to negative feedback regulation when the concentration of hormone is sufficiently high [251]. Additionally, circadian clock genes have been shown to play roles in dormancy control [252]. For instance, RVE1 and RVE2 TFs accumulate during seed development to promote seed dormancy but their transcription is repressed by light-mediated activation of PhyB to allow germination [253]. This regulation is counteracted by the ability of RVE1 to interact with RGL2, reducing its interaction with the SLY1 F-box protein and increasing RGL2 stability. In return, RGL2 enhances RVE1 transcriptional activity, which directly represses GA3ox2 expression [253,254].
Besides the hormonal control of dormancy, other pathways have been linked to this process, like those involving DOG1 and RDO5. These dormancy-specific genes were identified by quantitative trait loci analyses (QTL) of natural variation in Arabidopsis [255,256,257]. DOG1 was identified as a key effector of dormancy and has therefore been intensely characterized in recent years [258,259,260]. Its expression in seeds is controlled at four different levels, namely, transcriptional elongation [150], alternative splicing [261,262], alternative polyadenylation [263], and transcriptional suppression by a non-coding cis antisense transcript [264]. Different genetic and transcriptomic analyses suggested that DOG1 exerts its dormancy function in parallel to, but independent of ABA function. Among other pieces of evidence, it has been observed that the non-dormant mutant dog1 has a normal germination sensitivity to treatments with exogenous ABA [256] and that a high accumulation of ABA or DOG1 protein in the seed cannot compensate for the absence of function of DOG1 or ABA metabolism genes, respectively [265]. These results indicate that both routes are required for an efficient block of dormancy release [265]. Besides dormancy, a genetic interaction between DOG1 and ABI3 has been described during seed maturation, as well as the control of ABI5 expression by DOG1, revealing a dual role for DOG1 in dormancy and seed development [266].
Lately, new convergences have emerged between ABA and DOG1 routes regarding dormancy [260]. DOG1 binds to AHG1 and AHG3, two clade A protein phosphatases 2C (PP2Cs) [267,268] which negatively regulate of ABA signaling and dormancy [269,270,271]. Both phosphatases have redundant roles in dormancy release and are epistatic to DOG1 [267,268]. It seems that DOG1 has a role in increasing ABA sensitivity through an AHG1 and AHG3 sequestration mechanism, analogous to the perception and initiation of ABA signaling [260,267]. Unlike AHG3 and other members of PP2C, AHG1 is resistant to inhibition by PYR/PYL/RCAR receptors [272]. Some authors have argued that by sequestering this phosphatase, DOG1 might play a role in safeguarding ABA signaling to ensure dormancy until it is inactivated after AR. This would explain why overexpressing DOG1 in ABA-deficient mutants or increasing ABA levels in the dog1 mutant provoke reduced dormancy [265]. Likewise, the activity of non-sequestered PP2Cs in these ABA-deficient mutants would be sufficient to promote germination [260,267]. It has been recently discovered that DOG1 is a heme-binding protein and such binding is essential for its functionality in dormancy [268]. This may establish a role for DOG1 as an integrator of environmental signals, since heme-binding proteins act as oxygen and NO sensors [260,268].
In addition to the roles in dormancy and seed maturation, other functions have been identified for DOG1. One of them relies on the regulation of DOG1 expression by temperature with an impact on flowering time and dormancy release. Thus, DOG1 is able to modify the levels of two antagonistic miRNAs: miR156, which delays flowering and dormancy release, and miR172, which produces the opposite effect [273]. This coordinated regulation of two developmental phase transitions seems to conform a plant strategy to adapt its life cycle to seasonal environmental conditions [265,273,274,275,276,277]. Finally, in addition to its functionality in reproductive and germinative growth, a role has been proposed for DOG1 in vegetative growth based on the drought-sensitive phenotypes observed in its loss-of-function mutants [278]. Dormancy and drought responses show many similarities at the molecular level regarding ABA signaling. The antisense of DOG1, asDOG1/1GOD, silences DOG1 expression in seeds and leaves, causing dormancy release [264] and drought responses [278], respectively. Upon ABA accumulation, DOG1 transcript levels increase due to suppression of asDOG1 expression [274,278,279].
RDO5/DOG18/IBO is, together with DOG1, another dormancy-specific gene identified by QTL analyses [257,280,281]. As dog1, the rdo5 mutant shows loss of dormancy, identifying RDO5 as a positive regulator of this process [280]. RDO5 encodes a PP2C without phosphatase activity which probably controls phosphorylation levels by hampering dephosphorylation during imbibition [281,282]. In addition, RDO5 seems to act independently of ABA, since its loss of function does not affect ABA levels or sensitivity to the hormone [280], and does not produce changes in phosphorylation levels of ABA signaling regulators [282].
In addition to the main regulatory routes (ABA, DOG1, RDO5), other hormones and their associated molecular mechanisms have been involved in dormancy control. Ethylene reduces dormancy and improves seed germination in several plant species by counteracting ABA effects through the regulation of ABA metabolism and signaling pathways [283,284,285]. Recently, one of the mutants previously identified for exhibiting a reduced dormancy phenotype, rdo3 [286], was shown to be a loss of function of the ETR1 ethylene receptor [287]. Although ETR1 does not require the canonical ethylene signaling pathway to act in this process, it is involved in the induction of ABA signaling genes [288]. Thus, when ETR1 function is lost, ERF12 TF is upregulated by an unknown transduction pathway which probably involves MAP kinases, and forms a repression complex with TPL that binds the DOG1 promoter and represses its expression [287].
Auxins also have a role in dormancy in an ABA-dependent manner, since treatment with exogenous indole-3-acetic acid (IAA) in combination with ABA enhances dormancy. In addition, ARF TF mutants, components of the auxin response, have reduced dormancy levels and are less sensitive to ABA treatments [289]. ARF10 and ARF16 have been identified as positive regulators of dormancy through indirect regulation of ABI3 expression [289]. These results seem contradictory with those obtained by another study, which describes that in dormant seeds of near-isogenic Arabidopsis lines carrying the Cvi-0 DOG1 loci introgressed in a Landsberg erecta (Ler) genetic background, the latter being a less dormant accession than Cvi-0, tryptophan-dependent auxin biosynthesis and related pathways are strongly repressed compared to germinating seeds [216].
One interesting aspect of dormancy is the fact that forest fires generate chemical signals that can stimulate the germination of certain dormant seeds in the soil. These compounds include cyanohydrins [290] and karrikins [291,292,293]. Arabidopsis and other seeds of Brassicaceae respond to karrikins [294] by using the KAI2 receptor, a paralogue of the D14 strigolactone receptor [295]. The binding of karrikins to KAI2 leads to their interaction with the MAX2 F-box protein, which targets TFs such as SMAX1 for degradation [296]. Consequently, the loss-of-function mutants kai2 and max2 show enhanced dormancy [295,297,298,299], probably by negatively affecting CYP707A expression [300]. Such phenotype is reverted by the loss of SMAX1 function [296]. Interestingly, while GAs or nitrate always stimulate dormancy release, karrikins promote dormancy when combined with abiotic stresses such as NaCl, mannitol and elevated temperature [298]. This dual role of karrikins, as dormancy enhancers or repressors depending on environmental conditions, suggests that karrikin signaling factors may function as safeguards to prevent germination until conditions are optimal.
Effects on dormancy by jasmonate (JA) had rarely been reported. The best studied case is that of the JA precursor 12-oxo-phytodienoic acid (OPDA), which produces an increase in dormancy upon accumulation in the comatose (cts) mutant through a positive feedback with other positive dormancy regulators (i.e., ABA, RGL2 and MFT) [301,302]. Additionally, the maternal herbivory, defined as the maternal experience of herbivore feeding during flowering and seed development seems to have an effect on dormancy through JA pathway regulation [303]. The accumulation of JA-isoleucin (JA-Ile) during seed development, either by maternal herbivory or overexpression of the AOS JA biosynthesis enzyme, produces a reduction in dormancy. This phenotype is associated with increased GA content and reduced ABA sensitivity, a response absent in a JA-Ile-deficient (jar1-1) mutant [303].
Herbivory is not the only process that regulates dormancy maternally. The level of seed dormancy is highly influenced too by the environmental conditions experienced by the mother plant. In this case, FLC TF [304,305,306], RGL2 and the phosphatidylethanolamine-binding protein FT [305,307] play important roles, unlike other key dormancy proteins such as DOG1 [305]. In siliques, the expression of FT and FLC responds to temperature in the maternal tissues but not in those of the seed. Accordingly, under cold conditions, the maternal FT protein expressed in the silique phloem controls dormancy through inhibition of proanthocyanidin synthesis in the seed, thus altering the levels of testa tannins [305,308]. Maternal inheritance is also evidenced during genomic imprinting, the preferential expression of a given parental allele over the other [309]. The importance of this process in the maternal inheritance of seed dormancy has recently been described. A set of genes were found to be imprinted in endosperm cells and the maternal alleles were preferentially expressed upon seed imbibition [310]. For instance, in the case of the ALN gene, cold stimulates the differential methylation of the promoter of the paternal allele to promote dormancy [311].
3.2. After-ripening and Longevity
In dry seeds, the cytoplasm reaches a highly viscous glassy state which severely limits molecular diffusion and the occurrence of chemical and enzymatic reactions, but maximizes seed survival [312,313]. Despite apparent inactivity, seeds continue to undergo physiological changes such as loss of dormancy, or even loss of longevity, defined as the total time span during which seeds remain viable.
Primary dormancy is acquired during seed maturation and is gradually lost during dry storage (or AR), a process that depends on the relationship between seed moisture content and temperature [212,314,315,316,317]. Loss of dormancy has been associated with an accumulation of reactive oxygen species (ROS), which would lead to oxidation of proteins and mRNAs [212,318,319,320]. Non-enzymatic oxidative reactions have been associated with low moisture content (below ~0.12 g H2O g dw–1) while metabolic reactions would prevail under high moisture conditions [314]. No active transcription is required in Arabidopsis during the AR process [321]. Like in wheat and sunflower, there is hardly any change in the abundance of transcripts between dry dormant and AR non-dormant seeds [204,322,323]. In addition, although a clear correlation between transcriptome and translatome upon seed imbibition has not been found, there is a selective recruitment of mRNAs into polysomes [213,214]. Thus, in Arabidopsis, one-third of the polysome-associated transcripts are similar at 16 and 24 hai in dormant seeds, while only around 4% are common in nondormant seeds between 16 and 24 hai [214]. It has also been suggested that oxidation of specific mRNAs during AR might reduce their translation during seed imbibition, even if they are still present in the transcriptome [212,322,323,324]. In the same line, specific recruitment of mRNAs is thought to be based on features of the 5’-UTR [214,325]. These findings, along with the fact that the germination program is activated in non-dormant seeds after 8–24 hai, suggest that there is a developmental checkpoint during the first hours of imbibition [325]. In this way, a selective translation of mRNA during imbibition will maintain or not the inhibition of germination, depending on the oxidative imprinting of the seed. Certain components may be particularly sensitive to oxidation as part of such imprinting.
When environmental conditions impose a long block on germination, the viability of dry seeds, and therefore their longevity, can be reduced due to excessive oxidation-derived damage of molecular components. Seeds have different mechanisms to favor longevity and some genetic factors involved in this process have been identified by QTL analysis using natural variation in Arabidopsis [326,327,328]. Many of these factors have functions related to protection of different biomolecules from ROS [329,330,331,332]. Among these factors are vitamin E (tocopherols and tocotrienols), which prevents the non-enzymatic oxidation of lipids [333], protein L-isoaspartyl methyltransferase, with a role in the repair of damaged proteins [334], metallothioneins [335], methionine sulfoxide reductase [336], lipoxygenase [337], the glycosylase/apurinic/apyrimidinic lyase DNA [338], and the prolyl isomerases rotamase FKBP 1 and 2 [339]. Noteworthy are the mitochondrial NADH dehydrogenase ferric-chelate reductase 1 [326] and the NADP-malic enzyme 1, whose function losses produce a reduction in longevity, and, for the latter mutant, enhanced protein carbonylation in aged seeds [340,341]. Likewise, it has been observed that seed-storage proteins buffer seed biomolecules from oxidative stress [328]. One of the cellular components damaged during prolonged dry state is DNA, with double-strand breaks (DSBs) being rate-limiting for germination [342]. Plants have a specific response to integrate the germination progress of aged seeds with the monitoring of genome integrity. This mechanism is the DNA damage signaling or DNA damage response, in which the checkpoint kinases ATM and ATR play key roles [343,344]. Aged atr and atm mutant seeds germinate faster than aged WT seeds and show earlier activation of DNA replication and extensive chromosomal abnormalities. Thus, ATM and ATR contribute to the control of germination by inhibition of DNA replication in aged seeds upon imbibition, partly through the transcriptional up-regulation of the SMR5 cell cycle inhibitor by ATM [343].
Other regulatory proteins with an impact on these processes have also been identified. This is the case for PhyA and PhyB phytochromes [345], the RSL1 E3 ubiquitin ligase [346], and the TFs CDF4/DOF2.3 [345], ATHB25 [347] and COG1/DOF1.5 [345]. The composition and structure of the seed coat are critical factors for seed longevity by providing chemical and mechanical protection [348]. The longevity effects of mutations in ATHB25, as well as in phytochromes and COG1, correlate with accumulation of mucilage and suberine, respectively [345,347]. The loss of TT10 laccase function, which is involved in seed coat lignin biosynthesis [349,350], also produces a reduction in longevity [351]. Additionally, because longevity is induced during seed maturation [352,353,354], mutations altering seed development (e.g., lec1, lec2, fus3, abi3 [326]), and DOG1 (e.g., dog1 [256,266]), also produce longevity defects.
3.3. Seed Bank and Secondary Dormancy
Most studies on germination are made under controlled laboratory conditions with minimal environmental variation. However, seeds shed in the field are exposed to a journey of uncertain duration under shifting environmental conditions. During the year, seeds in the soil (seed bank) are repeatedly imbibed and dried, suffering temperature shifts during variable periods of time. Under this state of continuous change, the seed bank must be capable of sensing external conditions and adjusting their germination potential accordingly. That is why non-dormant seeds can retrieve the dormancy program as a protective measure upon encountering inadequate conditions. This ability is called secondary dormancy and can be lost and gained repeatedly until germination occurs or viability is lost. The molecular processes underlying this dormancy cycling in the seed bank have been less studied than those involved in primary or secondary dormancy in the laboratory. However, several publications have started shedding light on this ecological process from a physiological [355] and a molecular standpoint [316,356,357]. For a winter annual plant, the beginning of winter coincides with an increase in dormancy. This dormancy correlates with higher ABA/GA ratios supported by enhanced expression of ABA biosynthetic and GA catabolic genes, followed by a subsequent increase of gene expression related to ABA signaling [358,359]. This leads to a stage of deep dormancy, reinforced by the increased expression of DOG1 and MFT [358,359,360]. Reversion of these events is linked with dormancy release, starting in spring and leading to a shallow dormancy state [358,359]. This cycle of transitions between deep and shallow dormancy corresponds to a temporal sensing of seasonal changes. Germination is also fine-tuned during shallow dormancy stages by increasing sensitivity to light and nitrate and upregulating DELLA expression (RGL2/RGA). In this way seeds can couple spatial sensing information with the onset of germination [358,359,360].
