Contributions and Limitations of Biophysical Approaches to Study of the Interactions between Amphiphilic Molecules and the Plant Plasma Membrane
Abstract
1. Introduction
2. Specific Aspects of the Plant Plasma Membrane
3. Involvement of the Plant Plasma Membrane in Triggering the Immunity Signaling Process
4. Interaction of Amphiphilic Elicitors with the Plant Plasma Membrane
5. Biophysical Studies of Amphiphiles and Plant Plasma Membrane Interactions
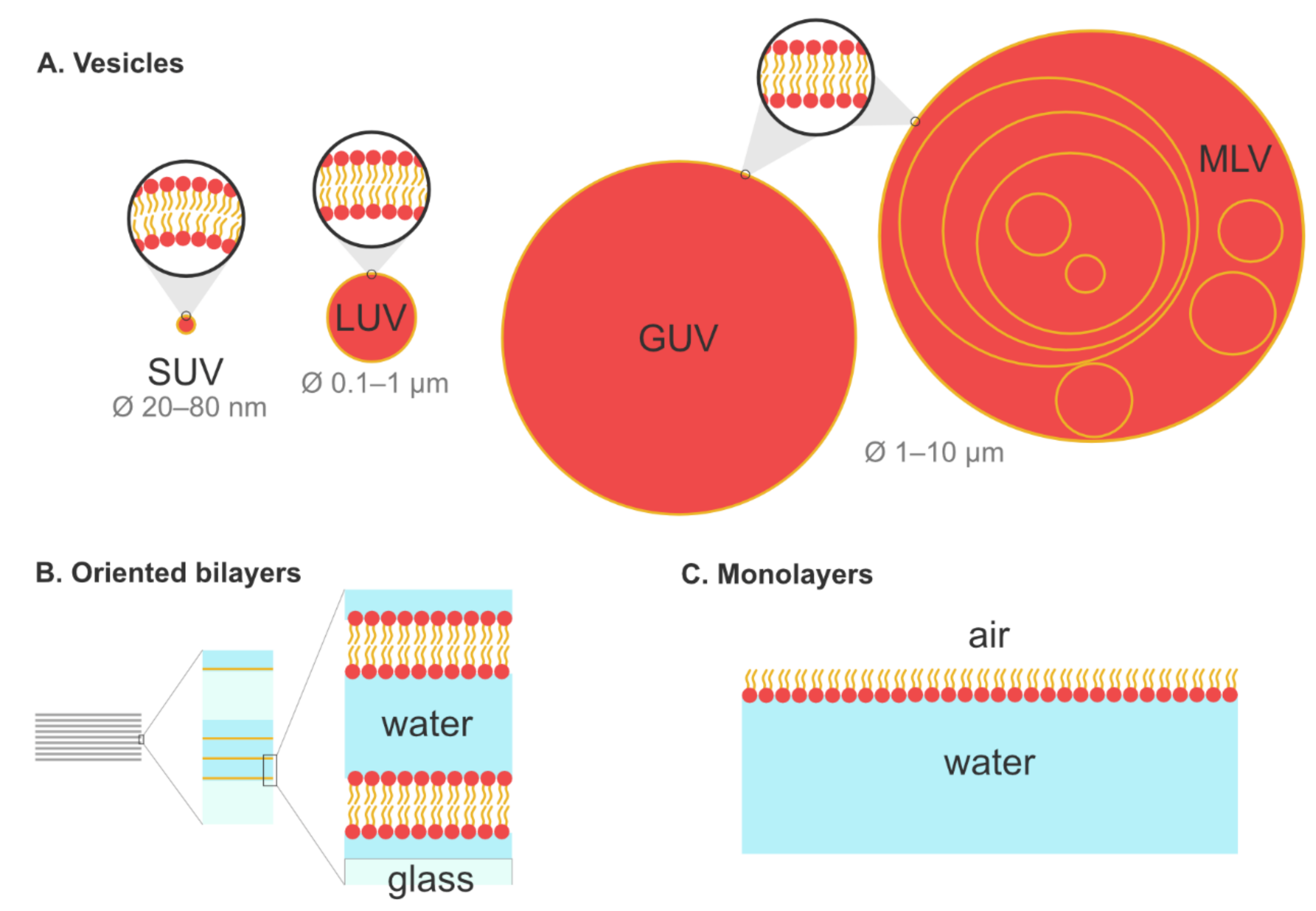
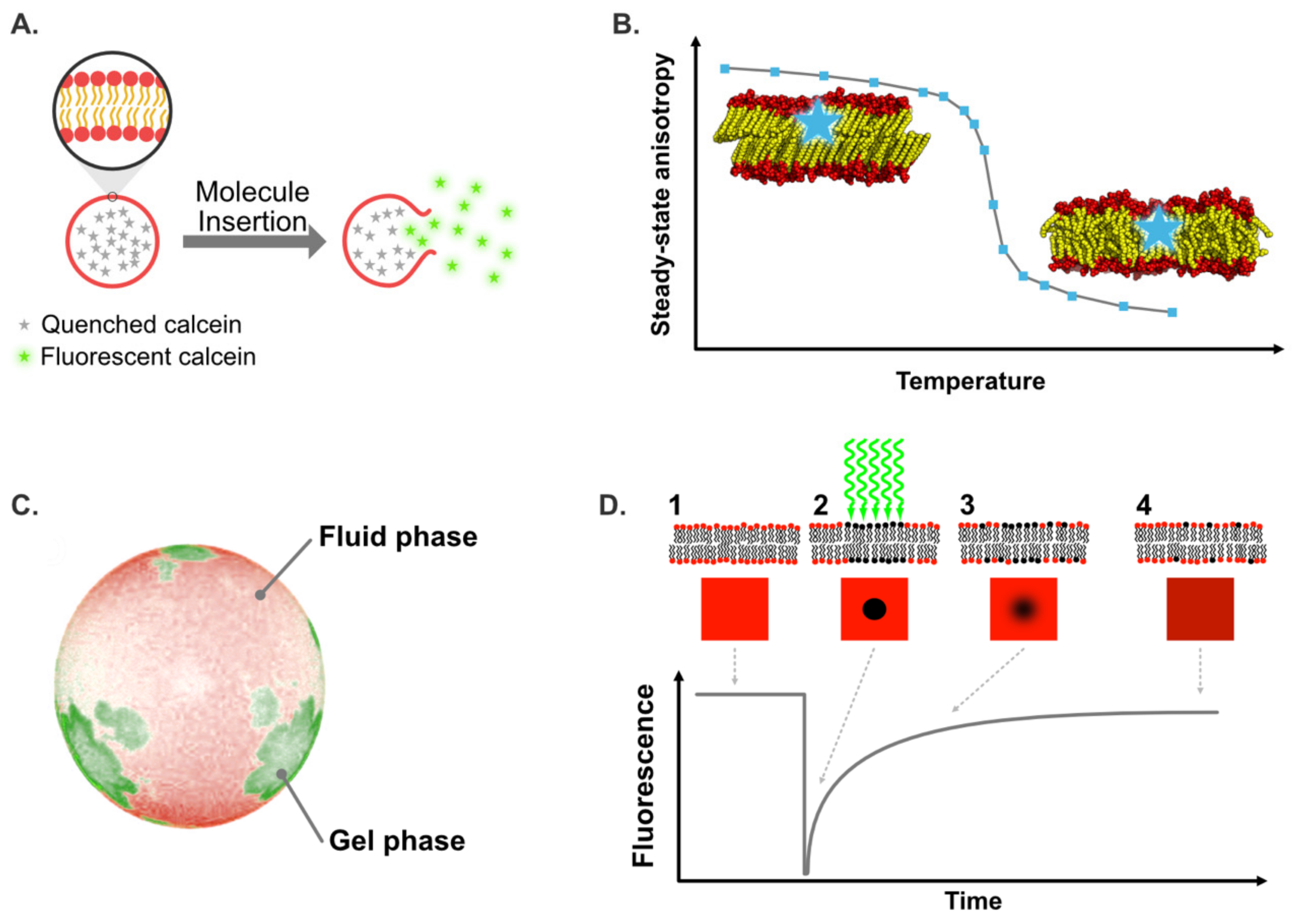
5.1. Biomimetic Membrane Models
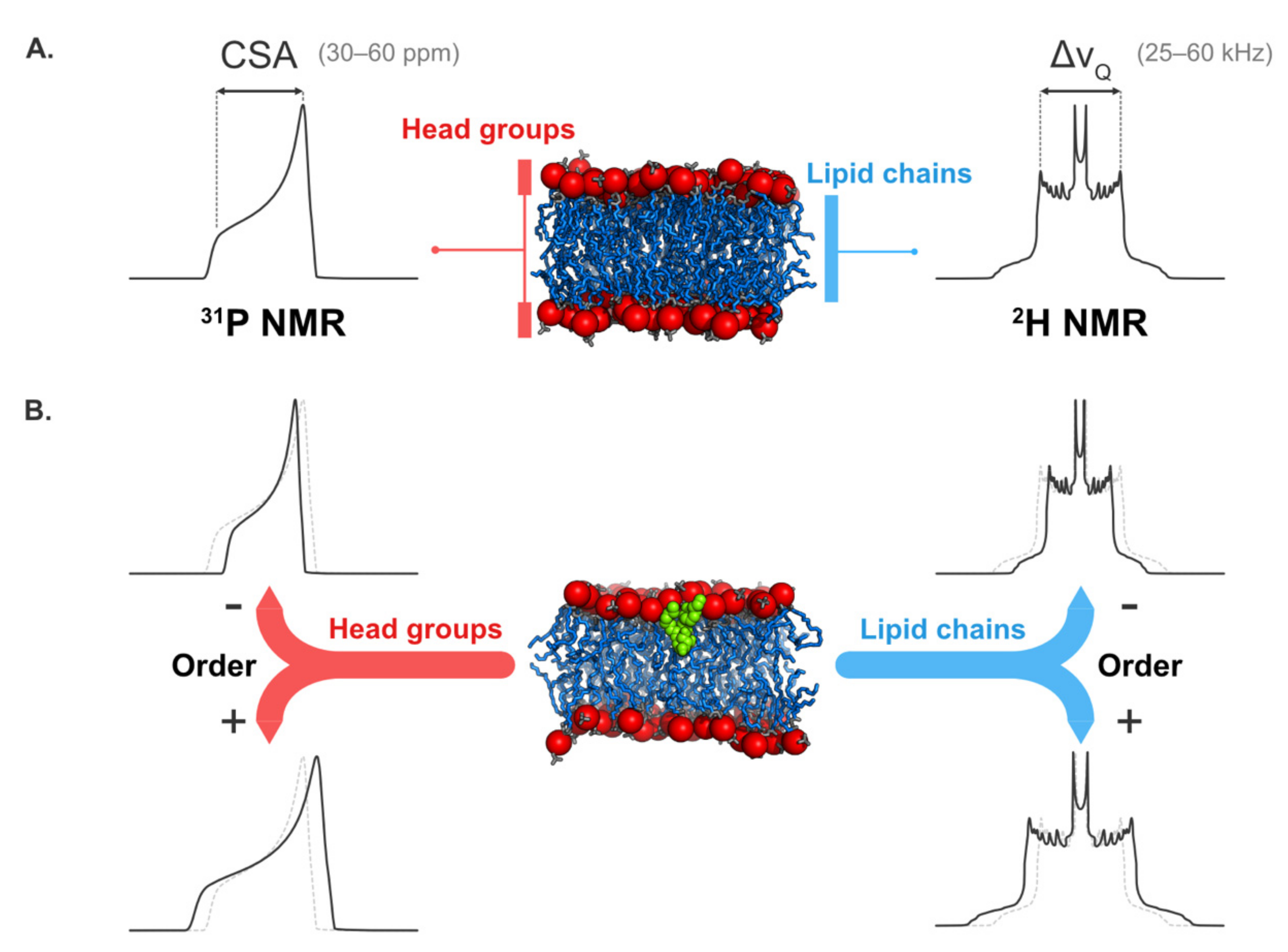
5.2. Solid-State NMR Spectroscopy
5.2.1. Structural Information
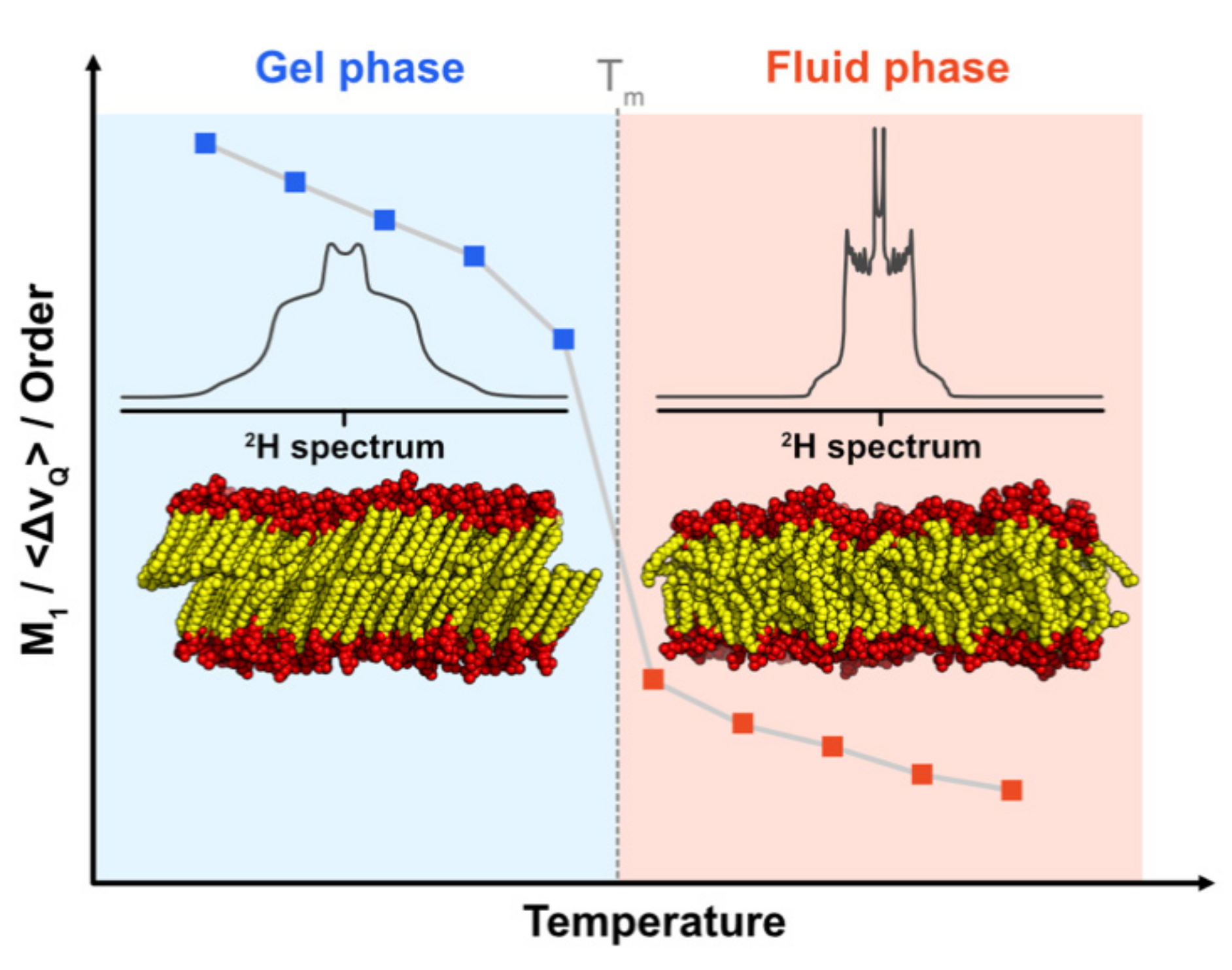
5.2.2. Information on Lipid Dynamics
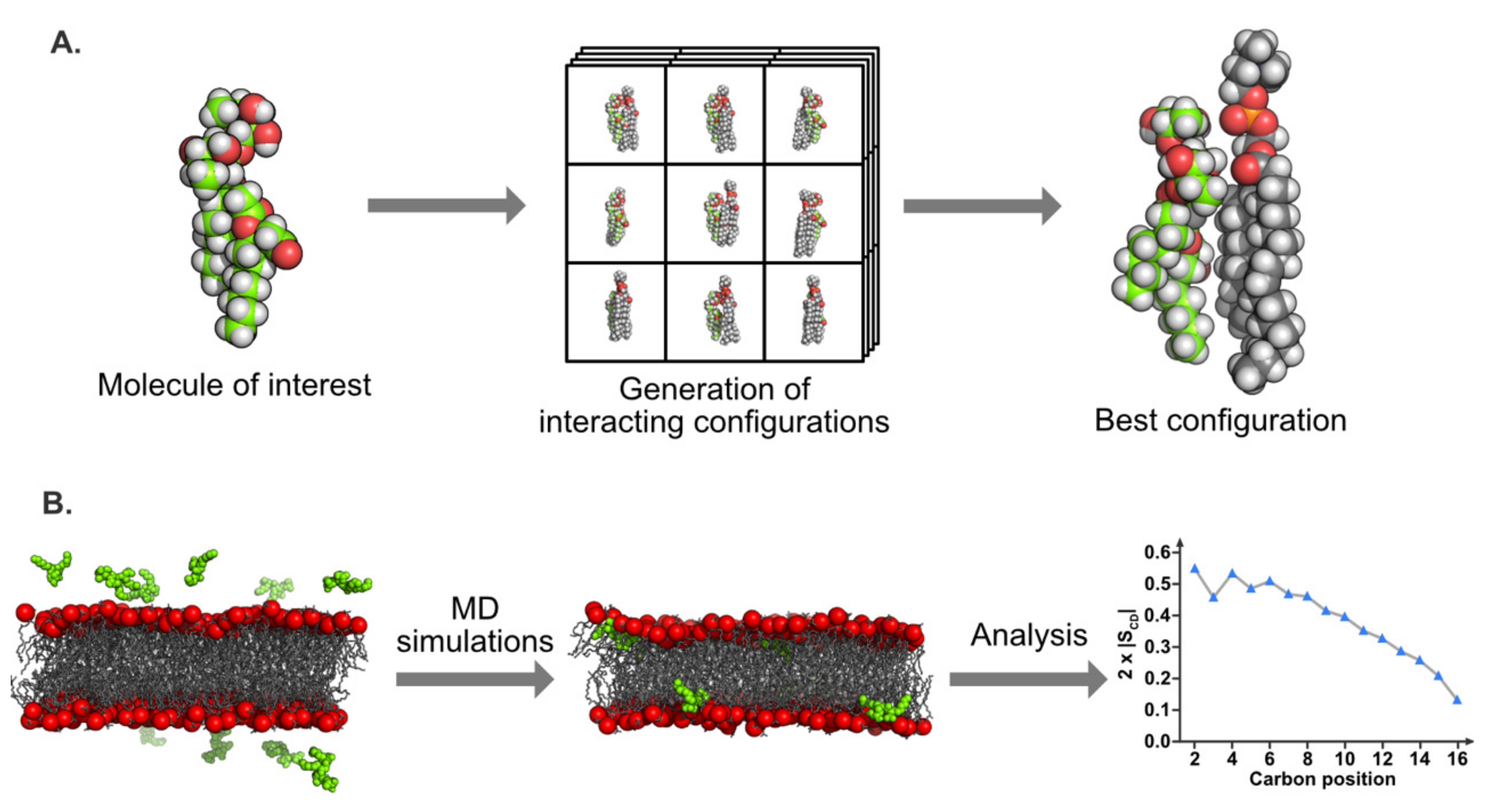
5.3. Molecular Modeling
5.3.1. Molecular Docking
5.3.2. Molecular Dynamics Simulations
5.4. Fluorescence Spectroscopy and Imaging
5.4.1. Membrane Permeabilization
5.4.2. Information on Lipid Dynamics
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lee, H.-A.; Lee, H.-Y.; Seo, E.; Lee, J.; Kim, S.-B.; Oh, S.; Choi, E.; Choi, E.; Lee, S.E.; Choi, D. Current Understandings of Plant Nonhost Resistance. Mol. Plant Microbe Interact. 2017, 30, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Schellenberger, R.; Touchard, M.; Clément, C.; Baillieul, F.; Cordelier, S.; Crouzet, J.; Dorey, S. Apoplastic invasion patterns triggering plant immunity: Plasma membrane sensing at the frontline. Mol. Plant Pathol. 2019, 20, 1602–1616. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.J.; Nicolson, G.L. The Fluid Mosaic Model of the Structure of Cell Membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef] [PubMed]
- van Meer, G. Cellular lipidomics. EMBO J. 2005, 24, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, K.; Mongrand, S.; Beney, L.; Simon-Plas, F.; Gerbeau-Pissot, P. Differential Effect of Plant Lipids on Membrane Organization. J. Biol. Chem. 2015, 290, 5810–5825. [Google Scholar] [CrossRef] [PubMed]
- Cacas, J.-L.; Buré, C.; Grosjean, K.; Gerbeau-Pissot, P.; Lherminier, J.; Rombouts, Y.; Maes, E.; Bossard, C.; Gronnier, J.; Furt, F.; et al. Revisiting Plant Plasma Membrane Lipids in Tobacco: A Focus on Sphingolipids. Plant Physiol. 2016, 170, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Bayer, E.; Lafarge, D.; Cluzet, S.; German Retana, S.; Boubekeur, T.; Leborgne-Castel, N.; Carde, J.-P.; Lherminier, J.; Noirot, E.; et al. Remorin, a Solanaceae Protein Resident in Membrane Rafts and Plasmodesmata, Impairs Potato virus X Movement. Plant Cell 2009, 21, 1541–1555. [Google Scholar] [CrossRef]
- Gerbeau-Pissot, P.; Der, C.; Thomas, D.; Anca, I.-A.; Grosjean, K.; Roche, Y.; Perrier-Cornet, J.-M.; Mongrand, S.; Simon-Plas, F. Modification of Plasma Membrane Organization in Tobacco Cells Elicited by Cryptogein. Plant Physiol. 2014, 164, 273–286. [Google Scholar] [CrossRef]
- Gronnier, J.; Gerbeau-Pissot, P.; Germain, V.; Mongrand, S.; Simon-Plas, F. Divide and Rule: Plant Plasma Membrane Organization. Trends Plant Sci. 2018, 23, 899–917. [Google Scholar] [CrossRef]
- Jaillais, Y.; Ott, T. The Nanoscale Organization of the Plasma Membrane and Its Importance in Signaling: A Proteolipid Perspective. Plant Physiol. 2020, 182, 1682–1696. [Google Scholar] [CrossRef]
- Tjellström, H.; Hellgren, L.I.; Wieslander, Å.; Sandelius, A.S. Lipid asymmetry in plant plasma membranes: Phosphate deficiency-induced phospholipid replacement is restricted to the cytosolic leaflet. FASEB J. 2010, 24, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Platre, M.P.; Noack, L.C.; Doumane, M.; Bayle, V.; Simon, M.L.A.; Maneta-Peyret, L.; Fouillen, L.; Stanislas, T.; Armengot, L.; Pejchar, P.; et al. A Combinatorial Lipid Code Shapes the Electrostatic Landscape of Plant Endomembranes. Dev. Cell 2018, 45, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 2009, 12, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005, 6, 973–979. [Google Scholar] [CrossRef]
- Boutrot, F.; Zipfel, C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Cook, D.E.; Mesarich, C.H.; Thomma, B.P.H.J. Understanding Plant Immunity as a Surveillance System to Detect Invasion. Annu. Rev. Phytopathol. 2015, 53, 541–563. [Google Scholar] [CrossRef]
