Non-Destructive Measurement of Three-Dimensional Plants Based on Point Cloud
Abstract
1. Introduction
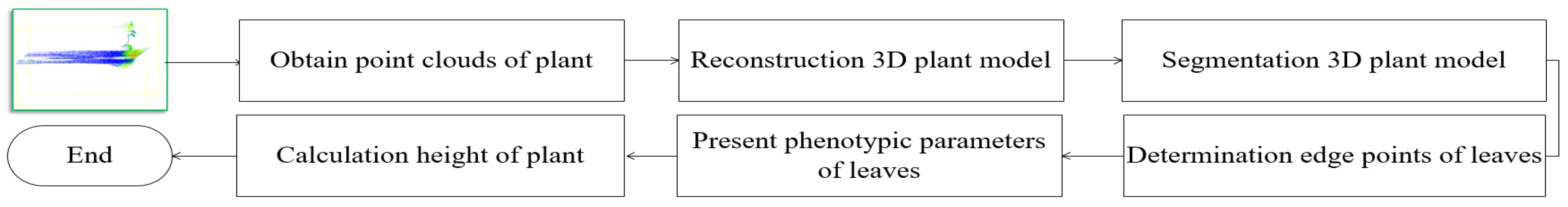
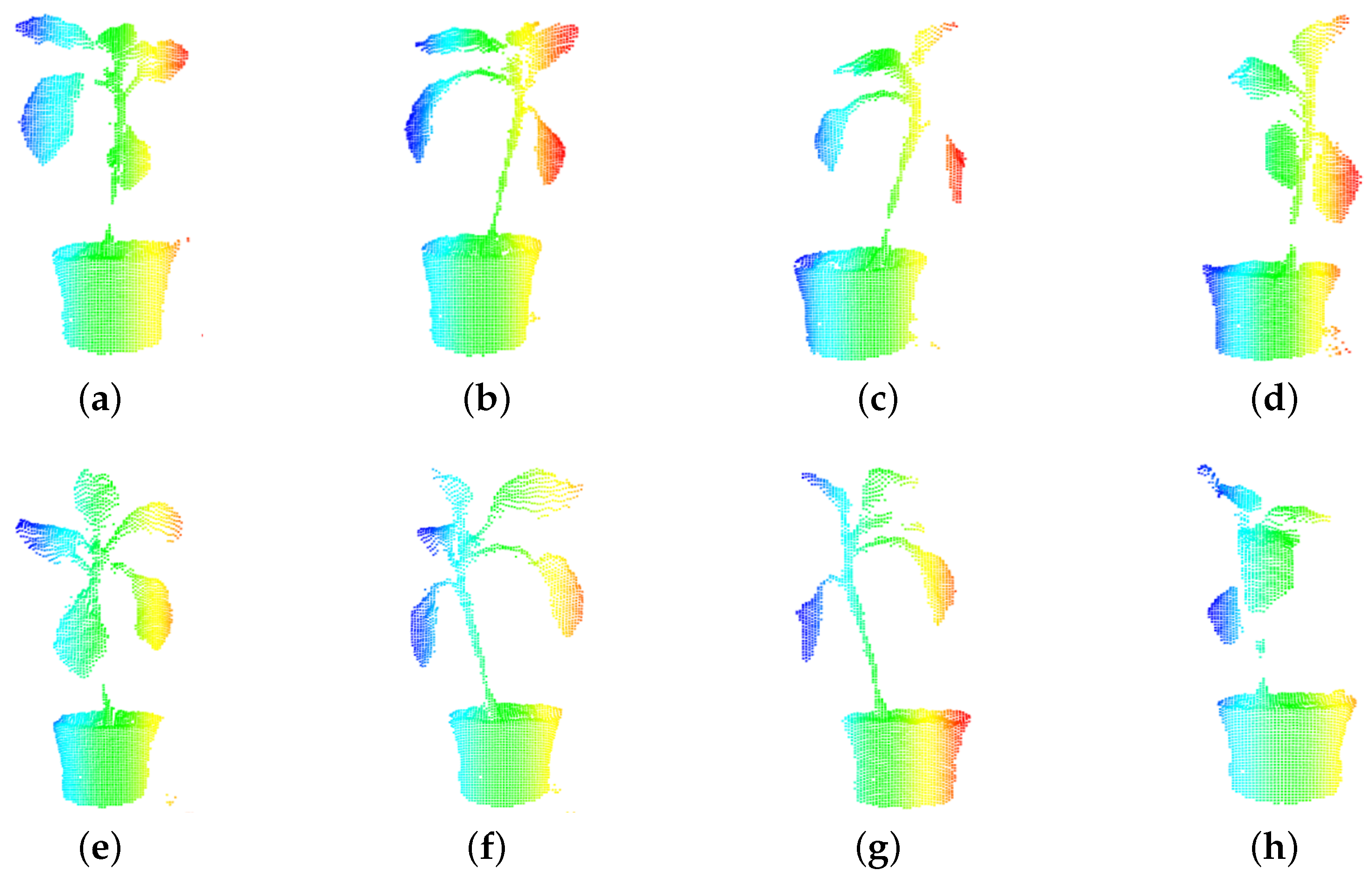
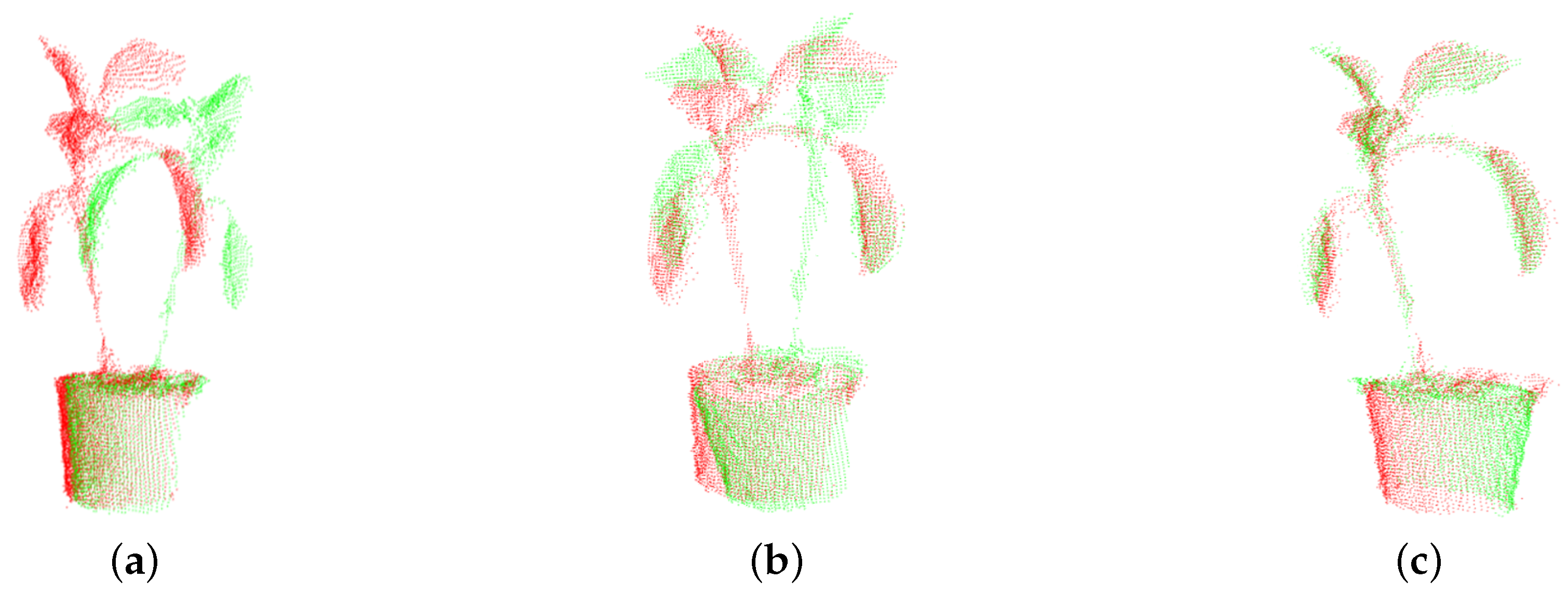