3.4. Regulation of Germination from a Spatial and Mechanical Perspective
Another important aspect of seed germination relates to the interplay of mechanical forces between seed tissues. The ability of seeds to germinate is thought to result from a balance between physical restrictions imposed by the embryo-surrounding tissues (testa and endosperm) and the ability of the embryo to grow and protrude [3,361]. The decline in the mechanical resistance of the micropylar endosperm, which covers the radicle tip, leads to endosperm weakening and appears to be a general prerequisite for radicle protrusion (germination sensu stricto) [362,363,364]. Previous studies with non-dormant seeds have shown that the expression of many Cell Wall Remodeling Enzymes (CWREs) are upregulated by GAs, in correlation with endosperm weakening and embryo radicle protrusion. These findings point at the composition of cell walls and their mechanical properties as relevant targets to control germination [3,363,364,365].
Xyloglucans (XyGs), the major components of hemicelluloses in the primary cell walls, have been found to play a role in wall remodeling. Mutant seeds lacking functional XYL1, an α-xylosidase involved in XyG biosynthesis, were able to germinate on PAC and had reduced dormancy, thermoinhibition-resistant germination and alterations in specific genes involved in ABA/GA metabolism, all characteristics resembling ABA-deficient mutants [366,367]. In addition, the mutants showed changes in the composition of endosperm cell walls resulting in reduced strength, which supports the notion that XYL1 is a negative regulator of germination [366]. Moreover, different results have shown a localization of XyGs compatible with a role in germination. Thus, immunolocalization experiments in germinating seeds indicated a reduction of xyloglucans (XyG) in the elongation zone of the embryonic axis but not in the cotyledons or root tips [366]. A promoter:GUS fusion also revealed low XYL1 expression in endosperm, as expected for a tissue required to reduce mechanical resistance to embryo growth upon imbibition [367]. XTH endotransglycosylases/hydrolases are another type of XyG-related enzymes that can cleave and reconnect XyG chains and several of them are upregulated upon seed imbibition of non-dormant seeds [3,365]. One of them, XTH31, is thought to reinforce endosperm cell walls, since its loss of function led to faster germination [365].
Other important components of cell walls, pectins and pectin methylesterases (PMEs), have been associated with promoting seed germination mainly acting on testa permeability [368]. Although PME activity is usually linked to enhanced pectin de-esterification and increased wall rigidity, it can produce the opposite effect in combination with appropriate enzymatic activities [369]. Genetic redundancy may be a problem when ascribing roles to PME members in the regulation of germination. One example is the pme58 mutant that shows normal germination despite having altered seed-coat mucilage structure [369].
Wherever cells are growing or modifying their walls, one or more expansin genes are usually active, promoting primary cell wall relaxation [370]. These proteins are thought to specifically modify the interactions between XyG and cellulose microfibrils. EXPA2 encodes a GA-induced endosperm-specific α-expansin with a proven genetic role in enhancing germination [371].
In addition to cell walls, a thick cuticle layer was discovered to be tightly associated with the outer surface of the endosperm cell layer of Arabidopsis seeds [372]. It was later found that this cuticle had a maternal origin, deriving from a specific layer of the ovule integuments that becomes associated to the endosperm at late stages of seed development [373]. This cuticle regulates permeability and has an impact on seed dormancy and viability. Mutants defective in cutin biosynthesis (e.g., lacs2, bdg1) are unable to block endosperm cell expansion and testa rupture under adverse conditions [372]. It has been suggested that this cuticle could prevent integument-endosperm fusion during seed development, keeping unwanted developmental signals from entering the endosperm [373]. Indeed, it has been shown that the endosperm plays an important role in linking the perception of environmental signals to the control of dormancy and its cycling by modulating ABA released to the embryo and controlling gene expression [63,64,357].
Several publications have established a link between germination and cell wall remodeling and cell expansion (Figure 2). It was found that GA signaling in the Arabidopsis embryo epidermis along the embryonic axis is required for proper germination. A DNA sequence (L1 box) conserved in the promoters of epidermis-specific genes is bound by two homeodomain (HD-ZIP) TFs (ATML1 and PDF2) and mediates GA-induced transcription of these genes. Since the function of these TFs is blocked by physical interactions with DELLA proteins, increased GA levels produced upon imbibition would cause DELLA degradation. Subsequent release of these TFs will enhance cell elongation and germination mediated by CWRE genes like EXP8 [59]. The elongation of the epidermal cells is likely to be coordinated with those of inner tissues by activating additional HD-ZIP target genes involved in the biosynthesis of very-long-chain fatty acids (VLCFAs) [59], a mechanism demonstrated for vegetative stages [374]. Indeed, imaging studies on germinating embryos demonstrated that cell surface area increases mainly in the epidermis and moves progressively towards inner layers [60]. Firstly, cell expansion in the lower embryonic axis contributes to testa rupture and then expansion of its upper part promotes protrusion of the radicle through the seed coverings [60]. By combining spatio-temporal gene expression information with promoter analyses, another homeodomain TF (ATHB5) was found to control the expression of the expansin gene EXPA3. This control takes place mainly in cortical cell layers of the upper embryonic axis, suggesting a tissue-specificity and partially overlapping roles of homeodomain genes [59,60]. Many morphological features related to elongation events and cell wall remodeling in the embryo seem to be conserved between species [375,376,377]. Moreover, the epidermal HD-ZIP-DELLA-L1 box regulatory module was found to be conserved in cotton where it controls fiber cell elongation [378], indicating that it has been recruited by other developmental stages. Another study demonstrated that expansion-promoting gene expression in embryo radicle tips, including GA biosynthetic genes, is induced very early after seed imbibition (1–3 hai). However, due to mechanical constraints cell expansion is observed mainly in the upper limits of the radicle, which extends along the embryonic axis during subsequent stages of germination. These results are a clear indication that cell geometry and the interplay of mechanical forces between cells have an influence on genetically specified growth [379]. The embryo radicle tip was also found to be an important place where temperature has an impact on gene expression by influencing tissue/cell-specific ABA/GA balance. This dynamic regulation largely determines whether a seed germinates or remains dormant in the soil [61].
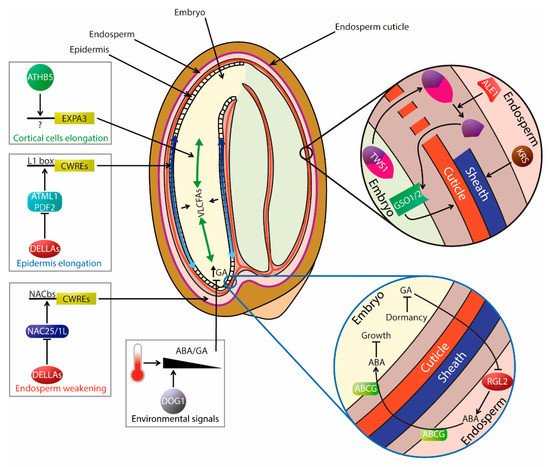
Figure 2.
Spatial and mechanical regulation of seed germination. The seed coat protects living tissues from mechanical and oxidative damage. In addition, a cuticle layer associated with the outer surface of the endosperm regulates permeability, modulating seed physiology. Despite these seed coverings, embryo inner cells are able to continuously sense the environment and decide when to germinate. A specific area within the embryonic radicle acts as a decision-making center inducing changes in ABA/GA in response to variable temperature. In the endosperm, DOG1 couples temperature with the regulation of GA metabolism to control CWRE gene expression required for the weakening of cell walls. Endosperm also controls embryo growth in dormant seeds by RGL2-dependent release of ABA and seed specific ABCG transporters. Once germination is triggered, there is an interplay of mechanical forces as the embryo pushes against its surrounding tissues. GA biosynthetic and expansion-promoting gene expression is induced very early in the radicle tip upon imbibition. Due to mechanical constraints, cell expansion is observed mainly in the upper limits of the radicle, extending afterward along the embryonic axis. This expansion is required for germination and depends on GA-responsive epidermis-specific gene expression mediated by two HD-ZIP proteins, ATML1 and PDF2. They activate CWRE and VLCFA genes to coordinate epidermal cell expansion with that of inner tissues. ATHB5 also controls cell expansion, but mainly in cortical cell layers of the upper embryonic axis. Cell expansion along the embryonic axis contributes to testa rupture and germination. Endosperm cells elongate at different rates to accommodate embryo growth. This process is controlled mainly by GA signaling mediated by NAC25 and NAC1L, which upon the perception of an unknown embryonic signal activates CWRE expression. Communication between embryo and endosperm to coordinate germination also occurs during seed development by two mechanisms: (1) A peptide-mediated bidirectional signaling controls the deposition of an embryo cuticle to minimize water loss (embryo secreted TWS1 peptide; endosperm-specific ALE1 subtilase; GSO1/GSO2 receptor-like kinases); (2) An endosperm-derived peptide triggers deposition of the embryo sheath, which facilitates coat shedding and seedling establishment (KRS endosperm-specific peptide).
Although the structure of cell walls differs between the endosperm and the embryo, cell wall architecture of Brassicaceae and Solanaceae species is similar in the micropylar endosperm [380]. Differential gene expression in the endosperm is concentrated in the micropylar end and involves key genes for cell wall function, many of them induced by GAs [3,58]. Genetic manipulation of specific cell wall components or cell wall-related enzymes in the endosperm is known to have an impact on seed germination [362,365,372,380,381]. DOG1 impacts germination by coupling temperature with GA metabolism to control the CWRE gene activity required for the biomechanical weakening of the endosperm [382]. Endosperm expansion during imbibition was also identified as a necessary key step in the regulation of germination and is controlled by an embryo-initiated gene network [62,382]. By using 3D geometry cell reconstruction, it was observed that all endosperm cells expand during imbibition, but at different rates, to accommodate embryo growth and to facilitate germination. A molecular mechanism underlying endosperm cell expansion was found to be controlled by two NAC TFs (NAC25 and NAC1L) that, when released from repression by the RGL2 DELLA protein, perceived appropriate signals from embryo and activated the expression of a cohort of CWREs [62].
Although the regulatory signals that move from embryo to endosperm remain elusive, endosperm-derived signals triggering a deposition of hydrophobic and anti-adhesive barriers on the embryo surface have been recently discovered. Peptide-mediated bidirectional signaling controls the deposition of the embryo cuticle that prevents organ fusion during seed development and excessive water loss in young plants to maximize seedling survival at germination [383,384,385,386,387,388]. Finally, the ZOU/RGE1 TF is also responsible for the production of the KRS endosperm-specific peptide, which triggers deposition of the embryo sheath, a layer of endosperm-derived material rich in extensin-like molecules [389]. This sheath is deposited outside the embryonic cuticle reducing the adhesion to the endosperm during germination and thus facilitating seed-coat shedding and rapid seedling establishment [390].
4. Future Directions
Despite our extensive knowledge of seed biology, many important questions remain to be answered. For example, the contribution of cell division between seed imbibition and seed germination is still unclear. Whereas in Arabidopsis and most species no cell division is observed before germination, genome duplications and activation of cell cycle genes are known to occur at late germination stages, contributing to germination speed [391,392,393,394]. This may be a strategy to guarantee genome integrity by allowing DNA damage responses to scan and repair the genome, but more studies are needed. Another interesting question is how the described transport activities are coordinated and linked to hormone metabolism and signaling. In this respect, the study and development of hormone sensors and tagged hormones [395,396,397] should provide helpful information. Novel transporters may participate in the precise control of hormonal balances in seeds (probably not only for ABA/GA). The fact that mutations in proteins involved in transcriptional elongation only seem to alter seed dormancy and flowering remains a puzzling issue. A link between these two developmental stages has been underscored by mutations in DOG1 and FT, two genes that independently regulate dormancy [273,305]. Despite increasing data on the regulation of germination by mechanisms affecting RNA dynamics, this topic has received much less attention than other molecular layers of regulation. In particular, RNA modifications with a profound impact on RNA activity, such as N6 or N1 adenosine methylation, have not yet been reported to play a role in seeds. Finally, the discovery of tissue-specific signaling mechanisms and supra-cellular structures is providing a more complex picture of the control of seed germination. Such a picture will be fine-tuned in the near future with studies at the single-cell level.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/6/703/s1, Table S1: Compilation of selected recent reviews on specific aspects of seed biology, Table S2: List of complete gene names in alphabetical order according to their acronyms.
Author Contributions
G.C.-C. and L.O.-S. drafted and wrote the manuscript; J.C.-C. draw the figures and revised the manuscript; M.P. and L.G. contributed in drafting and revising the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported with a grant to L.O.-S. by the Spanish Ministry of Science and Universities of Spain (BIO2016- 77840-R) and to M.P. by FP7, FACCE-JPI-ERA-NET+CLIMATE SMART AGRICULTURE (ERA46-SYBRACLYM). J.C.-C. was supported by a PhD contract (FPI) funded by grant SEV-2016-0672 to the CBGP (Centre of Excellence Severo Ochoa Program of the Agencia Estatal de Investigación, Spain). G.C.-C. was supported by a PhD contract by Universidad Politécnica de Madrid (UPM; programa propio).