- Ali, G.S.; Prasad, K.V.S.K.; Day, I.; Reddy, A.S.N. Ligand-Dependent Reduction in the Membrane Mobility of FLAGELLIN SENSITIVE2, an Arabidopsis Receptor-Like Kinase. Plant Cell Physiol. 2007, 48, 1601–1611. [Google Scholar] [CrossRef]
- Bücherl, C.A.; Jarsch, I.K.; Schudoma, C.; Segonzac, C.; Mbengue, M.; Robatzek, S.; MacLean, D.; Ott, T.; Zipfel, C. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 2017, 6. [Google Scholar] [CrossRef]
- Sandor, R.; Der, C.; Grosjean, K.; Anca, I.; Noirot, E.; Leborgne-Castel, N.; Lochman, J.; Simon-Plas, F.; Gerbeau-Pissot, P. Plasma membrane order and fluidity are diversely triggered by elicitors of plant defence. J. Exp. Bot. 2016, 67, 5173–5185. [Google Scholar] [CrossRef]
- Rippa, S.; Eid, M.; Formaggio, F.; Toniolo, C.; Béven, L. Hypersensitive-Like Response to the Pore-Former Peptaibol Alamethicin in Arabidopsis Thaliana. ChemBioChem 2010, 11, 2042–2049. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.-S.; Kim, W.; Lee, C.; Oh, C.-S. Harpins, Multifunctional Proteins Secreted by Gram-Negative Plant-Pathogenic Bacteria. Mol. Plant Microbe Interact. 2013, 26, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Lenarčič, T.; Albert, I.; Böhm, H.; Hodnik, V.; Pirc, K.; Zavec, A.B.; Podobnik, M.; Pahovnik, D.; Žagar, E.; Pruitt, R.; et al. Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 2017, 358, 1431–1434. [Google Scholar] [CrossRef] [PubMed]
- Derevnina, L.; Dagdas, Y.F.; De la Concepcion, J.C.; Bialas, A.; Kellner, R.; Petre, B.; Domazakis, E.; Du, J.; Wu, C.-H.; Lin, X.; et al. Nine things to know about elicitins. New Phytol. 2016, 212, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Deleu, M.; Paquot, M.; Nylander, T. Effect of Fengycin, a Lipopeptide Produced by Bacillus subtilis, on Model Biomembranes. Biophys. J. 2008, 94, 2667–2679. [Google Scholar] [CrossRef]
- Nasir, M.N.; Thawani, A.; Kouzayha, A.; Besson, F. Interactions of the natural antimicrobial mycosubtilin with phospholipid membrane models. Colloids Surf. B Biointerfaces 2010, 78, 17–23. [Google Scholar] [CrossRef]
- Henry, G.; Deleu, M.; Jourdan, E.; Thonart, P.; Ongena, M. The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cell. Microbiol. 2011, 13, 1824–1837. [Google Scholar] [CrossRef]
- Deleu, M.; Lorent, J.; Lins, L.; Brasseur, R.; Braun, N.; El Kirat, K.; Nylander, T.; Dufrêne, Y.F.; Mingeot-Leclercq, M.-P. Effects of surfactin on membrane models displaying lipid phase separation. Biochim. Biophys. Acta Biomembr. 2013, 1828, 801–815. [Google Scholar] [CrossRef] [PubMed]
- Buchoux, S.; Lai-Kee-Him, J.; Garnier, M.; Tsan, P.; Besson, F.; Brisson, A.; Dufourc, E.J. Surfactin-triggered small vesicle formation of negatively charged membranes: A novel membrane-lysis mechanism. Biophys. J. 2008, 95, 3840–3849. [Google Scholar] [CrossRef] [PubMed]
- Vatsa, P.; Sanchez, L.; Clément, C.; Baillieul, F.; Dorey, S. Rhamnolipid Biosurfactants as New Players in Animal and Plant Defense against Microbes. Int. J. Mol. Sci. 2010, 11, 5095–5108. [Google Scholar] [CrossRef] [PubMed]
- Pashynska, V.A. Mass spectrometric study of rhamnolipid biosurfactants and their interactions with cell membrane phospholipids. Biopolym. Cell 2009, 25, 504–508. [Google Scholar] [CrossRef]
- Ortiz, A.; Teruel, J.A.; Espuny, M.J.; Marqués, A.; Manresa, Á.; Aranda, F.J. Effects of dirhamnolipid on the structural properties of phosphatidylcholine membranes. Int. J. Pharm. 2006, 325, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Aranda, F.J.; Espuny, M.J.; Marqués, A.; Teruel, J.A.; Manresa, Á.; Ortiz, A. Thermodynamics of the Interaction of a Dirhamnolipid Biosurfactant Secreted by Pseudomonas aeruginosa with Phospholipid Membranes. Langmuir 2007, 23, 2700–2705. [Google Scholar] [CrossRef]
- Sánchez, M.; Aranda, F.J.; Teruel, J.A.; Ortiz, A. Interaction of a bacterial dirhamnolipid with phosphatidylcholine membranes: A biophysical study. Chem. Phys. Lipids 2009, 161, 51–55. [Google Scholar] [CrossRef]
- Sánchez, M.; Aranda, F.J.; Teruel, J.A.; Espuny, M.J.; Marqués, A.; Manresa, Á.; Ortiz, A. Permeabilization of biological and artificial membranes by a bacterial dirhamnolipid produced by Pseudomonas aeruginosa. J. Colloid Interface Sci. 2010, 341, 240–247. [Google Scholar] [CrossRef]