2. Preprocessing of a Three-Dimensional Plant Model
3. Measurement the Phenotypic Parameters of a Plant
3.1. Extraction of Boundary Points on a Plant Leaf
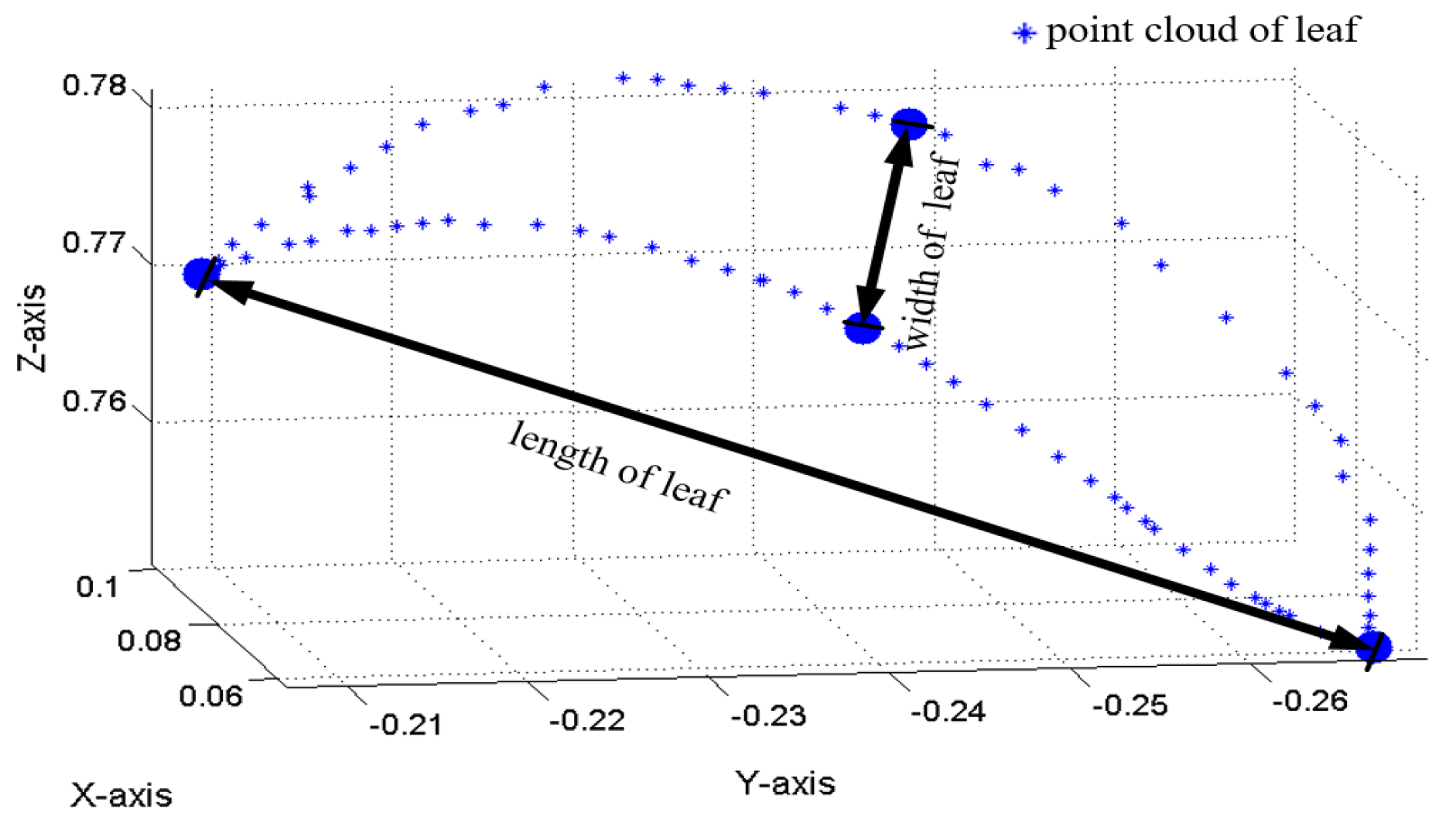
3.2. Calculation the Length, Width and Surface Area of Leaf