Acknowledgments
We apologize in advance for those contributions that might have been left out due to space limitations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bewley, J.D.; Bradford, K.; Hilhorst, H.; Nonogaki, H. Dormancy and the control of germination. In Seeds, Physiology of Development, Germination and Dormancy, 3rd ed.; Springer: New York, NY, USA, 2013; pp. 247–297. [Google Scholar]
- Willis, C.G.; Baskin, C.; Baskin, J.M.; Auld, J.R.; Venable, D.L.; Cavender-Bares, J.; Donohue, K.; De Casas, R.R. The nescent germination working group the evolution of seed dormancy: Environmental cues, evolutionary hubs, and diversification of the seed plants. New Phytol. 2014, 203, 300–309. [Google Scholar] [CrossRef]
- Holdsworth, M.; Bentsink, L.; Soppe, W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef]
- Koorneef, M.; Elgersma, A.; Hanhart, C.J.; Rijn, L.; Zeevaart, J.A.D.; Loenen-Martinet, E.P. A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 1985, 65, 33–39. [Google Scholar] [CrossRef]
- Koornneef, M.; Van Der Veen, J.H. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 1980, 58, 257–263. [Google Scholar] [CrossRef]
- Koornneef, M.; Jorna, M.L.; Der Swan, D.L.C.B.-V.; Karssen, C.M.; Der Swan, D.L.B.-V. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 1982, 61, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; Reuling, G.; Karssen, C.M. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 1984, 61, 377–383. [Google Scholar] [CrossRef]
- Steber, C.M.; E Cooney, S.; McCourt, P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 1998, 149, 509–521. [Google Scholar] [PubMed]
- Claeys, H.; De Bodt, S.; Inzé, D. Gibberellins and DELLAs: Central nodes in growth regulatory networks. Trends Plant Sci. 2014, 19, 231–239. [Google Scholar] [CrossRef]
- Vishal, B.; Kumar, P.P. Regulation of Seed Germination and Abiotic Stresses by Gibberellins and Abscisic Acid. Front. Plant Sci. 2018, 9, 838. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Ogawa, M.; Kuwahara, A.; Hanada, A.; Kamiya, Y.; Yamaguchi, S. Activation of Gibberellin Biosynthesis and Response Pathways by Low Temperature during Imbibition of Arabidopsis thaliana Seeds. Plant Cell 2004, 16, 367–378. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Takeda-Kamiya, N.; Hanada, A.; Ogawa, M.; Kuwahara, A.; Seo, M.; Kamiya, Y.; Yamaguchi, S. Contribution of Gibberellin Deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 2007, 48, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Mitchum, M.G.; Yamaguchi, S.; Hanada, A.; Kuwahara, A.; Yoshioka, Y.; Kato, T.; Tabata, S.; Kamiya, Y.; Sun, T.-P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006, 45, 804–818. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, K.M.; Thomas, S.G.; Soule, J.D.; Strader, L.C.; Zale, J.M.; Sun, T.-P.; Steber, C.M. The Arabidopsis SLEEPY1 Gene Encodes a Putative F-Box Subunit of an SCF E3 Ubiquitin Ligase. Plant Cell 2003, 15, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Richards, D.E.; Fleck, B.; Xie, D.; Burton, N.; Harberd, N.P. The Arabidopsis Mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the scfsly1 e3 ubiquitin ligase for della protein substrates. Plant Cell 2004, 16, 1406–1418. [Google Scholar] [CrossRef]
- Strader, L.C.; Ritchie, S.; Soule, J.D.; McGinnis, K.M.; Steber, C.M. Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc. Natl. Acad. Sci. USA 2004, 101, 12771–12776. [Google Scholar] [CrossRef]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genome Res. 1997, 11, 3194–3205. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T.-P. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155. [Google Scholar] [CrossRef]
- Davière, J.-M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef]
- Davière, J.-M.; Achard, P. A pivotal role of dellas in regulating multiple hormone signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef]
- Nelson, S.K.; Steber, C.M. Gibberellin hormone signal perception: Down-regulating DELLA repressors of plant growth and development. In Annual Plant Reviews: The Gibberellins; Hedden, P., Thomas, S.G., Eds.; John Wiley & Sons: Chichester, UK, 2016; pp. 153–188. [Google Scholar]
- Vera-Sirera, F.; Gomez, M.D.; Perez-Amador, M.A. Chapter 20—Della proteins, a group of Gras transcription regulators that mediate gibberellin signaling. In Plant Transcription Factors; Gonzalez, D.H., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 313–328. [Google Scholar]
- Griffiths, J.; Murase, K.; Rieu, I.; Zentella, R.; Zhang, Z.-L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.-P.; et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in arabidopsis. Plant Cell 2006, 18, 3399–3414. [Google Scholar] [CrossRef]
- Willige, B.C.; Ghosh, S.; Nill, C.; Zourelidou, M.; Dohmann, E.M.; Maier, A.; Schwechheimer, C. The della domain of ga insensitive mediates the interaction with the ga insensitive dwarf1a gibberellin receptor of arabidopsis. Plant Cell 2007, 19, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Locascio, A.; Blázquez, M.A.; Alabadí, D. Genomic Analysis of DELLA Protein Activity. Plant Cell Physiol. 2013, 54, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Ueguchi-Tanaka, M. Della and SCL3 balance gibberellin feedback regulation by utilizing indeterminate domain proteins as transcriptional scaffolds. Plant Signal. Behav. 2014, 9, e29726. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, N.M.-D.; Sotillo, B.; Miskolczi, P.; Gibbs, D.J.; Vicente, J.; Carbonero, P.; Oñate-Sánchez, L.; Holdsworth, M.; Bhalerao, R.P.; Alabadí, D.; et al. Large-scale identification of gibberellin-related transcription factors defines group vii ethylene response factors as functional della partners. Plant Physiol. 2014, 166, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Sarnowska, E.A.; Rolicka, A.; Bucior, E.; Cwiek, P.; Tohge, T.; Fernie, A.R.; Jikumaru, Y.; Kamiya, Y.; Franzen, R.; Schmelzer, E.; et al. Della-interacting swi3c core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in arabidopsis. Plant Physiol. 2013, 163, 305–317. [Google Scholar] [CrossRef]
- Lee, S.; Cheng, H.; King, K.E.; Wang, W.; He, Y.; Hussain, A.; Lo, J.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genome Res. 2002, 16, 646–658. [Google Scholar] [CrossRef]
- Tyler, L.; Thomas, S.G.; Hu, J.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.-P. Della proteins and gibberellin-regulated seed germination and floral development in arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef]
- Ariizumi, T.; Lawrence, P.K.; Steber, C.M. The role of two f-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol. 2010, 155, 765–775. [Google Scholar] [CrossRef]
- Voegele, A.; Linkies, A.; Müller, K.; Leubner-Metzger, G. Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J. Exp. Bot. 2011, 62, 5131–5147. [Google Scholar] [CrossRef]
- Hauvermale, A.L.; Tuttle, K.M.; Takebayashi, Y.; Seo, M.; Steber, C.M. Loss of Arabidopsis thaliana seed dormancy is associated with increased accumulation of the gid1 ga hormone receptors. Plant Cell Physiol. 2015, 56, 1773–1785. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Boil. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of ArabidopsisNCED6andNCED9genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef]
- Millar, A.A.; Jacobsen, J.V.; Ross, J.J.; Helliwell, C.; Poole, A.T.; Scofield, G.; Reid, J.B.; Gubler, F. Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 2006, 45, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, Which Encode Abscisic Acid 8′-Hydroxylases, Are Indispensable for Proper Control of Seed Dormancy and Germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Boil. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Hubbard, K.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Choi, H.-I.; Hong, J.-H.; Ha, J.-O.; Kang, J.-Y.; Kim, S.Y. ABFs, a Family of ABA-responsive element binding factors. J. Boil. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.-H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Li, Y.; Gilday, A.D.; Graham, S.; Graham, I.A. Arabidopsis aba insensitive4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 2006, 18, 1887–1899. [Google Scholar] [CrossRef] [PubMed]
- Bossi, F.; Cordoba, E.; Dupré, P.; Mendoza, M.S.; Román, C.S.; León, P. The arabidopsis aba-insensitive (abi) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction ofABI5andSBE2.2 genesduring sugar signaling. Plant J. 2009, 59, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and role of the arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Sponsel, V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef]
- Binenbaum, J.; Weinstain, R.; Shani, E. Gibberellin localization and transport in plants. Trends Plant Sci. 2018, 23, 410–421. [Google Scholar] [CrossRef]
- Kuromori, T.; Seo, M.; Shinozaki, K. ABA Transport and plant water stress responses. Trends Plant Sci. 2018, 23, 513–522. [Google Scholar] [CrossRef]
- Rizza, A.; Jones, A. The makings of a gradient: Spatiotemporal distribution of gibberellins in plant development. Curr. Opin. Plant Boil. 2019, 47, 9–15. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Kamiya, Y.; Sun, T.-P. Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 2001, 28, 443–453. [Google Scholar] [CrossRef]
- Ogawa, M.; Hanada, A.; Yamauchi, Y.; Kuwahara, A.; Kamiya, Y.; Yamaguchi, S. Gibberellin Biosynthesis and Response during Arabidopsis Seed Germination. Plant Cell 2003, 15, 1591–1604. [Google Scholar] [CrossRef]
- Penfield, S.; Rylott, E.L.; Gilday, A.D.; Graham, S.; Larson, T.; Graham, I.A. Reserve mobilization in the arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires phosphoenolpyruvate carboxykinase. Plant Cell 2004, 16, 2705–2718. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Hanada, A.; Kuwahara, A.; Endo, A.; Okamoto, M.; Yamauchi, Y.; North, H.; Marion-Poll, A.; Sun, T.-P.; Koshiba, T.; et al. Regulation of hormone metabolism in arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006, 48, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Piskurewicz, U.; Jikumaru, Y.; Kinoshita, N.; Nambara, E.; Kamiya, Y.; Lopez-Molina, L. The gibberellic acid signaling repressor rgl2 inhibits arabidopsis seed germination by stimulating abscisic acid synthesis and abi5 activity. Plant Cell 2008, 20, 2729–2745. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Andújar, C.; Pluskota, W.; Bassel, G.W.; Asahina, M.; Pupel, P.; Nguyen, T.T.; Takeda-Kamiya, N.; Toubiana, D.; Bai, B.; Górecki, R.; et al. Mechanisms of hormonal regulation of endosperm cap-specific gene expression in tomato seeds. Plant J. 2012, 71, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, B.J.W.; Pearce, S.P.; Van Bolderen-Veldkamp, R.P.; Marshall, A.; Widera, P.; Gilbert, J.; Drost, H.; Bassel, G.W.; Müller, K.; King, J.R.; et al. Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol. 2013, 163, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Rombolá-Caldentey, B.; Rueda-Romero, P.; Iglesias-Fernández, R.; Carbonero, P.; Oñate-Sánchez, L. Arabidopsis DELLA and Two HD-ZIP transcription factors regulate GA Signaling in the Epidermis through the L1 Box cis-Element. Plant Cell 2014, 26, 2905–2919. [Google Scholar] [CrossRef]
- Stamm, P.; Topham, A.; Mukhtar, N.K.; Jackson, M.D.B.; Tomé, D.F.; Beynon, J.; Bassel, G.W. The transcription factor ATHB5 affects GA-mediated plasticity in hypocotyl cell growth during seed germination. Plant Physiol. 2016, 173, 907–917. [Google Scholar] [CrossRef]
- Topham, A.T.; Taylor, R.E.; Yan, D.; Nambara, E.; Johnston, I.G.; Bassel, G.W. Temperature variability is integrated by a spatially embedded decision-making center to break dormancy in Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 2017, 114, 6629–6634. [Google Scholar] [CrossRef]
- Sánchez-Montesino, R.; Bouza-Morcillo, L.; Marquez, J.; Ghita, M.; Duran-Nebreda, S.; Gómez, L.; Holdsworth, M.; Bassel, G.W.; Oñate-Sánchez, L. A regulatory module controlling GA-mediated endosperm cell expansion is critical for seed germination in arabidopsis. Mol. Plant 2019, 12, 71–85. [Google Scholar] [CrossRef]
- Lee, K.P.; Piskurewicz, U.; Turečková, V.; Strnad, M.; Lopez-Molina, L. A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. USA 2010, 107, 19108–19113. [Google Scholar] [CrossRef]
- Kang, J.; Yim, S.; Choi, H.; Kim, A.; Lee, K.P.; Lopez-Molina, L.; Martinoia, E.; Lee, Y. Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 2015, 6, 8113. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. At SWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Hou, B.-H.; LaLonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2011, 335, 207–211. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Regnault, T.; Davière, J.-M.; Achard, P. Long-distance transport of endogenous gibberellins in Arabidopsis. Plant Signal. Behav. 2015, 11, e1110661. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tal, I.; Zhang, Y.; Jørgensen, M.E.; Pisanty, O.; Barbosa, I.C.R.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Olsen, C.E.; Weinstain, R.; et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef]
- David, L.C.; Berquin, P.; Kanno, Y.; Seo, M.; Daniel-Vedele, F.; Ferrario-Méry, S. N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta 2016, 244, 1315–1328. [Google Scholar] [CrossRef]
- Léran, S.; Varala, K.; Boyer, J.C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant. Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef]
- Corratgé-Faillie, C.; Lacombe, B. Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef]
- Huang, N.C.; Liu, K.H.; Lo, H.J.; Tsay, Y.F. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 1999, 11, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Emergence of a new step towards understanding the molecular mechanisms underlying nitrate-regulated gene expression. J. Exp. Bot. 2014, 65, 5589–5600. [Google Scholar] [CrossRef] [PubMed]