- Abbasi, H.; Noghabi, K.A.; Ortiz, A. Interaction of a bacterial monorhamnolipid secreted by Pseudomonas aeruginosa MA01 with phosphatidylcholine model membranes. Chem. Phys. Lipids 2012, 165, 745–752. [Google Scholar] [CrossRef]
- Nasir, M.N.; Lins, L.; Crowet, J.-M.; Ongena, M.; Dorey, S.; Dhondt-Cordelier, S.; Clément, C.; Bouquillon, S.; Haudrechy, A.; Sarazin, C.; et al. Differential Interaction of Synthetic Glycolipids with Biomimetic Plasma Membrane Lipids Correlates with the Plant Biological Response. Langmuir 2017, 33, 9979–9987. [Google Scholar] [CrossRef]
- Monnier, N.; Furlan, A.L.; Buchoux, S.; Deleu, M.; Dauchez, M.; Rippa, S.; Sarazin, C. Exploring the Dual Interaction of Natural Rhamnolipids with Plant and Fungal Biomimetic Plasma Membranes through Biophysical Studies. Int. J. Mol. Sci. 2019, 20, 1009. [Google Scholar] [CrossRef]
- Deboever, E.; Deleu, M.; Mongrand, S.; Lins, L.; Fauconnier, M.-L. Plant–Pathogen Interactions: Underestimated Roles of Phyto-oxylipins. Trends Plant Sci. 2020, 25, 22–34. [Google Scholar] [CrossRef]
- Mamode Cassim, A.; Gouguet, P.; Gronnier, J.; Laurent, N.; Germain, V.; Grison, M.; Boutté, Y.; Gerbeau-Pissot, P.; Simon-Plas, F.; Mongrand, S. Plant lipids: Key players of plasma membrane organization and function. Prog. Lipid Res. 2019, 73, 1–27. [Google Scholar] [CrossRef]
- Deleu, M.; Crowet, J.-M.; Nasir, M.N.; Lins, L. Complementary biophysical tools to investigate lipid specificity in the interaction between bioactive molecules and the plasma membrane: A review. Biochim. Biophys. Acta Biomembr. 2014, 1838, 3171–3190. [Google Scholar] [CrossRef]
- Peetla, C.; Stine, A.; Labhasetwar, V. Biophysical Interactions with Model Lipid Membranes: Applications in Drug Discovery and Drug Delivery. Mol. Pharm. 2009, 6, 1264–1276. [Google Scholar] [CrossRef]
- Uemura, M.; Joseph, R.A.; Steponkus, P.L. Cold Acclimation of Arabidopsis thaliana (Effect on Plasma Membrane Lipid Composition and Freeze-Induced Lesions). Plant Physiol. 1995, 109, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Funnekotter, B.; Kaczmarczyk, A.; Turner, S.R.; Bunn, E.; Zhou, W.; Smith, S.; Flematti, G.; Mancera, R.L. Acclimation-induced changes in cell membrane composition and influence on cryotolerance of in vitro shoots of native plant species. Plant Cell Tissue Organ Cult. 2013, 114, 83–96. [Google Scholar] [CrossRef]
- Minami, A.; Fujiwara, M.; Furuto, A.; Fukao, Y.; Yamashita, T.; Kamo, M.; Kawamura, Y.; Uemura, M. Alterations in Detergent-Resistant Plasma Membrane Microdomains in Arabidopsis thaliana During Cold Acclimation. Plant Cell Physiol. 2009, 50, 341–359. [Google Scholar] [CrossRef] [PubMed]
- Yeagle, P.L. Laboratory Membrane Systems. In The Membranes of Cells; Elsevier: Amsterdam, The Netherlands, 2016; pp. 95–114. [Google Scholar]
- Siontorou, C.; Nikoleli, G.-P.; Nikolelis, D.; Karapetis, S. Artificial Lipid Membranes: Past, Present, and Future. Membranes 2017, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Israelachvili, J.N.; Marčelja, S.; Horn, R.G. Physical principles of membrane organization. Q. Rev. Biophys. 1980, 13, 121–200. [Google Scholar] [CrossRef]
- Traïkia, M.; Warschawski, D.E.; Recouvreur, M.; Cartaud, J.; Devaux, P.F. Formation of unilamellar vesicles by repetitive freeze-thaw cycles: Characterization by electron microscopy and 31P-nuclear magnetic resonance. Eur. Biophys. J. 2000, 29, 184–195. [Google Scholar] [CrossRef]
- Morales-Penningston, N.F.; Wu, J.; Farkas, E.R.; Goh, S.L.; Konyakhina, T.M.; Zheng, J.Y.; Webb, W.W.; Feigenson, G.W. GUV preparation and imaging: Minimizing artifacts. Biochim. Biophys. Acta Biomembr. 2010, 1798, 1324–1332. [Google Scholar] [CrossRef]
- Stein, H.; Spindler, S.; Bonakdar, N.; Wang, C.; Sandoghdar, V. Production of Isolated Giant Unilamellar Vesicles under High Salt Concentrations. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef]
- Opella, S.J.; Ma, C.; Marassi, F.M. Nuclear Magnetic Resonance of Membrane-Associated Peptides and Proteins. In Methods in Enzymolog; Elsevier: Amsterdam, The Netherlands, 2001; pp. 285–313. [Google Scholar]