- Alboresi, A.; Gestin, C.; Leydecker, M.-T.; Bédu, M.; Meyer, C.; Truong, H.-N. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant, Cell Environ. 2005, 28, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Matakiadis, T.; Alboresi, A.; Jikumaru, Y.; Tatematsu, K.; Pichon, O.; Renou, J.-P.; Kamiya, Y.; Nambara, E.; Truong, H.-N. The arabidopsis abscisic acid catabolic gene cyp707a2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2008, 149, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Easwaran, V.; Chau, V.; Okamoto, M.; Ierullo, M.; Kimura, M.; Endo, A.; Yano, R.; Pasha, A.; Gong, Y.; et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 2016, 7, 13179. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 506. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; Neff, M.M.; Hong, S.-W.; Zhang, H.; Deng, X.-W.; Xiong, L. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. USA 2008, 105, 4495–4500. [Google Scholar] [CrossRef]
- Yu, L.-H.; Wu, J.; Meng, X.; Miao, Z.-Q.; Zhao, P.-X.; Wang, Z.; Xiang, C.-B. Arabidopsis MADS-Box transcription factor AGL21 acts as environmental surveillance of seed germination by regulating ABI5 expression. Mol. Plant 2017, 10, 834–845. [Google Scholar] [CrossRef]
- Yang, B.; Song, Z.; Li, C.; Jiang, J.; Zhou, Y.; Wang, R.; Wang, Q.; Ni, C.; Liang, Q.; Chen, H.; et al. RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 2018, 14, e1007839. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, J.; Gangappa, S.; Hettiarachchi, C.; Lin, F.; Andersson, M.X.; Jiang, Y.; Deng, X.-W.; Holm, M. Convergence of Light and ABA Signaling on the ABI5 Promoter. PLoS Genet. 2014, 10, e1004197. [Google Scholar] [CrossRef]
- Jing, Y.; Lin, R. The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiol. 2015, 169, 371–378. [Google Scholar] [CrossRef]
- Pan, J.; Wang, H.; Hu, Y.; Yu, D. Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination. Plant J. 2018, 95, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, S.-H.; Seo, D.H.; Chung, S.; Kim, S.-W.; Lee, J.-S.; Kim, W.T.; Lee, J.-H. ABA-Hypersensitive BTB/POZ Protein 1 functions as a negative regulator in ABA-mediated inhibition of germination in Arabidopsis. Plant Mol. Boil. 2015, 90, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.-Z.; Chen, Y.; Wang, C.; Kong, Y.-H.; Wu, W.-H.; Chen, Y.-F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression ofABI3, ABI4, andABI5during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, C.; Miao, J.; Lei, Y.; Zhao, D.; Sun, D.; Yang, G.; Huang, J.; Zheng, C. Arabidopsis SAG protein containing the MDN1 domain participates in seed germination and seedling development by negatively regulating ABI3 and ABI5. J. Exp. Bot. 2013, 65, 35–45. [Google Scholar] [CrossRef]
- Shang, Y.; Yan, L.; Liu, Z.-Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.-Q.; Wang, X.-F.; Du, S.-Y.; Jiang, T.; et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef]
- Liu, Z.-Q.; Yan, L.; Wu, Z.; Mei, C.; Lu, K.; Yu, Y.-T.; Liang, S.; Zhang, X.-F.; Wang, X.-F.; Zhang, D.-P. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 2012, 63, 6371–6392. [Google Scholar] [CrossRef]
- Kim, J.H.; Hyun, W.Y.; Nguyen, H.N.; Jeong, C.Y.; Xiong, L.; Hong, S.-W.; Lee, H. AtMyb7, a subgroup 4 R2R3 Myb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ. 2014, 38, 559–571. [Google Scholar] [CrossRef]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC–RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-H.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genome Res. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, X.; Yang, M.; Zhang, M.; Pan, J.; Yu, D. The transcription factor inducer of cbf expression1 interacts with abscisic acid insensitive5 and della proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yu, D. Brassinosteroid insensitive2 interacts with abscisic acid insensitive5 to Mediate the Antagonism of Brassinosteroids to Abscisic Acid during Seed Germination in Arabidopsis. Plant Cell 2014, 26, 4394–4408. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Dou, L.; Gong, Z.; Wang, X.; Liu, X. BES 1 hinders ABSCISIC ACID INSENSITIVE 5 and promotes seed germination in Arabidopsis. New Phytol. 2018, 221, 908–918. [Google Scholar] [CrossRef]
- Steber, C.M. A Role for Brassinosteroids in Germination in Arabidopsis. Plant Physiol. 2001, 125, 763–769. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Ye, T.; Zhao, S.; Liu, Z.; Feng, Y.-Q.; Wu, Y. Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J. 2011, 68, 249–261. [Google Scholar] [CrossRef]
- Guan, C.; Wang, X.; Feng, J.; Hong, S.; Liang, Y.; Ren, B.; Zuo, J. Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of abscisic acid insensitive5 protein in arabidopsis. Plant Physiol. 2014, 164, 1515–1526. [Google Scholar] [CrossRef]
- Chahtane, H.; Nogueira Füller, T.; Allard, P.-M.; Marcourt, L.; Ferreira Queiroz, E.; Shanmugabalaji, V.; Falquet, J.; Wolfender, J.-L.; Lopez-Molina, L. The plant pathogen Pseudomonas aeruginosa triggers a DELLA-dependent seed germination arrest in Arabidopsis. eLife 2018, 7, e37082. [Google Scholar] [CrossRef]
- Kim, N.H.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. Somnus, a CCCH-type zinc finger protein in arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 2008, 20, 1260–1277. [Google Scholar] [CrossRef]
- Park, J.; Lee, N.; Kim, W.; Lim, S.; Choi, G. ABI3 and PIL5 collaboratively activate the expression of somnus by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 2011, 23, 1404–1415. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Park, J.; Lee, N.; Jeong, J.; Toh, S.; Watanabe, A.; Kim, J.; Kang, H.; Kim, N.H.; Kawakami, N.; et al. ABA-Insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of somnus and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 2013, 25, 4863–4878. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Kim, J.; Park, E.; Kim, J.-I.; Kang, C.; Choi, G. PIL5, a Phytochrome-Interacting Basic Helix-Loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.-I.; Choi, G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139. [Google Scholar] [CrossRef]
- Shen, H.; Zhu, L.; Castillon, A.; Majee, M.; Downie, B.; Huq, E. Light-Induced phosphorylation and degradation of the negative regulator Phytochrome-Interacting factor1 from arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 2008, 20, 1586–1602. [Google Scholar] [CrossRef]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef]
- Zhu, L.; Bu, Q.; Xu, X.; Paik, I.; Huang, X.; Hoecker, U.; Deng, X.W.; Huq, E. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 2015, 6, 7245. [Google Scholar] [CrossRef]
- Paik, I.; Chen, F.; Pham, V.N.; Zhu, L.; Kim, J.-I.; Huq, E. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 2019, 10, 4216–4217. [Google Scholar] [CrossRef]
- Dirk, L.M.A.; Kumar, S.; Majee, M.; Downie, A.B. Phytochrome Interacting Factor1 interactions leading to the completion or prolongation of seed germination. Plant Signal. Behav. 2018, 13, e1525999. [Google Scholar] [CrossRef]
- Majee, M.; Kumar, S.; Kathare, P.K.; Wu, S.; Gingerich, D.; Nayak, N.R.; Salaita, L.; Dinkins, R.; Martin, K.; Goodin, M.; et al. KELCH F-BOX protein positively influences Arabidopsis seed germination by targeting PHYTOCHROME-INTERACTING FACTOR1. Proc. Natl. Acad. Sci. USA 2018, 115, E4120–E4129. [Google Scholar] [CrossRef]
- Shi, H.; Zhong, S.; Mo, X.; Liu, N.; Nezames, C.D.; Deng, X.-W. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 2013, 25, 3770–3784. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Mo, X.; Tang, C.; Zhong, S.; Deng, X.-W. Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc. Natl. Acad. Sci. USA 2015, 112, 3817–3822. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.-S.; Sun, T.-P.; Kamiya, Y.; Choi, G. PIL5, a Phytochrome-Interacting bHLH Protein, Regulates Gibberellin Responsiveness by Binding Directly to the GAI and RGA Promoters in Arabidopsis Seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef] [PubMed]
- Gabriele, S.; Rizza, A.; Martone, J.; Circelli, P.; Costantino, P.; Vittorioso, P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 2009, 61, 312–323. [Google Scholar] [CrossRef]
- Boccaccini, A.; Santopolo, S.; Capauto, D.; Lorrai, R.; Minutello, E.; Serino, G.; Costantino, P.; Vittorioso, P. The DOF protein dag1 and the della protein gai cooperate in negatively regulating the atga3ox1 gene. Mol. Plant 2014, 7, 1486–1489. [Google Scholar] [CrossRef]
- Ruta, V.; Longo, C.; Lepri, A.; De Angelis, V.; Occhigrossi, S.; Costantino, P.; Vittorioso, P. The DOF Transcription Factors in Seed and Seedling Development. Plants 2020, 9, 218. [Google Scholar] [CrossRef] [PubMed]
- Huq, E.; Al-Sady, B.; Hudson, M.; Kim, C.; Apel, K.; Quail, P.H. Phytochrome-interacting factor 1 is a critical bhlh regulator of chlorophyll biosynthesis. Science 2004, 305, 1937–1941. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, H.; Park, J.; Kim, W.; Yoo, J.; Lee, N.; Kim, J.; Yoon, T.-Y.; Choi, G. Pif1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of pif1. Plant Cell 2016, 28, 1388–1405. [Google Scholar] [CrossRef]
- Shinomura, T.; Nagatani, A.; Chory, J.; Furuya, M. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome b and secondarily by Phytochrome A. Plant Physiol. 1994, 104, 363–371. [Google Scholar] [CrossRef]
- Seo, M.; Nambara, E.; Choi, G.; Yamaguchi, S. Interaction of light and hormone signals in germinating seeds. Plant Mol. Boil. 2008, 69, 463–472. [Google Scholar] [CrossRef]
- Lee, K.P.; Piskurewicz, U.; Turečková, V.; Carat, S.; Chappuis, R.; Strnad, M.; Fankhauser, C.; Lopez-Molina, L. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 2012, 26, 1984–1996. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, S.E.; Auge, G.; Sánchez, R.A.; Botto, J. Transcriptional programs related to phytochrome a function in arabidopsis seed germination. Mol. Plant 2013, 6, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Zeljković, S. Ćavar; Piskurewicz, U.; Megies, C.; Tarkowski, P.; Lopez-Molina, L. Polyamine uptake transporter 2 (put2) and decaying seeds enhance phyA-mediated germination by overcoming PIF1 repression of germination. PLoS Genet. 2019, 15, e1008292. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Park, E.; Song, K.; Bae, G.; Choi, G. Phytochrome interacting factor8 inhibits phytochrome a-mediated far-red light responses in arabidopsis. Plant Cell 2019, 32, 186–205. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Josse, E.-M.; Halliday, K. A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol. Boil. 2009, 73, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Hennig, L.; Stoddart, W.M.; Dieterle, M.; Whitelam, G.C.; Schäfer, E. Phytochrome E Controls Light-Induced Germination of Arabidopsis. Plant Physiol. 2002, 128, 194–200. [Google Scholar] [CrossRef]
- Arana, M.; Sanchez-Lamas, M.; Strasser, B.; Ibarra, S.E.; Cerdan, P.D.; Botto, J.; Sanchez, R.A. Functional diversity of phytochrome family in the control of light and gibberellin-mediated germination in Arabidopsis. Plant, Cell Environ. 2014, 37, 2014–2023. [Google Scholar] [CrossRef]
- Martel, C.; Blair, L.K.; Donohue, K. PHYD prevents secondary dormancy establishment of seeds exposed to high temperature and is associated with lower PIL5 accumulation. J. Exp. Bot. 2018, 69, 3157–3169. [Google Scholar] [CrossRef]
- Clack, T.; Shokry, A.; Moffet, M.; Liu, P.; Faul, M.; Sharrock, R. Obligate heterodimerization of arabidopsis phytochromes c and e and interaction with the pif3 basic helix-loop-helix transcription factor. Plant Cell 2009, 21, 786–799. [Google Scholar] [CrossRef]
- Hu, W.; Franklin, K.A.; Sharrock, R.A.; Jones, M.A.; Harmer, S.; Lagarias, J.C. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc. Natl. Acad. Sci. USA 2013, 110, 1542–1547. [Google Scholar] [CrossRef]
- Penfield, S.; Josse, E.-M.; Kannangara, R.; Gilday, A.D.; Halliday, K.; Graham, I.A. Cold and light control seed germination through the bhlh transcription factor spatula. Curr. Boil. 2005, 15, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Barros-Galvão, T.; Cole, A.F.; Gilday, A.D.; He, Z.; Li, Y.; Harvey, D.; Larson, T.R.; Graham, I.A. MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, 8442–8447. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Gan, Y.; Penfield, S.; Gilday, A.D.; Dave, A.; He, Z.; Josse, E.-M.; Choi, G.; Halliday, K.; Graham, I.A. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor spatula. Proc. Natl. Acad. Sci. USA 2013, 110, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. Mother of ft and tfl1 regulates seed germination through a negative feedback loop modulating aba signaling in arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Arc, E.; Galland, M.; Godin, B.; Cueff, G.; Rajjou, L. Nitric oxide implication in the control of seed dormancy and germination. Front. Plant Sci. 2013, 4, 346. [Google Scholar] [CrossRef]
- Sanz, L.; Albertos, P.; Mateos, I.; Sánchez-Vicente, I.; Lechón, T.; Fernández-Marcos, M.; Lorenzo, O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015, 66, 2857–2868. [Google Scholar] [CrossRef]
- Li, R.; Jia, Y.; Yu, L.; Yang, W.; Chen, Z.; Chen, H.; Hu, X.-Y. Nitric oxide promotes light-initiated seed germination by repressing PIF1 expression and stabilizing HFR1. Plant Physiol. Biochem. 2018, 123, 204–212. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, L.; Ye, N.; Liu, R.; Jia, W.; Zhang, J.-H. Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 2009, 183, 1030–1042. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Isa, N.M.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; La Rosa, N.M.-D.; Conde, J.V.; Correia, C.S.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015, 6, 8669. [Google Scholar] [CrossRef]