- Bechinger, B.; Skladnev, D.A.; Ogrel, A.; Li, X.; Rogozhkina, E.V.; Ovchinnikova, T.V.; O’Neil, J.D.J.; Raap, J. 15N and 31P Solid-State NMR Investigations on the Orientation of Zervamicin II and Alamethicin in Phosphatidylcholine Membranes. Biochemistry 2001, 40, 9428–9437. [Google Scholar] [CrossRef] [PubMed]
- Marassi, F.M.; Crowell, K.J. Hydration-optimized oriented phospholipid bilayer samples for solid-state NMR structural studies of membrane proteins. J. Magn. Reson. 2003, 161, 64–69. [Google Scholar] [CrossRef]
- Cheng, H.-T.; Megha; London, E. Preparation and Properties of Asymmetric Vesicles That Mimic Cell Membranes. J. Biol. Chem. 2009, 284, 6079–6092. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; London, E. Preparation of Artificial Plasma Membrane Mimicking Vesicles with Lipid Asymmetry. PLoS ONE 2014, 9, e87903. [Google Scholar] [CrossRef]
- Drechsler, C.; Markones, M.; Choi, J.-Y.; Frieling, N.; Fiedler, S.; Voelker, D.R.; Schubert, R.; Heerklotz, H. Preparation of Asymmetric Liposomes Using a Phosphatidylserine Decarboxylase. Biophys. J. 2018, 115, 1509–1517. [Google Scholar] [CrossRef]
- Markones, M.; Drechsler, C.; Kaiser, M.; Kalie, L.; Heerklotz, H.; Fiedler, S. Engineering Asymmetric Lipid Vesicles: Accurate and Convenient Control of the Outer Leaflet Lipid Composition. Langmuir 2018, 34, 1999–2005. [Google Scholar] [CrossRef]
- Doktorova, M.; Heberle, F.A.; Eicher, B.; Standaert, R.F.; Katsaras, J.; London, E.; Pabst, G.; Marquardt, D. Preparation of asymmetric phospholipid vesicles for use as cell membrane models. Nat. Protoc. 2018, 13, 2086–2101. [Google Scholar] [CrossRef]
- Kamiya, K.; Takeuchi, S. Giant liposome formation toward the synthesis of well-defined artificial cells. J. Mater. Chem. B 2017, 5, 5911–5923. [Google Scholar] [CrossRef]
- Enoki, T.A.; Feigenson, G.W. Asymmetric Bilayers by Hemifusion: Method and Leaflet Behaviors. Biophys. J. 2019, 117, 1037–1050. [Google Scholar] [CrossRef]
- Naito, A.; Matsumori, N.; Ramamoorthy, A. Dynamic membrane interactions of antibacterial and antifungal biomolecules, and amyloid peptides, revealed by solid-state NMR spectroscopy. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 307–323. [Google Scholar] [CrossRef]
- Matsuoka, S.; Murata, M. Structural Studies of Small Bioactive Compounds Interacting with Membranes and Proteins. In Advances in Biological Solid-State NMR: Proteins and Membrane-Active Peptides; Separovic, F., Naito, A., Eds.; Royal Society of Chemistry: London, UK, 2014; pp. 133–161. [Google Scholar]
- Nagao, T.; Mishima, D.; Javkhlantugs, N.; Wang, J.; Ishioka, D.; Yokota, K.; Norisada, K.; Kawamura, I.; Ueda, K.; Naito, A. Structure and orientation of antibiotic peptide alamethicin in phospholipid bilayers as revealed by chemical shift oscillation analysis of solid state nuclear magnetic resonance and molecular dynamics simulation. Biochim. Biophys. Acta Biomembr. 2015, 1848, 2789–2798. [Google Scholar] [CrossRef] [PubMed]
- Salnikov, E.S.; Friedrich, H.; Li, X.; Bertani, P.; Reissmann, S.; Hertweck, C.; O’Neil, J.D.J.; Raap, J.; Bechinger, B. Structure and Alignment of the Membrane-Associated Peptaibols Ampullosporin A and Alamethicin by Oriented 15N and 31P Solid-State NMR Spectroscopy. Biophys. J. 2009, 96, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Bie, X.; Lv, F.; Zhao, H.; Lu, Z. Antifungal activity and mechanism of fengycin in the presence and absence of commercial surfactin against Rhizopus stolonifer. J. Microbiol. 2011, 49, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Heerklotz, H.; Seelig, J. Leakage and lysis of lipid membranes induced by the lipopeptide surfactin. Eur. Biophys. J. 2007, 36, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.S.; Ladizhansky, V. Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy. Protein Sci. 2015, 24, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Sani, M.-A.; Separovic, F. Solid-State NMR Studies of Antimicrobial Peptide Interactions with Specific Lipid Environments. In Advances in Biological Solid-State NMR: Proteins and Membrane-Active Peptides; Separovic, F., Naito, A., Eds.; Royal Society of Chemistry: London, UK, 2014; pp. 287–303. [Google Scholar]
- Salnikov, E.S.; Raya, J.; De Zotti, M.; Zaitseva, E.; Peggion, C.; Ballano, G.; Toniolo, C.; Raap, J.; Bechinger, B. Alamethicin Supramolecular Organization in Lipid Membranes from 19F Solid-State NMR. Biophys. J. 2016, 111, 2450–2459. [Google Scholar] [CrossRef]
- Bertelsen, K.; Dorosz, J.; Hansen, S.K.; Nielsen, N.C.; Vosegaard, T. Mechanisms of Peptide-Induced Pore Formation in Lipid Bilayers Investigated by Oriented 31P Solid-State NMR Spectroscopy. PLoS ONE 2012, 7, e47745. [Google Scholar] [CrossRef]
- Bertelsen, K.; Paaske, B.; Thøgersen, L.; Tajkhorshid, E.; Schiøtt, B.; Skrydstrup, T.; Nielsen, N.C.; Vosegaard, T. Residue-Specific Information about the Dynamics of Antimicrobial Peptides from 1H−15N and 2H Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2009, 131, 18335–18342. [Google Scholar] [CrossRef]
- Kim, S.; Cross, T.A. 2D solid state NMR spectral simulation of 310, α, and π-helices. J. Magn. Reson. 2004, 168, 187–193. [Google Scholar] [CrossRef]
- Marassi, F.M. A Simple Approach to Membrane Protein Secondary Structure and Topology based on NMR Spectroscopy. Biophys. J. 2001, 80, 994–1003. [Google Scholar] [CrossRef][Green Version]
- Dufourc, E.J.; Mayer, C.; Stohrer, J.; Althoff, G.; Kothe, G. Dynamics of phosphate head groups in biomembranes. Comprehensive analysis using phosphorus-31 nuclear magnetic resonance lineshape and relaxation time measurements. Biophys. J. 1992, 61, 42–57. [Google Scholar] [CrossRef]
- Davis, J.H. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim. Biophys. Acta Rev. Biomembr. 1983, 737, 117–171. [Google Scholar] [CrossRef]
- Davis, J.H. Deuterium magnetic resonance study of the gel and liquid crystalline phases of dipalmitoyl phosphatidylcholine. Biophys. J. 1979, 27, 339–358. [Google Scholar] [CrossRef]
- Kouzayha, A.; Nasir, M.N.; Buchet, R.; Wattraint, O.; Sarazin, C.; Besson, F. Conformational and Interfacial Analyses of K 3 A 18 K 3 and Alamethicin in Model Membranes. J. Phys. Chem. B 2009, 113, 7012–7019. [Google Scholar] [CrossRef]
- Liu, J.; Zou, A.; Mu, B. Surfactin effect on the physicochemical property of PC liposome. Colloids Surf. A Physicochem. Eng. Asp. 2010, 361, 90–95. [Google Scholar] [CrossRef]
- Brasseur, R.; Killian, J.A.; De Kruijff, B.; Ruysschaert, J.M. Conformational analysis of gramicidin-gramicidin interactions at the air/water interface suggests that gramicidin aggregates into tube-like structures similar as found in the gramicidin-induced hexagonal HII phase. Biochim. Biophys. Acta Biomembr. 1987, 903, 11–17. [Google Scholar] [CrossRef]
- Lins, L.; Brasseur, R. The hydrophobic effect in protein folding. FASEB J. 1995, 9, 535–540. [Google Scholar] [CrossRef]
- Lins, L.; Thomas-Soumarmon, A.; Pillot, T.; Vandekerckhove, J.; Rosseneu, M.; Brasseur, R. Molecular Determinants of the Interaction Between the C-Terminal Domain of Alzheimer’s β-Amyloid Peptide and Apolipoprotein E α-Helices. J. Neurochem. 2002, 73, 758–769. [Google Scholar] [CrossRef]
- Razafindralambo, H.; Blecker, C.; Mezdour, S.; Deroanne, C.; Crowet, J.-M.; Brasseur, R.; Lins, L.; Paquot, M. Impacts of the Carbonyl Group Location of Ester Bond on Interfacial Properties of Sugar-Based Surfactants: Experimental and Computational Evidences. J. Phys. Chem. B 2009, 113, 8872–8877. [Google Scholar] [CrossRef]
- Gallet, X.; Brasseur, R.; Deleu, M.; Razafindralambo, H.; Paquot, M.; Jacques, P.; Thonart, P.; Razafindralambo, H.; Jacques, P.; Thonart, P.; et al. Computer Simulation of Surfactin Conformation at a Hydrophobic/Hydrophilic Interface. Langmuir 1999, 15, 2409–2413. [Google Scholar] [CrossRef]
- Deleu, M.; Bouffioux, O.; Razafindralambo, H.; Paquot, M.; Hbid, C.; Thonart, P.; Jacques, P.; Brasseur, R. Interaction of Surfactin with Membranes: A Computational Approach. Langmuir 2003, 19, 3377–3385. [Google Scholar] [CrossRef]
- Nasir, M.N.; Crowet, J.-M.; Lins, L.; Obounou Akong, F.; Haudrechy, A.; Bouquillon, S.; Deleu, M. Interactions of sugar-based bolaamphiphiles with biomimetic systems of plasma membranes. Biochimie 2016, 130, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Zakanda, F.N.; Lins, L.; Nott, K.; Paquot, M.; Lelo, G.M.; Deleu, M. Interaction of Hexadecylbetainate Chloride with Biological Relevant Lipids. Langmuir 2012, 28, 3524–3533. [Google Scholar] [CrossRef] [PubMed]