- Liu, H.; Stone, S.L. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 2010, 22, 2630–2641. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, J.-K.; Lang, Z. Nitric oxide suppresses the inhibitory effect of abscisic acid on seed germination by S-nitrosylation of SnRK2 proteins. Plant Signal. Behav. 2015, 10, e1031939. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Koornneef, M.; Soppe, W.J.J. The absence of histone h2b monoubiquitination in the arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 2007, 19, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Weake, V.; Workman, J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell 2008, 29, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Grasser, M.; Kane, C.M.; Merkle, T.; Melzer, M.; Emmersen, J.; Grasser, K.D. Transcript elongation factor tfiis is involved in arabidopsis seed dormancy. J. Mol. Boil. 2009, 386, 598–611. [Google Scholar] [CrossRef]
- Liu, Y.; Geyer, R.; Van Zanten, M.; Carles, A.; Li, Y.; Hörold, A.; Van Nocker, S.; Soppe, W.J.J. Identification of the Arabidopsis reduced Dormancy 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS ONE 2011, 6, e22241. [Google Scholar] [CrossRef]
- Mortensen, S.A.; Grasser, K.D. The seed dormancy defect ofArabidopsismutants lacking the transcript elongation factor TFIIS is caused by reduced expression of theDOG1gene. FEBS Lett. 2013, 588, 47–51. [Google Scholar] [CrossRef]
- Lee, N.; Kang, H.; Lee, D.; Choi, G. A histone methyltransferase inhibits seed germination by increasing PIF1 mRNA expression in imbibed seeds. Plant J. 2014, 78, 282–293. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, H.; Sun, Y.; Li, X.; Chen, F.; Carles, A.; Li, Y.; Ding, M.; Zhang, C.; Deng, X.; et al. Arabidopsis paired amphipathic helix proteins snl1 and snl2 redundantly regulate primary seed dormancy via abscisic acid–ethylene antagonism mediated by histone deacetylation. Plant Cell 2013, 25, 149–166. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, F.; Li, X.; Cao, H.; Ding, M.; Zhang, C.; Zuo, J.; Xu, C.; Xu, J.; Deng, X.; et al. Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nat. Commun. 2016, 7, 13412. [Google Scholar] [CrossRef]
- Piñeiro, M.; Gómez-Mena, C.; Schaffer, R.; Martínez, M.; Ángeles, L.; Coupland, G. EARLY BOLTING IN SHORT DAYS Is Related to Chromatin Remodeling Factors and Regulates Flowering in Arabidopsis by Repressing FT. Plant Cell 2003, 15, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Narro-Diego, L.; López-González, L.; Jarillo, J.A.; Piñeiro, M. The PHD-containing protein EARLY BOLTING IN SHORT DAYS regulates seed dormancy in Arabidopsis. Plant, Cell Environ. 2017, 40, 2393–2405. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Chen, C.-Y.; Zhao, M.; Zhao, L.; Duan, X.; Duan, J.; Wu, K.; Liu, X. Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 2017, 45, 7137–7150. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Golz, J.F. Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signal. Behav. 2012, 7, 86–92. [Google Scholar] [CrossRef]
- Lee, N.; Park, J.; Kim, K.; Choi, G. The Transcriptional Coregulator LEUNIG_HOMOLOG Inhibits Light-Dependent Seed Germination in Arabidopsis. Plant Cell 2015, 27, 2301–2313. [Google Scholar] [CrossRef]
- van Zanten, M.; Zöll, C.; Wang, Z.; Philipp, C.; Carles, A.; Li, Y.; Kornet, N.G.; Liu, Y.; Soppe, W.J.J. HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. Plant J. 2014, 80, 475–488. [Google Scholar] [CrossRef]
- Tanaka, M.; Kikuchi, A.; Kamada, H. The arabidopsis histone deacetylases hda6 and hda19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2007, 146, 149–161. [Google Scholar] [CrossRef]
- Molitor, A.M.; Bu, Z.; Yu, Y.; Shen, W. Arabidopsis al phd-prc1 complexes promote seed germination through h3k4me3-to-h3k27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014, 10, e1004091. [Google Scholar] [CrossRef]
- Xiao, J.; Jin, R.; Wagner, D. Developmental transitions: Integrating environmental cues with hormonal signaling in the chromatin landscape in plants. Genome Boil. 2017, 18, 88. [Google Scholar] [CrossRef]
- Tang, X.; Lim, M.-H.; Pelletier, J.; Tang, M.; Nguyen, V.; Keller, W.A.; Tsang, E.W.T.; Wang, A.; Rothstein, S.J.; Harada, J.J.; et al. Synergistic repression of the embryonic programme by SET domain group 8 and EMBRYONIC FLOWER 2 in Arabidopsis seedlings. J. Exp. Bot. 2011, 63, 1391–1404. [Google Scholar] [CrossRef]
- Feng, J.; Chen, D.; Berr, A.; Shen, W. ZRF1 chromatin regulators have polycomb silencing and independent roles in development. Plant Physiol. 2016, 172, 1746–1759. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Ji, R.; He, C.; Peng, T.; Zhang, M.; Duan, J.; Xiong, C.; Liu, X. Arabidopsis histone methyltransferase suvh5 is a positive regulator of light-mediated seed germination. Front. Plant Sci. 2019, 10, 841. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Yang, W.; He, Y.; Amasino, R.M. Arabidopsis relatives of the human lysine-specific demethylase1 repress the expression of fwa and flowering locus c and thus promote the floral transition. Plant Cell 2007, 19, 2975–2987. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, S.; Liu, X.; Wu, K. Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating delay of germination 1 and ABA signaling-related genes. Front. Plant Sci. 2015, 6, 159. [Google Scholar] [CrossRef] [PubMed]
- Yano, R.; Takebayashi, Y.; Nambara, E.; Kamiya, Y.; Seo, M. Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana. Plant J. 2013, 74, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Cho, H.; Bae, W.; Hwang, I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014, 5, 4138. [Google Scholar] [CrossRef]
- Gao, M.-J.; Li, X.; Huang, J.; Gropp, G.M.; Gjetvaj, B.; Lindsay, D.L.; Wei, S.; Coutu, C.; Chen, Z.; Wan, X.-C.; et al. SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nat. Commun. 2015, 6, 7243. [Google Scholar] [CrossRef]
- Chhun, T.; Chong, S.Y.; Park, B.S.; Wong, E.C.C.; Yin, J.-L.; Kim, M.; Chua, N.-H. HSI2 Repressor Recruits MED13 and HDA6 to Down-Regulate Seed Maturation Gene Expression Directly During Arabidopsis Early Seedling Growth. Plant Cell Physiol. 2016, 57, 1689–1706. [Google Scholar] [CrossRef]
- Bouyer, D.; Roudier, F.; Heese, M.; Andersen, E.D.; Gey, D.; Nowack, M.K.; Goodrich, J.; Renou, J.-P.; Grini, P.; Colot, V.; et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011, 7, e1002014. [Google Scholar] [CrossRef]
- Boccaccini, A.; Lorrai, R.; Ruta, V.; Frey, A.; Mercey-Boutet, S.; Marion-Poll, A.; Tarkowská, D.; Strnad, M.; Costantino, P.; Vittorioso, P. The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. BMC Plant Boil. 2016, 16, 198. [Google Scholar] [CrossRef]
- Chen, X.; Lu, L.; Mayer, K.; Scalf, M.; Qian, S.; Lomax, A.; Smith, L.M.; Zhong, X. Powerdress interacts with histone deacetylase 9 to promote aging in Arabidopsis. eLife 2016, 5, 6832. [Google Scholar] [CrossRef]
- Yang, W.; Chen, Z.; Huang, Y.; Chang, G.; Li, P.; Wei, J.; Yuan, X.; Huang, J.; Hu, X.-Y. Powerdress as the novel regulator enhances Arabidopsis seeds germination tolerance to high temperature stress by histone modification of SOM locus. Plant Sci. 2019, 284, 91–98. [Google Scholar] [CrossRef]
- Liu, K.; Xu, S.; Xuan, W.; Ling, T.; Cao, Z.; Huang, B.; Sun, Y.; Fang, L.; Liu, Z.; Zhao, N.; et al. Carbon monoxide counteracts the inhibition of seed germination and alleviates oxidative damage caused by salt stress in Oryza sativa. Plant Sci. 2007, 172, 544–555. [Google Scholar] [CrossRef]
- Wang, M.; Liao, W. Carbon Monoxide as a Signaling Molecule in Plants. Front. Plant Sci. 2016, 7, 259. [Google Scholar] [CrossRef]
- Jia, Y.; Li, R.; Yang, W.; Chen, Z.; Hu, X.-Y. Carbon monoxide signal regulates light-initiated seed germination by suppressing SOM expression. Plant Sci. 2018, 272, 88–98. [Google Scholar] [CrossRef]
- Cho, J.; Ryu, J.-Y.; Jeong, Y.-M.; Park, J.; Song, J.-J.; Amasino, R.M.; Noh, B.; Noh, Y.-S. Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 2012, 22, 736–748. [Google Scholar] [CrossRef]
- Han, S.-K.; Wu, M.-F.; Cui, S.; Wagner, D. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015, 83, 62–77. [Google Scholar] [CrossRef]
- Han, S.K.; Sang, Y.; Rodrigues, A.; Biol, F.; Wu, M.F.; Rodriguez, P.L.; Wagner, D. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 2012, 24, 4892–4906. [Google Scholar] [CrossRef]
- Li, H.-C.; Chuang, K.; Henderson, J.T.; Rider, S.D.; Bai, Y.; Zhang, H.; Fountain, M.; Gerber, J.; Ogas, J. PICKLE acts during germination to repress expression of embryonic traits. Plant J. 2005, 44, 1010–1022. [Google Scholar] [CrossRef]
- Archacki, R.; Buszewicz, D.; Sarnowski, T.J.; Sarnowska, E.; Rolicka, A.; Tohge, T.; Fernie, A.R.; Jikumaru, Y.; Kotliński, M.; Iwanicka-Nowicka, R.; et al. BRAHMA atpase of the swi/snf chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in arabidopsis. PLoS ONE 2013, 8, e58588. [Google Scholar] [CrossRef]
- Park, J.; Oh, D.-H.; Dassanayake, M.; Nguyen, K.T.; Ogas, J.; Choi, G.; Sun, T.-P. Gibberellin signaling requires chromatin remodeler pickle to promote vegetative growth and phase transitions. Plant Physiol. 2017, 173, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Leeggangers, H.A.C.F.; Folta, A.; Muras, A.; Nap, J.-P.; Mlynárová, L. Reduced seed germination in Arabidopsis over-expressing SWI/SNF2 ATPase genes. Physiol. Plant. 2014, 153, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Boil. 2017, 18, 171. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Gouil, Q.; Secco, D.; Srivastava, A.; Karpievitch, Y.V.; Liew, L.C.; Lister, R.; Lewsey, M.G.; Whelan, J. Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Boil. 2017, 18, 172. [Google Scholar] [CrossRef] [PubMed]
- Willmann, M.R.; Mehalick, A.J.; Packer, R.L.; Jenik, P. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 2011, 155, 1871–1884. [Google Scholar] [CrossRef] [PubMed]
- Das, S.S.; Karmakar, P.; Nandi, A.K.; Sanan-Mishra, N. Small RNA mediated regulation of seed germination. Front. Plant Sci. 2015, 6, 798. [Google Scholar] [CrossRef]
- Liu, Y.; El-Kassaby, Y.A. Regulatory crosstalk between microRNAs and hormone signalling cascades controls the variation on seed dormancy phenotype at Arabidopsis thaliana seed set. Plant Cell Rep. 2017, 36, 705–717. [Google Scholar] [CrossRef]
- Nonogaki, H. MicroRNA Gene Regulation cascades during early stages of plant development. Plant Cell Physiol. 2010, 51, 1840–1846. [Google Scholar] [CrossRef]
- Li, J.-F.; Chung, H.S.; Niu, Y.; Bush, J.; McCormack, M.; Sheen, J. Comprehensive protein-based artificial microrna screens for effective gene silencing in plants. Plant Cell 2013, 25, 1507–1522. [Google Scholar] [CrossRef]
- Das, S.S.; Yadav, S.; Singh, A.; Gautam, V.; Sarkar, A.K.; Nandi, A.K.; Karmakar, P.; Majee, M.; Sanan-Mishra, N. Expression dynamics of miRNAs and their targets in seed germination conditions reveals miRNA-ta-siRNA crosstalk as regulator of seed germination. Sci. Rep. 2018, 8, 1233. [Google Scholar] [CrossRef]
- Liu, P.-P.; A Montgomery, T.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef]
- Reyes, J.L.; Chua, N.-H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007, 49, 592–606. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.J.; Park, B.S.; Wang, H.; Liu, J.; Jang, I.-C.; Chua, N.-H. Light-inducible mir163 targets pxmt1 transcripts to promote seed germination and primary root elongation in arabidopsis. Plant Physiol. 2016, 170, 1772–1782. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Bian, S.; Tang, M.; Lu, Q.; Li, S.; Liu, X.; Tian, G.; Nguyen, V.; Tsang, E.W.T.; Wang, A.; et al. MicroRNA–mediated repression of the seed maturation program during vegetative development in arabidopsis. PLoS Genet. 2012, 8, e1003091. [Google Scholar] [CrossRef]
- A Mosher, R.; Melnyk, C.W.; Kelly, K.A.; Dunn, R.M.; Studholme, D.J.; Baulcombe, D.C. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 2009, 460, 283–286. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, C.; Baulcombe, D.C.; Chen, Z.J. Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 2012, 109, 5529–5534. [Google Scholar] [CrossRef]
- Erdmann, R.; Satyaki, P.R.; Klosinska, M.; Gehring, M. A small RNA pathway mediates allelic dosage in endosperm. Cell Rep. 2017, 21, 3364–3372. [Google Scholar] [CrossRef]
- Kirkbride, R.C.; Lu, J.; Zhang, C.; A Mosher, R.; Baulcombe, D.C.; Chen, Z.J. Maternal small RNAs mediate spatial-temporal regulation of gene expression, imprinting, and seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 2761–2766. [Google Scholar] [CrossRef]
- Axtell, M.J. Classification and comparison of small rnas from plants. Annu. Rev. Plant Boil. 2013, 64, 137–159. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005, 41, 697–709. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Cadman, C.S.C.; Toorop, P.E.; Lynn, J.R.; Hilhorst, H.W.M. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 2007, 51, 60–78. [Google Scholar] [CrossRef] [PubMed]
- Carrera, E.; Holman, T.; Medhurst, A.; Dietrich, D.; Footitt, S.; Theodoulou, F.; Holdsworth, M. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2007, 53, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Nambara, E. Stored and neosynthesized mRNA in Arabidopsis seeds: Effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol. Boil. 2010, 73, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Howell, K.; Millar, A.H.; O’Toole, N.; Small, I.D.; Whelan, J. Genome-wide analysis of mrna decay rates and their determinants in Arabidopsis thaliana. Plant Cell 2007, 19, 3418–3436. [Google Scholar] [CrossRef]
- Basbouss-Serhal, I.; Pateyron, S.; Cochet, F.; Leymarie, J.; Bailly, C. 5′ to 3′ mRNA decay contributes to the regulation of Arabidopsis seed germination by dormancy. Plant Physiol. 2017, 173, 1709. [Google Scholar] [CrossRef]