- Lins, L.; Charloteaux, B.; Heinen, C.; Thomas, A.; Brasseur, R. “De Novo” Design of Peptides with Specific Lipid-Binding Properties. Biophys. J. 2006, 90, 470–479. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crowet, J.-M.; Lins, L.; Dupiereux, I.; Elmoualija, B.; Lorin, A.; Charloteaux, B.; Stroobant, V.; Heinen, E.; Brasseur, R. Tilted properties of the 67-78 fragment of α-synuclein are responsible for membrane destabilization and neurotoxicity. Proteins Struct. Funct. Bioinform. 2007, 68, 936–947. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; Van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef]
- Ermilova, I.; Lyubartsev, A.P. Extension of the Slipids Force Field to Polyunsaturated Lipids. J. Phys. Chem. B 2016, 120, 12826–12842. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Lagardère, L.; Jolly, L.-H.; Lipparini, F.; Aviat, F.; Stamm, B.; Jing, Z.F.; Harger, M.; Torabifard, H.; Cisneros, G.A.; Schnieders, M.J.; et al. Tinker-HP: A massively parallel molecular dynamics package for multiscale simulations of large complex systems with advanced point dipole polarizable force fields. Chem. Sci. 2018, 9, 956–972. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [PubMed]
- Ingólfsson, H.I.; Melo, M.N.; van Eerden, F.J.; Arnarez, C.; López, C.A.; Wassenaar, T.A.; Periole, X.; De Vries, A.H.; Tieleman, D.P.; Marrink, S.J. Lipid Organization of the Plasma Membrane. J. Am. Chem. Soc. 2014. [Google Scholar] [CrossRef] [PubMed]
- Risselada, H.J.; Marrink, S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA 2008, 105, 17367–17372. [Google Scholar] [CrossRef]
- Javanainen, M.; Martinez-Seara, H.; Vattulainen, I. Nanoscale Membrane Domain Formation Driven by Cholesterol. Sci. Rep. 2017, 7, 1143. [Google Scholar] [CrossRef] [PubMed]
- Morra, G.; Razavi, A.M.; Pandey, K.; Weinstein, H.; Menon, A.K.; Khelashvili, G. Mechanisms of Lipid Scrambling by the G Protein-Coupled Receptor Opsin. Structure 2018, 26, 356–367. [Google Scholar] [CrossRef]
- Sodt, A.J.; Pastor, R.W. Molecular Modeling of Lipid Membrane Curvature Induction by a Peptide: More than Simply Shape. Biophys. J. 2014, 106, 1958–1969. [Google Scholar] [CrossRef]
- Marrink, S.J.; Corradi, V.; Souza, P.C.T.; Ingólfsson, H.I.; Tieleman, D.P.; Sansom, M.S.P. Computational Modeling of Realistic Cell Membranes. Chem. Rev. 2019, 119, 6184–6226. [Google Scholar] [CrossRef]
- Gronnier, J.; Crowet, J.-M.; Habenstein, B.; Nasir, M.N.; Bayle, V.; Hosy, E.; Platre, M.P.; Gouguet, P.; Raffaele, S.; Martinez, D.; et al. Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Deleu, M.; Deboever, E.; Nasir, M.N.; Crowet, J.-M.; Dauchez, M.; Ongena, M.; Jijakli, H.; Fauconnier, M.-L.; Lins, L. Linoleic and linolenic acid hydroperoxides interact differentially with biomimetic plant membranes in a lipid specific manner. Colloids Surf. B Biointerfaces 2019, 175, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Bobone, S.; De Zotti, M.; Bortolotti, A.; Biondi, B.; Ballano, G.; Palleschi, A.; Toniolo, C.; Formaggio, F.; Stella, L. The fluorescence and infrared absorption probe para -cyanophenylalanine: Effect of labeling on the behavior of different membrane-interacting peptides. Biopolymers 2015, 104, 521–532. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stella, L.; Burattini, M.; Mazzuca, C.; Palleschi, A.; Venanzi, M.; Coin, I.; Peggion, C.; Toniolo, C.; Pispisa, B. Alamethicin Interaction with Lipid Membranes: A Spectroscopic Study on Synthetic Analogues. Chem. Biodivers. 2007, 4, 1299–1312. [Google Scholar] [CrossRef]
- Rausch, J.M.; Wimley, W.C. A High-Throughput Screen for Identifying Transmembrane Pore-Forming Peptides. Anal. Biochem. 2001, 293, 258–263. [Google Scholar] [CrossRef]
- Nasir, M.N.; Laurent, P.; Flore, C.; Lins, L.; Ongena, M.; Deleu, M. Analysis of calcium-induced effects on the conformation of fengycin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 110, 450–457. [Google Scholar] [CrossRef]
- Zhao, X.; Li, R.; Lu, C.; Baluška, F.; Wan, Y. Di-4-ANEPPDHQ, a fluorescent probe for the visualisation of membrane microdomains in living Arabidopsis thaliana cells. Plant Physiol. Biochem. 2015, 87, 53–60. [Google Scholar] [CrossRef]
- D’Auria, L.; Deleu, M.; Dufour, S.; Mingeot-Leclercq, M.-P.; Tyteca, D. Surfactins modulate the lateral organization of fluorescent membrane polar lipids: A new tool to study drug:membrane interaction and assessment of the role of cholesterol and drug acyl chain length. Biochim. Biophys. Acta Biomembr. 2013, 1828, 2064–2073. [Google Scholar] [CrossRef]
- Kyrychenko, A. Using fluorescence for studies of biological membranes: A review. Methods Appl. Fluoresc. 2015, 3, 042003. [Google Scholar] [CrossRef]
- Patel, H.; Huynh, Q.; Bärlehner, D.; Heerklotz, H. Additive and Synergistic Membrane Permeabilization by Antimicrobial (Lipo)Peptides and Detergents. Biophys. J. 2014, 106, 2115–2125. [Google Scholar] [CrossRef]
- Uttlová, P.; Pinkas, D.; Bechyňková, O.; Fišer, R.; Svobodová, J.; Seydlová, G. Bacillus subtilis alters the proportion of major membrane phospholipids in response to surfactin exposure. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2965–2971. [Google Scholar] [CrossRef] [PubMed]
- González-Jaramillo, L.M.; Aranda, F.J.; Teruel, J.A.; Villegas-Escobar, V.; Ortiz, A. Antimycotic activity of fengycin C biosurfactant and its interaction with phosphatidylcholine model membranes. Colloids Surf. B Biointerfaces 2017, 156, 114–122. [Google Scholar] [CrossRef]
- Kikukawa, T.; Araiso, T. Changs in lipid mobility associated with alamethicin incorporation into membranes. Arch. Biochem. Biophys. 2002, 405, 214–222. [Google Scholar] [CrossRef]
- Fiedler, S.; Heerklotz, H. Vesicle Leakage Reflects the Target Selectivity of Antimicrobial Lipopeptides from Bacillus subtilis. Biophys. J. 2015, 109, 2079–2089. [Google Scholar] [CrossRef] [PubMed]
- Haba, E.; Pinazo, A.; Pons, R.; Pérez, L.; Manresa, A. Complex rhamnolipid mixture characterization and its influence on DPPC bilayer organization. Biochim. Biophys. Acta Biomembr. 2014, 1838, 776–783. [Google Scholar] [CrossRef]
- Faust, J.E.; Yang, P.-Y.; Huang, H.W. Action of Antimicrobial Peptides on Bacterial and Lipid Membranes: A Direct Comparison. Biophys. J. 2017, 112, 1663–1672. [Google Scholar] [CrossRef]
- Patel, H.; Tscheka, C.; Edwards, K.; Karlsson, G.; Heerklotz, H. All-or-none membrane permeabilization by fengycin-type lipopeptides from Bacillus subtilis QST713. Biochim. Biophys. Acta Biomembr. 2011, 1808, 2000–2008. [Google Scholar] [CrossRef]
- Haapalainen, M.; Engelhardt, S.; Küfner, I.; Li, C.-M.; Nürnberger, T.; Lee, J.; Romantschuk, M.; Taira, S. Functional mapping of harpin HrpZ of Pseudomonas syringae reveals the sites responsible for protein oligomerization, lipid interactions and plant defence induction. Mol. Plant Pathol. 2011, 12, 151–166. [Google Scholar] [CrossRef]
- do Canto, A.M.T.M.; Robalo, J.R.; Santos, P.D.; Carvalho, A.J.P.; Ramalho, J.P.P.; Loura, L.M.S. Diphenylhexatriene membrane probes DPH and TMA-DPH: A comparative molecular dynamics simulation study. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2647–2661. [Google Scholar] [CrossRef]
- Kaiser, R.D.; London, E. Location of Diphenylhexatriene (DPH) and Its Derivatives within Membranes: Comparison of Different Fluorescence Quenching Analyses of Membrane Depth. Biochemistry 1998, 37, 8180–8190. [Google Scholar] [CrossRef]
- Das, M.K.; Balaram, P. Interactions of the channel forming peptide alamethicin with artificial and natural membranes. J. Biosci. 1984, 6, 337–348. [Google Scholar] [CrossRef]
- Lentz, B.R. Use of fluorescent probes to monitor molecular order and motions within liposome bilayers. Chem. Phys. Lipids 1993, 64, 99–116. [Google Scholar] [CrossRef]
- Oliva, A.; Teruel, J.A.; Aranda, F.J.; Ortiz, A. Effect of a dirhamnolipid biosurfactant on the structure and phase behaviour of dimyristoylphosphatidylserine model membranes. Colloids Surf. B Biointerfaces 2020, 185, 110576. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, T.; Hunt, G.; Farkas, E.R.; Webb, W.W.; Feigenson, G.W. Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochim. Biophys. Acta Biomembr. 2007, 1768, 2182–2194. [Google Scholar] [CrossRef] [PubMed]
- Bagatolli, L.A.; Gratton, E. Direct observation of lipid domains in free-standing bilayers using two-photon excitation fluorescence microscopy. J. Fluoresc. 2001, 11, 141–160. [Google Scholar] [CrossRef]
- Parasassi, T.; Krasnowska, E.K.; Bagatolli, L.; Gratton, E. Laurdan and Prodan as Polarity-Sensitive Fluorescent Membrane Probes. J. Fluoresc. 1998, 8, 365–373. [Google Scholar] [CrossRef]