- Nelson, S.K.; Ariizumi, T.; Steber, C.M. Biology in the dry seed: Transcriptome changes associated with dry seed dormancy and dormancy loss in the arabidopsis GA-insensitive sleepy1-2 Mutant. Front. Plant Sci. 2017, 8, 2158. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, B.; Jia, J.; Yan, C.; Habaike, A.; Han, Y. RRP41L, a putative core subunit of the exosome, plays an important role in seed germination and early seedling growth in arabidopsis. Plant Physiol. 2012, 161, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Wawer, I.; Golisz, A.; Sulkowska, A.; Kawa, D.; Kulik, A.; Kufel, J. mRNA decapping and 5′-3′ decay contribute to the regulation of aba signaling in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 312. [Google Scholar] [CrossRef]
- Bazin, J.; Langlade, N.; Vincourt, P.; Arribat, S.; Balzergue, S.; El-Maarouf-Bouteau, H.; Bailly, C. Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 2011, 23, 2196–2208. [Google Scholar] [CrossRef]
- Layat, E.; Leymarie, J.; El-Maarouf-Bouteau, H.; Caius, J.; Langlade, N.; Bailly, C. Translatome profiling in dormant and nondormant sunflower (Helianthus annuus) seeds highlights post-transcriptional regulation of germination. New Phytol. 2014, 204, 864–872. [Google Scholar] [CrossRef]
- Basbouss-Serhal, I.; Soubigou-Taconnat, L.; Bailly, C.; Leymarie, J. Germination potential of dormant and nondormant arabidopsis seeds is driven by distinct recruitment of messenger rnas to polysomes. Plant Physiol. 2015, 168, 1049–1065. [Google Scholar] [CrossRef]
- Bai, B.; Peviani, A.; Van Der Horst, S.; Gamm, M.; Snel, B.; Bentsink, L.; Hanson, J. Extensive translational regulation during seed germination revealed by polysomal profiling. New Phytol. 2016, 214, 233–244. [Google Scholar] [CrossRef]
- Bai, B.; Novak, O.; Ljung, K.; Hanson, J.; Bentsink, L. Combined transcriptome and translatome analyses reveal a role for tryptophan-dependent auxin biosynthesis in the control of DOG1 -dependent seed dormancy. New Phytol. 2017, 217, 1077–1085. [Google Scholar] [CrossRef]
- Bai, B.; Van Der Horst, S.; Cordewener, J.; America, A.; Hanson, J.; Bentsink, L. Seed-Stored mRNAs that Are Specifically Associated to Monosomes Are Translationally Regulated during Germination. Plant Physiol. 2019, 182, 378–392. [Google Scholar] [CrossRef]
- Ingolia, N.T. Ribosome footprint profiling of translation throughout the genome. Cell 2016, 165, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Yang, P.; Sakata, K.; Komatsu, S. Quantitative proteomics reveals the role of protein phosphorylation in rice embryos during early stages of germination. J. Proteome Res. 2014, 13, 1766–1782. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Wang, K.; Yang, P. Gel-Based comparative phosphoproteomic analysis on rice embryo during germination. Plant Cell Physiol. 2014, 55, 1376–1394. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yin, X.; Sakata, K.; Yang, P.; Komatsu, S. Proteomic analysis of phosphoproteins in the rice nucleus during the early stage of seed germination. J. Proteome Res. 2015, 14, 2884–2896. [Google Scholar] [CrossRef]
- Dai, M.; Xue, Q.; McCray, T.; Margavage, K.; Chen, F.; Lee, J.-H.; Nezames, C.D.; Guo, L.; Terzaghi, W.; Wan, J.; et al. The PP6 phosphatase regulates abi5 phosphorylation and abscisic acid signaling in arabidopsis. Plant Cell 2012, 25, 517–534. [Google Scholar] [CrossRef]
- Lee, S.-J.; Lee, M.H.; Kim, J.-I.; Kim, S.Y. Arabidopsis putative map kinase kinase kinases raf10 and raf11 are positive regulators of seed dormancy and aba response. Plant Cell Physiol. 2014, 56, 84–97. [Google Scholar] [CrossRef]
- Nguyen, Q.T.C.; Lee, S.-J.; Choi, S.-W.; Na, Y.-J.; Song, M.-R.; Hoang, Q.T.N.; Sim, S.Y.; Kim, M.-S.; Kim, J.-I.; Soh, M.-S.; et al. Arabidopsis raf-like kinase raf10 is a regulatory component of core aba signaling. Mol. Cells 2019, 42, 646–660. [Google Scholar] [PubMed]
- Hu, R.; Zhu, Y.; Shen, G.; Zhang, H. TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiol. 2013, 164, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, K.; Ramadan, A.; Arimura, G.-I.; Imai, K.; Tomii, K.; Shinozaki, K.; Sawasaki, T. Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signalling by preventing GID1 degradation. Nat. Commun. 2017, 8, 1004. [Google Scholar] [CrossRef]
- Qin, Q.; Wang, W.; Guo, X.; Yue, J.; Huang, Y.; Xu, X.; Li, J.; Hou, S. Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet. 2014, 10, e1004464. [Google Scholar] [CrossRef]
- Nguyen, X.C.; Hoang, M.H.T.; Kim, H.S.; Lee, K.; Liu, X.-M.; Kim, S.H.; Bahk, S.; Park, H.C.; Chung, W.S. Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochem. Biophys. Res. Commun. 2012, 423, 703–708. [Google Scholar] [CrossRef]
- Sasaki, A.; Itoh, H.; Gomi, K.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Jeong, N.-H.; An, G.; Kitano, H.; Ashikari, M.; et al. Accumulation of phosphorylated repressor for gibberellin signaling in an f-box mutant. Science 2003, 299, 1896–1898. [Google Scholar] [CrossRef]
- Zhang, X.; Garreton, V.; Chua, N.-H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genome Res. 2005, 19, 1532–1543. [Google Scholar] [CrossRef]
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. KEEP ON GOING, a RING E3 ligase essential for arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 2006, 18, 3415–3428. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, W.; Wang, X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2016, 15, 4–14. [Google Scholar] [CrossRef]
- He, D.; Li, M.; Damaris, R.N.; Bu, C.; Xue, J.; Yang, P.; Chen, B. Quantitative ubiquitylomics approach for characterizing the dynamic change and extensive modulation of ubiquitylation in rice seed germination. Plant J. 2020, 101, 1430–1447. [Google Scholar] [CrossRef]
- Miura, K.; Lee, J.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Hasegawa, P.M. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 5418–5423. [Google Scholar] [CrossRef]
- Liu, H.; Stone, S.L. Regulation of ABI5 turnover by reversible post-translational modifications. Plant Signal. Behav. 2014, 9, 27577. [Google Scholar] [CrossRef]
- Zheng, Y.; Schumaker, K.S.; Guo, Y. Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 12822–12827. [Google Scholar] [CrossRef]
- Zentella, R.; Sui, N.; Barnhill, B.; Hsieh, W.-P.; Hu, J.; Shabanowitz, J.; Boyce, M.; E Olszewski, N.; Zhou, P.; Hunt, N.F.; et al. The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Methods 2017, 13, 479–485. [Google Scholar] [CrossRef]
- Zentella, R.; Hu, J.; Hsieh, W.-P.; Matsumoto, P.A.; Dawdy, A.; Barnhill, B.; Oldenhof, H.; Hartweck, L.M.; Maitra, S.; Thomas, S.G.; et al. O-GlcNAcylation of master growth repressor della by secret agent modulates multiple signaling pathways in Arabidopsis. Genes Dev. 2016, 30, 164–176. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; El-Esawi, M. Seed dormancy: The Complex Process Regulated by Abscisic Acid, Gibberellins, and Other Phytohormones that Makes Seed Germination Work. In Phytohormones—Signaling Mechanisms and Crosstalk in Plant Development and Stress Responses; El-Esawi, M., Ed.; IntechOpen: London, UK, 2017. [Google Scholar]
- Tuan, P.A.; Kumar, R.; Rehal, P.K.; Toora, P.K.; Ayele, B.T. Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 2018, 9, 668. [Google Scholar] [CrossRef]
- Rodríguez, M.V.; Barrero, J.; Corbineau, F.; Gubler, F.; Benech-Arnold, R.L. Dormancy in cereals (not too much, not so little): About the mechanisms behind this trait. Seed Sci. Res. 2015, 25, 99–119. [Google Scholar] [CrossRef]
- Shu, K.; Meng, Y.; Shuai, H.; Liu, W.; Du, J.; Liu, J.; Yang, W. Dormancy and germination: How does the crop seed decide? Plant Boil. 2015, 17, 1104–1112. [Google Scholar] [CrossRef]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.-Q.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.-D.; Xie, Q.; He, Z. Two Faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Yamagishi, K.; Tatematsu, K.; Yano, R.; Preston, J.; Kitamura, S.; Takahashi, H.; McCourt, P.; Kamiya, Y.; Nambara, E. CHOTTO1, a double ap2 domain protein of arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant Cell Physiol. 2008, 50, 330–340. [Google Scholar] [CrossRef]
- Yano, R.; Kanno, Y.; Jikumaru, Y.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Chotto1, a putative double apetala2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in arabidopsis. Plant Physiol. 2009, 151, 641–654. [Google Scholar] [CrossRef]
- Gil Lee, H.; Lee, K.; Seo, P.J. The arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Boil. 2015, 87, 371–381. [Google Scholar] [CrossRef]
- Rueda-Romero, P.; Sicilia, C.B.; Gómez-Cadenas, A.; Carbonero, P.; Oñate-Sánchez, L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 2011, 63, 1937–1949. [Google Scholar] [CrossRef]
- Ravindran, P.; Verma, V.; Stamm, P.; Kumar, P. A novel rgl2–dof6 complex contributes to primary seed dormancy in arabidopsis thaliana by regulating a gata transcription factor. Mol. Plant 2017, 10, 1307–1320. [Google Scholar] [CrossRef]
- Cao, H.; Han, Y.; Li, J.; Ding, M.; Li, Y.; Li, X.; Chen, F.; Soppe, W.J.J.; Liu, Y. Arabidopsis thaliana seed dormancy 4-like regulates dormancy and germination by mediating the gibberellin pathway. J. Exp. Bot. 2019, 71, 919–933. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY41 controls Arabidopsis seed dormancy via direct regulation ofABI3transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef]
- Penfield, S.; Hall, A. A Role for Multiple Circadian Clock Genes in the response to signals that break seed dormancy in arabidopsis. Plant Cell 2009, 21, 1722–1732. [Google Scholar] [CrossRef]
- Jiang, Z.; Xu, G.; Jing, Y.; Tang, W.; Lin, R. Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 2016, 7, 12377. [Google Scholar] [CrossRef]
- Yang, L.; Jiang, Z.; Liu, S.; Lin, R. Interplay between reveille1 and rga-like2 regulates seed dormancy and germination in arabidopsis. New Phytol. 2019, 225, 1593–1605. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Bentsink, L.; Hanhart, C.J.; Vries, H.B.-D.; Koornneef, M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 2003, 164, 711–729. [Google Scholar]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef]
- Bentsink, L.; Hanson, J.; Hanhart, C.J.; Vries, H.B.-D.; Coltrane, C.; Keizer, P.; El-Lithy, M.; Alonso-Blanco, C.; De Andres, M.T.; Reymond, M.; et al. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 4264–4269. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed dormancy and germination—emerging mechanisms and new hypotheses. Front. Plant Sci. 2014, 5, 233. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed biology updates—Highlights and new discoveries in seed dormancy and dermination research. Front. Plant. Sci. 2017, 8, 524. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed germination and dormancy: The classic story, new puzzles, and evolution. J. Integr. Plant Boil. 2019, 61, 541–563. [Google Scholar] [CrossRef]
- Dolata, J.; Guo, Y.; Kołowerzo, A.; Smolinski, D.J.; Brzyżek, G.; Jarmołowski, A.; Swiezewski, S. NTR 1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 2015, 34, 544–558. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Bartsch, M.; Ding, J.; Soppe, W.J. Seed dormancy in arabidopsis requires self-binding ability of dog1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genet. 2015, 11, e1005737. [Google Scholar] [CrossRef]
- Cyrek, M.; Fedak, H.; Ciesielski, A.; Guo, Y.; Sliwa, A.; Brzezniak, L.; Krzyczmonik, K.; Pietras, Z.; Kaczanowski, S.; Liu, F.; et al. Seed dormancy in arabidopsis is controlled by alternative polyadenylation of dog1. Plant Physiol. 2015, 170, 947–955. [Google Scholar] [CrossRef]
- Fedak, H.; Palusinska, M.; Krzyczmonik, K.; Brzezniak, L.; Yatusevich, R.; Pietras, Z.; Kaczanowski, S.; Swiezewski, S. Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci. USA 2016, 113, E7846–E7855. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Bartsch, M.; Xiang, Y.; Miatton, E.; Pellengahr, S.; Yano, R.; Seo, M.; Soppe, W.J. The time required for dormancy release in Arabidopsis is determined by delay of germination protein levels in freshly harvested seeds. Plant Cell 2012, 24, 2826–2838. [Google Scholar] [CrossRef]
- Dekkers, B.J.W.; He, H.; Hanson, J.; Willems, L.A.; Jamar, D.C.; Cueff, G.; Rajjou, L.; Hilhorst, H.W.M.; Bentsink, L. The arabidopsisdelay of germination 1 gene affectsabscisic acid insensitive 5 (abi5)expression and genetically interacts withabi3during Arabidopsis seed development. Plant J. 2016, 85, 451–465. [Google Scholar] [CrossRef]
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, I.J.R.; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 2132. [Google Scholar] [CrossRef]
- Nishimura, N.; Yoshida, T.; Murayama, M.; Asami, T.; Shinozaki, K.; Hirayama, T. Isolation and Characterization of Novel Mutants Affecting the Abscisic Acid Sensitivity of Arabidopsis Germination and Seedling Growth. Plant Cell Physiol. 2004, 45, 1485–1499. [Google Scholar] [CrossRef]
- Nishimura, N.; Yoshida, T.; Kitahata, N.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007, 50, 935–949. [Google Scholar] [CrossRef]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive germination3 encodes a protein phosphatase 2c (atpp2ca) that strongly regulates abscisic acid signaling during germination among arabidopsis protein phosphatase 2cs. Plant Physiol. 2005, 140, 115–126. [Google Scholar] [CrossRef]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Rodrigues, A.; Pizzio, G.; Rodriguez, P.L. Selective inhibition of clade a phosphatases type 2c by pyr/pyl/rcar abscisic acid receptors. Plant Physiol. 2011, 158, 970–980. [Google Scholar] [CrossRef]
- Huo, H.; Wei, S.; Bradford, K.J. DELAY OF GERMINATION1(DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 2016, 113, E2199–E2206. [Google Scholar] [CrossRef]