- Sezgin, E.; Schwille, P. Fluorescence Techniques to Study Lipid Dynamics. Cold Spring Harb. Perspect. Biol. 2011, 3, a009803. [Google Scholar] [CrossRef]
- Bagatolli, L.A. To see or not to see: Lateral organization of biological membranes and fluorescence microscopy. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1541–1556. [Google Scholar] [CrossRef]
- Jin, L.; Millard, A.C.; Wuskell, J.P.; Clark, H.A.; Loew, L.M. Cholesterol-Enriched Lipid Domains Can Be Visualized by di-4-ANEPPDHQ with Linear and Nonlinear Optics. Biophys. J. 2005, 89, L04–L06. [Google Scholar] [CrossRef]
- Dinic, J.; Biverståhl, H.; Mäler, L.; Parmryd, I. Laurdan and di-4-ANEPPDHQ do not respond to membrane-inserted peptides and are good probes for lipid packing. Biochim. Biophys. Acta Biomembr. 2011, 1808, 298–306. [Google Scholar] [CrossRef]
- Amaro, M.; Reina, F.; Hof, M.; Eggeling, C.; Sezgin, E. Laurdan and Di-4-ANEPPDHQ probe different properties of the membrane. J. Phys. D Appl. Phys. 2017, 50, 134004. [Google Scholar] [CrossRef]
- Kahya, N.; Scherfeld, D.; Bacia, K.; Poolman, B.; Schwille, P. Probing Lipid Mobility of Raft-exhibiting Model Membranes by Fluorescence Correlation Spectroscopy. J. Biol. Chem. 2003, 278, 28109–28115. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xing, J.; Qiu, Z.; He, Q.; Lin, J. Quantification of Membrane Protein Dynamics and Interactions in Plant Cells by Fluorescence Correlation Spectroscopy. Mol. Plant 2016, 9, 1229–1239. [Google Scholar] [CrossRef]
- Elson, E.L. Fluorescence Correlation Spectroscopy: Past, Present, Future. Biophys. J. 2011, 101, 2855–2870. [Google Scholar] [CrossRef] [PubMed]
- Schwille, P.; Meyer-Almes, F.J.; Rigler, R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997, 72, 1878–1886. [Google Scholar] [CrossRef]

| Fluorescent Probes | λexcitation (nm) | λemission (nm) | Membrane Location | Information | Refs |
|---|---|---|---|---|---|
| Calcein | 495 | 515 | Aqueous core | Permeabilization/solubilization | [67,114] |
| Carboxyfluorescein | 490–500 | 515–520 | Aqueous core | Permeabilization/solubilization | [115] |
| Di-4-ANEPPDHQ | 488 | 560–570 (Lβ′ phase) 610–630 (Lα phase) | Membrane surface | Lipid order, lipid phases, membrane dynamics | [8,20] |
| DPH | 358 | 430 | Hydrophobic core | Membrane dynamics, gel-to-fluid transition temperature | [34,116] |
| DPH-PC (“lipid-like”) | 350 | 430 | Hydrophobic core | Lipid dynamics | [117] |
| Laurdan | 340 | 440 (Lβ′ phase) 490 (Lα phase) | Membrane surface | Lipid order, lipid phases, membrane dynamics | [28] |
| NBD-PE (“lipid-like”) | 450 | 560 | Lipid/water interface | Lipid dynamics | [117] |
| TMA-DPH | 360 | 435 | Lipid/water interface | Membrane dynamics, gel-to-fluid transition temperature | [34] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furlan, A.L.; Laurin, Y.; Botcazon, C.; Rodríguez-Moraga, N.; Rippa, S.; Deleu, M.; Lins, L.; Sarazin, C.; Buchoux, S. Contributions and Limitations of Biophysical Approaches to Study of the Interactions between Amphiphilic Molecules and the Plant Plasma Membrane. Plants 2020, 9, 648. https://doi.org/10.3390/plants9050648
Furlan AL, Laurin Y, Botcazon C, Rodríguez-Moraga N, Rippa S, Deleu M, Lins L, Sarazin C, Buchoux S. Contributions and Limitations of Biophysical Approaches to Study of the Interactions between Amphiphilic Molecules and the Plant Plasma Membrane. Plants. 2020; 9(5):648. https://doi.org/10.3390/plants9050648
Chicago/Turabian StyleFurlan, Aurélien L., Yoann Laurin, Camille Botcazon, Nely Rodríguez-Moraga, Sonia Rippa, Magali Deleu, Laurence Lins, Catherine Sarazin, and Sébastien Buchoux. 2020. "Contributions and Limitations of Biophysical Approaches to Study of the Interactions between Amphiphilic Molecules and the Plant Plasma Membrane" Plants 9, no. 5: 648. https://doi.org/10.3390/plants9050648
APA StyleFurlan, A. L., Laurin, Y., Botcazon, C., Rodríguez-Moraga, N., Rippa, S., Deleu, M., Lins, L., Sarazin, C., & Buchoux, S. (2020). Contributions and Limitations of Biophysical Approaches to Study of the Interactions between Amphiphilic Molecules and the Plant Plasma Membrane. Plants, 9(5), 648. https://doi.org/10.3390/plants9050648