- Chiang, G.C.K.; Bartsch, M.; Barua, D.; Nakabayashi, K.; Debieu, M.; Kronholm, I.; Koornneef, M.; Soppe, W.J.; Donohue, K.; De Meaux, J. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol. Ecol. 2011, 20, 3336–3349. [Google Scholar] [CrossRef]
- Kendall, S.L.; Hellwege, A.; Marriot, P.; Whalley, C.; Graham, I.A.; Penfield, S. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 2011, 23, 2568–2580. [Google Scholar] [CrossRef]
- Murphey, M.; Kovach, K.; Elnacash, T.; He, H.; Bentsink, L.; Donohue, K.; Elnaccash, T. DOG1-imposed dormancy mediates germination responses to temperature cues. Environ. Exp. Bot. 2015, 112, 33–43. [Google Scholar] [CrossRef]
- He, H.; Willems, L.; Batushansky, A.; Fait, A.; Hanson, J.; Nijveen, H.; Hilhorst, H.W.M.; Bentsink, L. Effects of parental temperature and nitrate on seed performance are reflected by partly overlapping genetic and metabolic pathways. Plant Cell Physiol. 2016, 57, 473–487. [Google Scholar] [CrossRef]
- Yatusevich, R.; Fedak, H.; Ciesielski, A.; Krzyczmonik, K.; Kulik, A.; Dobrowolska, G.; Swiezewski, S. Antisense transcription represses Arabidopsis seed dormancy QTL DOG 1 to regulate drought tolerance. EMBO Rep. 2017, 18, 2186–2196. [Google Scholar] [CrossRef]
- Teng, S.; Rognoni, S.; Bentsink, L.; Smeekens, S. The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA-mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. Plant J. 2008, 55, 372–381. [Google Scholar] [CrossRef]
- Xiang, Y.; Nakabayashi, K.; Ding, J.; He, F.; Bentsink, L.; Soppe, W.J. Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 2014, 26, 4362–4375. [Google Scholar] [CrossRef]
- Xiang, Y.; Song, B.; Née, G.; Krämer, K.; Finkemeier, I.; Soppe, W.J. Sequence polymorphisms at the reduced Dormancy5 pseudophosphatase underlie natural variation in arabidopsis dormancy. Plant Physiol. 2016, 171, 2659–2670. [Google Scholar] [CrossRef]
- Amiguet-Vercher, A.; Santuari, L.; Gonzalez-Guzman, M.; Depuydt, S.; Rodriguez, P.L.; Hardtke, C.S. TheIBOgermination quantitative trait locus encodes a phosphatase 2C-related variant with a nonsynonymous amino acid change that interferes with abscisic acid signaling. New Phytol. 2014, 205, 1076–1082. [Google Scholar] [CrossRef]
- Linkies, A.; Leubner-Metzger, G. Beyond gibberellins and abscisic acid: How ethylene and jasmonates control seed germination. Plant Cell Rep. 2011, 31, 253–270. [Google Scholar] [CrossRef]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef]
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 2014, 5, 539. [Google Scholar] [CrossRef]
- Peeters, A.J.M.; Vries, H.B.-D.; Hanhart, C.; Léon-Kloosterziel, K.M.; Zeevaart, J.A.D.; Koornneef, M. Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol. Plant. 2002, 115, 604–612. [Google Scholar] [CrossRef]
- Li, X.; Chen, T.; Li, Y.; Wang, Z.; Cao, H.; Chen, F.; Li, Y.; Soppe, W.J.J.; Li, W.; Liu, Y. ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on delay of germination1 expression. Plant Cell 2019, 31, 832. [Google Scholar] [CrossRef]
- Bakshi, A.; Piya, S.; Fernandez, J.C.; Chervin, C.; Hewezi, T.; Binder, B.M. Ethylene receptors signal via a noncanonical pathway to regulate abscisic acid responses. Plant Physiol. 2017, 176, 910–929. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.-Q.; Luan, S.; Li, J.; He, Z. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–1548590. [Google Scholar] [CrossRef]
- Flematti, G.R.; Merritt, D.J.; Piggott, M.J.; Trengove, R.; Smith, S.M.; Dixon, K.W.; Ghisalberti, E.L. Burning vegetation produces cyanohydrins that liberate cyanide and stimulate seed germination. Nat. Commun. 2011, 2, 360. [Google Scholar] [CrossRef]
- Chiwocha, S.D.; Dixon, K.W.; Flematti, G.R.; Ghisalberti, E.L.; Merritt, D.J.; Nelson, D.C.; Riseborough, J.-A.M.; Smith, S.M.; Stevens, J. Karrikins: A new family of plant growth regulators in smoke. Plant Sci. 2009, 177, 252–256. [Google Scholar] [CrossRef]
- Flematti, G.R.; Dixon, K.W.; Smith, S.M. What are karrikins and how were they ’discovered’ by plants? BMC Boil. 2015, 13, 108. [Google Scholar] [CrossRef]
- Nelson, D.C.; Flematti, G.R.; Ghisalberti, E.L.; Dixon, K.W.; Smith, S.M. Regulation of Seed Germination and Seedling Growth by Chemical Signals from Burning Vegetation. Annu. Rev. Plant Boil. 2012, 63, 107–130. [Google Scholar] [CrossRef]
- Long, R.L.; Stevens, J.; Griffiths, E.M.; Adamek, M.; Gorecki, M.J.; Powles, S.; Merritt, D.J. Seeds of Brassicaceae weeds have an inherent or inducible response to the germination stimulant karrikinolide. Ann. Bot. 2011, 108, 933–944. [Google Scholar] [CrossRef]
- Waters, M.T.; Nelson, D.C.; Scaffidi, A.; Flematti, G.R.; Sun, Y.K.; Dixon, K.W.; Smith, S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Dev. 2012, 139, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Stanga, J.; Smith, S.M.; Briggs, W.R.; Nelson, D.C. Suppressor of more axillary Growth2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 2013, 163, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Scaffidi, A.; Dun, E.A.; Waters, M.T.; Flematti, G.R.; Dixon, K.W.; Beveridge, C.; Ghisalberti, E.L.; Smith, S.M. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 8897–8902. [Google Scholar] [CrossRef]
- Wang, S.; Waters, M.T.; Smith, S.M. Karrikin-KAI2 signalling provides Arabidopsis seeds with tolerance to abiotic stress and inhibits germination under conditions unfavourable to seedling establishment. New Phytol. 2018, 219, 605–618. [Google Scholar] [CrossRef]
- Waters, M.T.; Scaffidi, A.; Moulin, S.; Sun, Y.K.; Flematti, G.R.; Smith, S.M. A Selaginella moellendorffii ortholog of karrikin insensitive2 functions in arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 2015, 27, 1925–1944. [Google Scholar] [CrossRef]
- Brun, G.; Thoiron, S.; Braem, L.; Pouvreau, J.; Montiel, G.; Lechat, M.; Simier, P.; Gevaert, K.; Goormachtig, S.; Delavault, P.; et al. CYP707As are effectors of karrikin and strigolactone signalling pathways in Arabidopsis thaliana and parasitic plants. Plant, Cell Environ. 2019, 42, 2612–2626. [Google Scholar] [CrossRef]
- Dave, A.; Hernandez, L.; He, Z.; Andriotis, V.; Vaistij, F.E.; Larson, T.; Graham, I.A. 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 2011, 23, 583–599. [Google Scholar] [CrossRef]
- Dave, A.; Vaistij, F.E.; Gilday, A.D.; Penfield, S.; Graham, I.A. Regulation of Arabidopsis thaliana seed dormancy and germination by 12-oxo-phytodienoic acid. J. Exp. Bot. 2016, 67, 2277–2284. [Google Scholar] [CrossRef]
- Singh, P.; Dave, A.; Vaistij, F.E.; Worrall, D.; Holroyd, G.H.; Wells, J.G.; Kaminski, F.; Graham, I.A.; Roberts, M.R. Jasmonic acid-dependent regulation of seed dormancy following maternal herbivory in Arabidopsis. New Phytol. 2017, 214, 1702–1711. [Google Scholar] [CrossRef]
- Chiang, G.C.K.; Barua, D.; Kramer, E.M.; Amasino, R.M.; Donohue, K. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 11661–11666. [Google Scholar] [CrossRef]
- Chen, M.; MacGregor, D.R.; Dave, A.; Florance, H.; Moore, K.; Paszkiewicz, K.; Smirnoff, N.; Graham, I.A.; Penfield, S. Maternal temperature history activates Flowering Locus T in fruits to control progeny dormancy according to time of year. Proc. Natl. Acad. Sci. USA 2014, 111, 18787–18792. [Google Scholar] [CrossRef]
- Blair, L.; Auge, G.; Donohue, K. Effect of FLOWERING LOCUS C on seed germination depends on dormancy. Funct. Plant Boil. 2017, 44, 493. [Google Scholar] [CrossRef]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; MacGregor, D.R. Effects of environmental variation during seed production on seed dormancy and germination. J. Exp. Bot. 2016, 68, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Batista, R.A.; Köhler, C. Genomic imprinting in plants—Revisiting existing models. Genome Res. 2020, 34, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Piskurewicz, U.; Iwasaki, M.; Susaki, D.; Megies, C.; Kinoshita, T.; Lopez-Molina, L. Dormancy-specific imprinting underlies maternal inheritance of seed dormancy in Arabidopsis thaliana. eLife 2016, 5, 479. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Hyvärinen, L.; Piskurewicz, U.; Lopez-Molina, L. Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife 2019, 8, 37434. [Google Scholar] [CrossRef]
- Buitink, J.; Leprince, O. Intracellular glasses and seed survival in the dry state. Comptes Rendus Boil. 2008, 331, 788–795. [Google Scholar] [CrossRef]
- Leprince, O.; Pellizzaro, A.; Berriri, S.; Buitink, J. Late seed maturation: Drying without dying. J. Exp. Bot. 2016, 68, 827–841. [Google Scholar] [CrossRef]
- Basbouss-Serhal, I.; Leymarie, J.; Bailly, C. Fluctuation of Arabidopsis seed dormancy with relative humidity and temperature during dry storage. J. Exp. Bot. 2015, 67, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Oracz, K.; Karpiński, S. Phytohormones signaling pathways and ros involvement in seed germination. Front. Plant Sci. 2016, 7, 980. [Google Scholar] [CrossRef] [PubMed]
- Née, G.; Xiang, Y.; Soppe, W.J. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Boil. 2017, 35, 8–14. [Google Scholar] [CrossRef]
- Chahtane, H.; Kim, W.; Lopez-Molina, L. Primary seed dormancy: A temporally multilayered riddle waiting to be unlocked. J. Exp. Bot. 2016, 68, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Oracz, K.; Bouteau, H.E.-M.; Farrant, J.M.; Cooper, K.; Belghazi, M.; Job, C.; Job, M.; Corbineau, F.; Bailly, C. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007, 50, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. From intracellular signaling networks to cell death: The dual role of reactive oxygen species in seed physiology. Comptes Rendus Boil. 2008, 331, 806–814. [Google Scholar] [CrossRef]
- El-Maarouf-Bouteau, H.; Meimoun, P.; Job, C.; Job, M.; Bailly, C. Role of protein and mRNA oxidation in seed dormancy and germination. Front. Plant Sci. 2013, 4, 77. [Google Scholar] [CrossRef]
- Rajjou, L.; Gallardo, K.; Debeaujon, I.; Vandekerckhove, J.; Job, C.; Job, D. The effect of α-amanitin on the arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mrnas during germination. Plant Physiol. 2004, 134, 1598–1613. [Google Scholar] [CrossRef]
- Gao, F.; Rampitsch, C.; Chitnis, V.R.; Humphreys, G.D.; Jordan, M.C.; Ayele, B.T. Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivumL.). Plant Biotechnol. J. 2013, 11, 921–932. [Google Scholar] [CrossRef]
- Meimoun, P.; Mordret, E.; Langlade, N.; Balzergue, S.; Arribat, S.; Bailly, C.; El-Maarouf-Bouteau, H. Is gene transcription involved in seed dry after-ripening? PLoS ONE 2014, 9, e86442. [Google Scholar] [CrossRef]
- Sano, N.; Rajjou, L.; North, H.M. Lost in translation: Physiological roles of stored mrnas in seed germination. Plants 2020, 9, 347. [Google Scholar] [CrossRef]
- Galland, M.; Huguet, R.; Arc, E.; Cueff, G.; Job, M.; Rajjou, L. Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Mol. Cell. Proteom. 2013, 13, 252–268. [Google Scholar] [CrossRef] [PubMed]
- Clerkx, E.J.; El-Lithy, M.E.; Vierling, E.; Ruys, G.J.; Vries, H.B.-D.; Groot, S.P.; Vreugdenhil, D.; Koornneef, M. Analysis of natural allelic variation of arabidopsis seed germination and seed longevity traits between the accessions landsberg erecta and shakdara, using a new recombinant inbred line population. Plant Physiol. 2004, 135, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.-P.; Keizer, P.; Van Eeuwijk, F.; Smeekens, S.; Bentsink, L. Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis. Plant Physiol. 2012, 160, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.-P.; Cueff, G.; Hegedus, D.D.; Rajjou, L.; Bentsink, L. A role for seed storage proteins in Arabidopsis seed longevity. J. Exp. Bot. 2015, 66, 6399–6413. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-Q.; Liu, S.-J.; Song, S.-Q.; Møller, I.M. Proteomics of seed development, desiccation tolerance, germination and vigor. Plant Physiol. Biochem. 2015, 86, 1–15. [Google Scholar] [CrossRef]
- Kumar, S.P.J.; Prasad, S.R.; Banerjee, R.; Thammineni, C. Seed birth to death: Dual functions of reactive oxygen species in seed physiology. Ann. Bot. 2015, 116, 663–668. [Google Scholar] [CrossRef]
- Wojtyla, Ł.; Lechowska, K.; Kubala, S.; Garnczarska, M. Different Modes of Hydrogen Peroxide Action During Seed Germination. Front. Plant Sci. 2016, 7, 75. [Google Scholar] [CrossRef]
- Kurek, K.; Plitta-Michalak, B.; Ratajczak, E. Reactive Oxygen Species as Potential Drivers of the Seed Aging Process. Plants 2019, 8, 174. [Google Scholar] [CrossRef]
- E Sattler, S.; Gilliland, L.U.; Magallanes-Lundback, M.; Pollard, M.; DellaPenna, D. Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 2004, 16, 1419–1432. [Google Scholar] [CrossRef]
- Ogé, L.; Bourdais, G.; Bove, J.; Collet, B.; Godin, B.; Granier, F.; Boutin, J.-P.; Job, M.; Jullien, M.; Grappin, P. Protein repair l-isoaspartyl methyltransferase1 is involved in both seed longevity and germination vigor in arabidopsis. Plant Cell 2008, 20, 3022–3037. [Google Scholar] [CrossRef]
- Zhou, C.; Tokuhisa, J.G.; Bevan, D.R.; Esen, A. Properties of beta-thioglucoside hydrolases (TGG1 and TGG2) from leaves of Arabidopsis thaliana. Plant Sci. 2012, 191–192, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Chatelain, E.; Satour, P.; Laugier, E.; Vu, B.L.; Payet, N.; Rey, P.; Montrichard, F. Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proc. Natl. Acad. Sci. USA 2013, 110, 3633–3638. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wei, Y.; Zhu, Y.; Lian, L.; Xie, H.; Cai, Q.; Chen, Q.; Lin, Z.; Wang, Z.; Xie, H.; et al. Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity. Plant Biotechnol. J. 2014, 13, 526–539. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tsang, E.W.; Chu, P.; Zhou, Y.; Li, Y.; Liu, J.; Ding, Y.; Jiang, L.; Wu, K.; Huang, S. Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis. J. Exp. Bot. 2012, 63, 4107–4121. [Google Scholar] [CrossRef] [PubMed]
- Bissoli, G.; Niñoles, R.; Fresquet, S.; Palombieri, S.; Bueso, E.; Rubio, L.; García-Sánchez, M.J.; Fernández, J.A.; Mulet, J.M.; Serrano, R. Peptidyl-prolyl cis-trans isomerase ROF2 modulates intracellular pH homeostasis in Arabidopsis. Plant J. 2012, 70, 704–716. [Google Scholar] [CrossRef]
- Yazdanpanah, F.; Hanson, J.; Hilhorst, H.W.M.; Bentsink, L. Differentially expressed genes during the imbibition of dormant and after-ripened sedes—A reverse genetics approach. BMC Plant Boil. 2017, 17, 151. [Google Scholar] [CrossRef]
- Yazdanpanah, F.; Maurino, V.G.; Mettler-Altmann, T.; Buijs, G.; Bailly, M.; Jashni, M.K.; Willems, L.; I Sergeeva, L.; Rajjou, L.; Hilhorst, H.W.M.; et al. NADP-MALIC ENZYME 1 Affects Germination after Seed Storage in Arabidopsis thaliana. Plant Cell Physiol. 2018, 60, 318–328. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Bray, C.M.; West, C.E. The importance of safeguarding genome integrity in germination and seed longevity. J. Exp. Bot. 2015, 66, 3549–3558. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Footitt, S.; Bray, C.M.; Finch-Savage, W.E.; West, C.E. DNA damage checkpoint kinase ATM regulates germination and maintains genome stability in seeds. Proc. Natl. Acad. Sci. USA 2016, 113, 9647–9652. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Bray, C.M.; West, C.E. Seeds and the art of genome maintenance. Front. Plant Sci. 2019, 10, 706. [Google Scholar] [CrossRef]
- Bueso, E.; Muñoz-Bertomeu, J.; Campos, F.; Martínez, C.; Tello, C.; Martínez-Almonacid, I.; Ballester, P.; Simón-Moya, M.; Brunaud, V.; Yenush, L.; et al. Arabidopsis COGWHEEL1 links light perception and gibberellins with seed tolerance to deterioration. Plant J. 2016, 87, 583–596. [Google Scholar] [CrossRef]
- Bueso, E.; Ibáñez, C.; Sayas, E.; Muñoz-Bertomeu, J.; Gonzalez-Guzman, M.; Rodriguez, P.L.; Serrano, R. A forward genetic approach in Arabidopsis thaliana identifies a RING-type ubiquitin ligase as a novel determinant of seed longevity. Plant Sci. 2014, 215, 110–116. [Google Scholar] [CrossRef]
- Bueso, E.; Muñoz-Bertomeu, J.; Campos, F.; Brunaud, V.; Martínez, L.; Sayas, E.; Ballester, P.; Yenush, L.; Serrano, R. Arabidopsis Thaliana HOMEOBOX25 uncovers a role for gibberellins in seed longevity. Plant Physiol. 2013, 164, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Rajjou, L.; North, H.M.; Debeaujon, I.; Marion-Poll, A.; Seo, M. Staying alive: Molecular aspects of seed longevity. Plant Cell Physiol. 2015, 57, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Haroldsen, V.; Cai, X.; Wu, Y. Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress. Plant, Cell Environ. 2006, 29, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-C.; Yü, Y.; Wang, C.-Y.; Li, Z.-Y.; Liu, Q.; Xu, J.; Liao, J.-Y.; Wang, X.; Qu, L.-H.; Chen, F.; et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848–852. [Google Scholar] [CrossRef]
- Debeaujon, I. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000, 122, 403–414. [Google Scholar] [CrossRef]
- Chatelain, E.; Le Gall, S.; Deligny-Penninck, S.; Hundertmark, M.; Leprince, O.; Satour, P.; Rogniaux, H.; Buitink, J. Temporal profiling of the heat? Stable proteome during late maturation of medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant, Cell Environ. 2012, 35, 1440–1455. [Google Scholar] [CrossRef]
- Probert, R.J.; Daws, M.; Hay, F.R. Ecological correlates of ex situ seed longevity: A comparative study on 195 species. Ann. Bot. 2009, 104, 57–69. [Google Scholar] [CrossRef]
- Righetti, K.; Vu, J.L.; Pelletier, S.; Vu, B.L.; Glaab, E.; Lalanne, D.; Pasha, A.; Patel, R.V.; Provart, N.J.; Verdier, J.; et al. Inference of longevity-related genes from a robust coexpression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. Plant Cell 2015, 27, 2692–2708. [Google Scholar] [CrossRef]
- Long, R.L.; Gorecki, M.J.; Renton, M.; Scott, J.; Colville, L.; Goggin, D.E.; Commander, L.E.; Westcott, D.A.; Cherry, H.; Finch-Savage, W.E. The ecophysiology of seed persistence: A mechanistic view of the journey to germination or demise. Boil. Rev. 2014, 90, 31–59. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Footitt, S. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 2017, 68, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Buijs, G.; Vogelzang, A.; Nijveen, H.; Bentsink, L. Dormancy cycling: Translation-related transcripts are the main difference between dormant and non-dormant seeds in the field. Plant J. 2020, 102, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Soler, I.D.; Clay, H.; Finch-Savage, W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 20236–20241. [Google Scholar] [CrossRef]
- Footitt, S.; Clay, H.A.; Dent, K.; Finch-Savage, W.E. Environment sensing in spring-dispersed seeds of a winter annual Arabidopsis influences the regulation of dormancy to align germination potential with seasonal changes. New Phytol. 2014, 202, 929–939. [Google Scholar] [CrossRef]
- Footitt, S.; Huang, Z.; Clay, H.A.; Mead, A.; Finch-Savage, W.E. Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. Plant J. 2013, 74, 1003–1015. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Yan, D.; Duermeyer, L.; Leoveanu, C.; Nambara, E. The functions of the endosperm during seed germination. Plant Cell Physiol. 2014, 55, 1521–1533. [Google Scholar] [CrossRef]
- Steinbrecher, T.; Leubner-Metzger, G. The biomechanics of seed germination. J. Exp. Bot. 2016, 68, 765–783. [Google Scholar] [CrossRef]
- Steinbrecher, T.; Leubner-Metzger, G. Tissue and cellular mechanics of seeds. Curr. Opin. Genet. Dev. 2018, 51, 1–10. [Google Scholar] [CrossRef]
- Endo, A.; Tatematsu, K.; Hanada, K.; Duermeyer, L.; Okamoto, M.; Yonekura-Sakakibara, K.; Saito, K.; Toyoda, T.; Kawakami, N.; Kamiya, Y.; et al. Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis and defense responses are activated in the endosperm of germinating Arabidopsis thaliana seeds. Plant Cell Physiol. 2011, 53, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Sechet, J.; Frey, A.; Effroy-Cuzzi, D.; Berger, A.; Perreau, F.; Cueff, G.; Charif, D.; Rajjou, L.; Mouille, G.; North, H.M.; et al. Xyloglucan metabolism differentially impacts the cell wall characteristics of the endosperm and embryo during arabidopsis seed germination. Plant Physiol. 2016, 170, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- Shigeyama, T.; Watanabe, A.; Tokuchi, K.; Toh, S.; Sakurai, N.; Shibuya, N.; Kawakami, N. α-Xylosidase plays essential roles in xyloglucan remodelling, maintenance of cell wall integrity, and seed germination in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 5615–5629. [Google Scholar] [CrossRef] [PubMed]
- Scheler, C.; Weitbrecht, K.; Pearce, S.P.; Hampstead, A.; Büttner-Mainik, A.; Lee, K.J.; Voegele, A.; Oracz, K.; Dekkers, B.J.; Wang, X.; et al. Promotion of testa rupture during garden cress germination involves seed compartment-specific expression and activity of pectin methylesterases. Plant Physiol. 2014, 167, 200–215. [Google Scholar] [CrossRef]
- Turbant, A.; Fournet, F.; Lequart, M.; Zabijak, L.; Pageau, K.; Bouton, S.; Van Wuytswinkel, O. PME58 plays a role in pectin distribution during seed coat mucilage extrusion through homogalacturonan modification. J. Exp. Bot. 2016, 67, 2177–2190. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Boil. 2015, 25, 162–172. [Google Scholar] [CrossRef]
- Yan, A.; Wu, M.; Yan, L.; Hu, R.; Ali, I.; Gan, Y. AtEXP2 Is involved in seed germination and abiotic stress response in arabidopsis. PLoS ONE 2014, 9, e85208. [Google Scholar] [CrossRef]
- De Giorgi, J.; Piskurewicz, U.; Loubéry, S.; Utz-Pugin, A.; Bailly, C.; Mène-Saffrané, L.; Lopez-Molina, L. An endosperm-associated cuticle is required for arabidopsis seed viability, dormancy and early control of germination. PLoS Genet. 2015, 11, e1005708. [Google Scholar] [CrossRef]
- Loubéry, S.; De Giorgi, J.; Utz-Pugin, A.; Demonsais, L.; Lopez-Molina, L. A Maternally Deposited Endosperm Cuticle Contributes to the Physiological Defects of transparent testa Seeds. Plant Physiol. 2018, 177, 1218–1233. [Google Scholar] [CrossRef]
- Nobusawa, T.; Okushima, Y.; Nagata, N.; Kojima, M.; Sakakibara, H.; Umeda, M. Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Boil. 2013, 11, e1001531. [Google Scholar] [CrossRef]
- Silva, E.A.; Toorop, P.E.; Van Lammeren, A.A.M.; Hilhorst, H.W.M. ABA inhibits embryo cell expansion and early cell division events during coffee (coffea arabica ‘rubi’) seed germination. Ann. Bot. 2008, 102, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Gimeno-Gilles, C.; Lelièvre, E.; Viau, L.; Malik-Ghulam, M.; Ricoult, C.; Niebel, A.; LeDuc, N.; Limami, A.M. ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture: Modifying enzymes and structural proteins in medicago truncatula embryo axis. Mol. Plant 2009, 2, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Souza, N.M.; Topham, A.T.; Bassel, G.W. Quantitative analysis of the 3D cell shape changes driving soybean germination. J. Exp. Bot. 2017, 68, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.-M.; Shangguan, X.-X.; Zhao, B.; Zhang, X.-F.; Chao, L.; Yang, C.; Wang, L.-J.; Zhu, H.-Y.; Zeng, Y.-D.; Guo, W.-Z.; et al. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 2014, 5, 5519. [Google Scholar] [CrossRef] [PubMed]
- Bassel, G.W.; Stamm, P.; Mosca, G.; De Reuille, P.B.; Gibbs, D.J.; Winter, R.; Janka, A.; Holdsworth, M.; Smith, R.S. Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc. Natl. Acad. Sci. USA 2014, 111, 8685–8690. [Google Scholar] [CrossRef]
- Lee, K.J.; Dekkers, B.J.W.; Steinbrecher, T.; Walsh, C.T.; Bacic, A.; Bentsink, L.; Leubner-Metzger, G.; Knox, J.P. Distinct cell wall architectures in seed endosperms in representatives of the Brassicaceae and Solanaceae. Plant Physiol. 2012, 160, 1551–1566. [Google Scholar] [CrossRef]
- Iglesias-Fernandez, R.; Barrero-Sicilia, C.; Carrillo-Barral, N.; Onate-Sanchez, L.; Carbonero, P. Arabidopsis thaliana bZIP44: A transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7. Plant J. 2013, 74, 767–780. [Google Scholar] [CrossRef]
- Graeber, K.; Linkies, A.; Steinbrecher, T.; Mummenhoff, K.; Tarkowská, D.; Turečková, V.; Ignatz, M.; Sperber, K.; Voegele, A.; De Jong, H.; et al. DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. USA 2014, 111, E3571–E3580. [Google Scholar] [CrossRef]
- Fiume, E.; Guyon, V.; Remoué, C.; Magnani, E.; Miquel, M.; Grain, D.; Lepiniec, L. TWS1, a Novel Small Protein, Regulates Various Aspects of Seed and Plant Development. Plant Physiol. 2016, 172, 1732–1745. [Google Scholar] [CrossRef]
- Tsuwamoto, R.; Fukuoka, H.; Takahata, Y. GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J. 2007, 54, 30–42. [Google Scholar] [CrossRef]
- Doll, N.M.; Royek, S.; Fujita, S.; Okuda, S.; Chamot, S.; Stintzi, A.; Widiez, T.; Hothorn, M.; Schaller, A.; Geldner, N.; et al. A two-way molecular dialogue between embryo and endosperm is required for seed development. Sci. 2020, 367, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Onouchi, H.; Kondo, M.; Hara-Nishimura, I.; Nishimura, M.; Machida, C.; Machida, Y. A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 2002, 128, 4681. [Google Scholar] [CrossRef]
- Xing, Q.; Creff, A.; Waters, A.; Tanaka, H.; Goodrich, J.; Ingram, G.C. ZHOUPI controls embryonic cuticle formation via a signalling pathway involving the subtilisin protease ABNORMAL LEAF-SHAPE1 and the receptor kinases GASSHO1 and GASSHO2. Development 2013, 140, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Creff, A.; Brocard, L.; Joubès, J.; Taconnat, L.; Doll, N.M.; Marsollier, A.-C.; Pascal, S.; Galletti, R.; Boeuf, S.; Moussu, S.; et al. A stress-response-related inter-compartmental signalling pathway regulates embryonic cuticle integrity in Arabidopsis. PLoS Genet. 2019, 15, e1007847. [Google Scholar] [CrossRef]
- Moussu, S.; Doll, N.M.; Chamot, S.; Brocard, L.; Creff, A.; Fourquin, C.; Widiez, T.; Nimchuk, Z.L.; Ingram, G.C. Zhoupi and kerberos mediate embryo/endosperm separation by promoting the formation of an extracuticular sheath at the embryo surface. Plant Cell 2017, 29, 1642–1656. [Google Scholar] [CrossRef]
- Doll, N.M.; Bovio, S.; Gaiti, A.; Marsollier, A.-C.; Chamot, S.; Moussu, S.; Widiez, T.; Ingram, G. The endosperm-derived embryo sheath is an anti-adhesive structure that facilitates cotyledon emergence during germination in arabidopsis. Curr. Boil. 2020, 30, 909–915.e4. [Google Scholar] [CrossRef]
- Barrôco, R.M.; Van Poucke, K.; Bergervoet, J.H.; De Veylder, L.; Groot, S.P.; Inzé, D.; Engler, G. The role of the cell cycle machinery in resumption of postembryonic development. Plant Physiol. 2005, 137, 127–140. [Google Scholar] [CrossRef]
- Masubelele, N.H.; Dewitte, W.; Menges, M.; Maughan, S.; Collins, C.; Huntley, R.P.; Nieuwland, J.; Scofield, S.; Murray, J.A.H. D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 15694–15699. [Google Scholar] [CrossRef]
- Sliwinska, E.; Bassel, G.W.; Bewley, J.D. Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocoty. J. Exp. Bot. 2009, 60, 3587–3594. [Google Scholar] [CrossRef]
- Resentini, F.; Felipo-Benavent, A.; Colombo, L.; Blázquez, M.A.; Alabadí, D.; Masiero, S. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 2015, 8, 482–485. [Google Scholar] [CrossRef]
- Shani, E.; Weinstain, R.; Zhang, Y.; Castillejo, C.; Kaiserli, E.; Chory, J.; Tsien, R.Y.; Estelle, M. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. USA 2013, 110, 4834–4839. [Google Scholar] [CrossRef] [PubMed]
- Rizza, A.; Walia, A.; Lanquar, V.; Frommer, W.B.; Jones, A. In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat. Plants 2017, 3, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Khakhar, A.; Leydon, A.R.; Lemmex, A.C.; Klavins, E.; Nemhauser, J.L. Synthetic hormone-responsive transcription factors can monitor and re-program plant development. eLife 2018, 7, e34702. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).