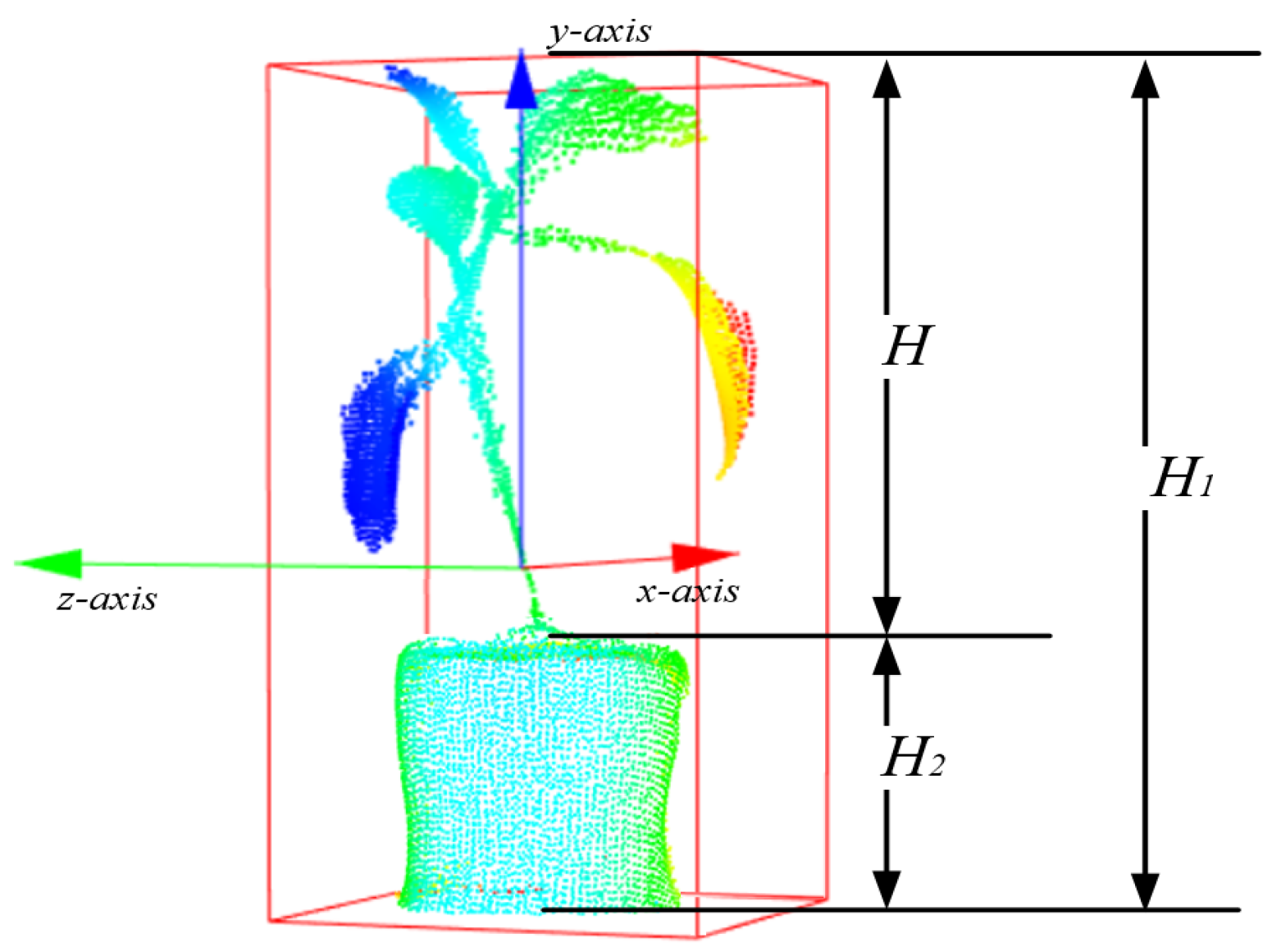
3.3. Measurement the Height of a Plant
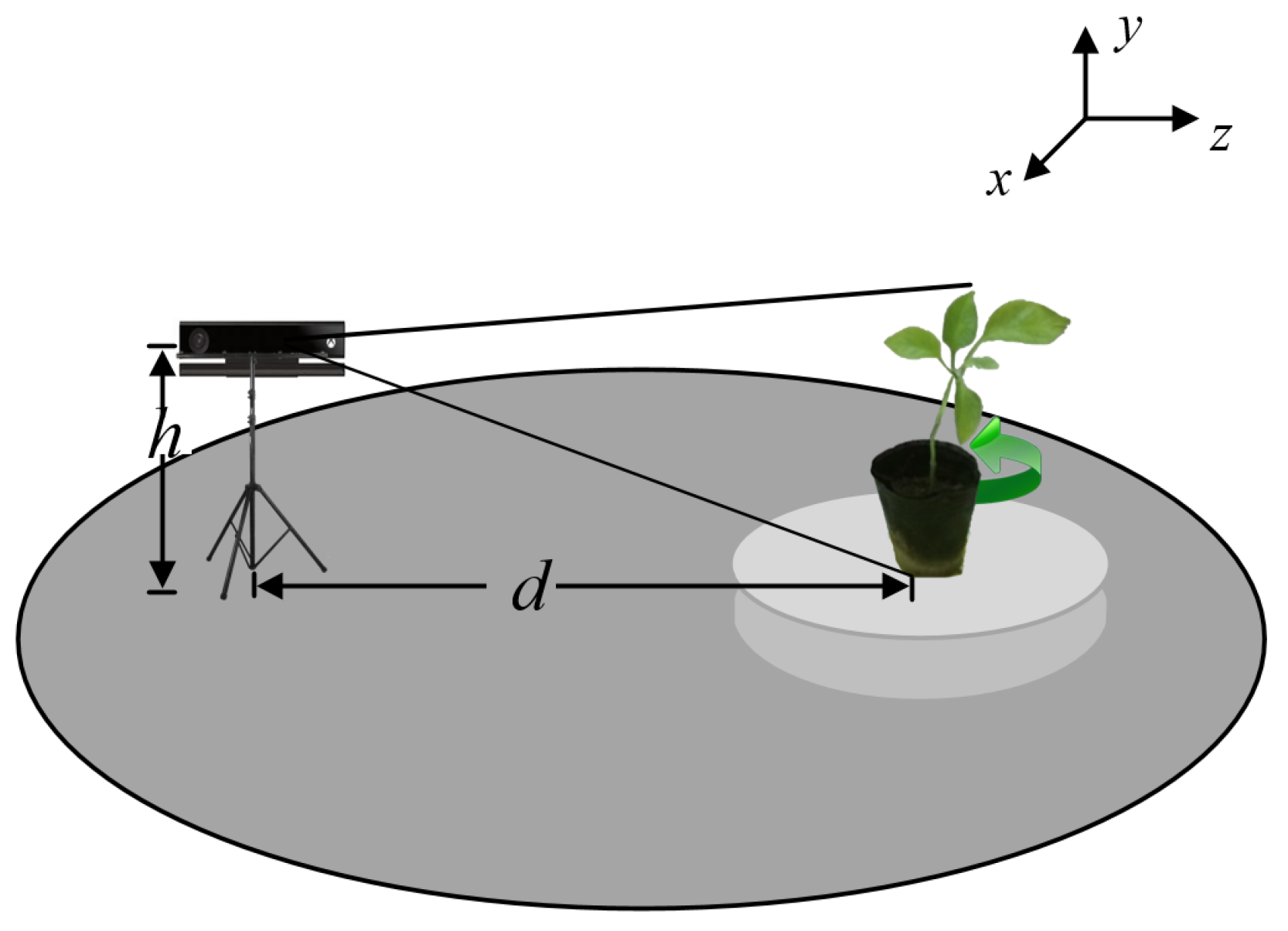
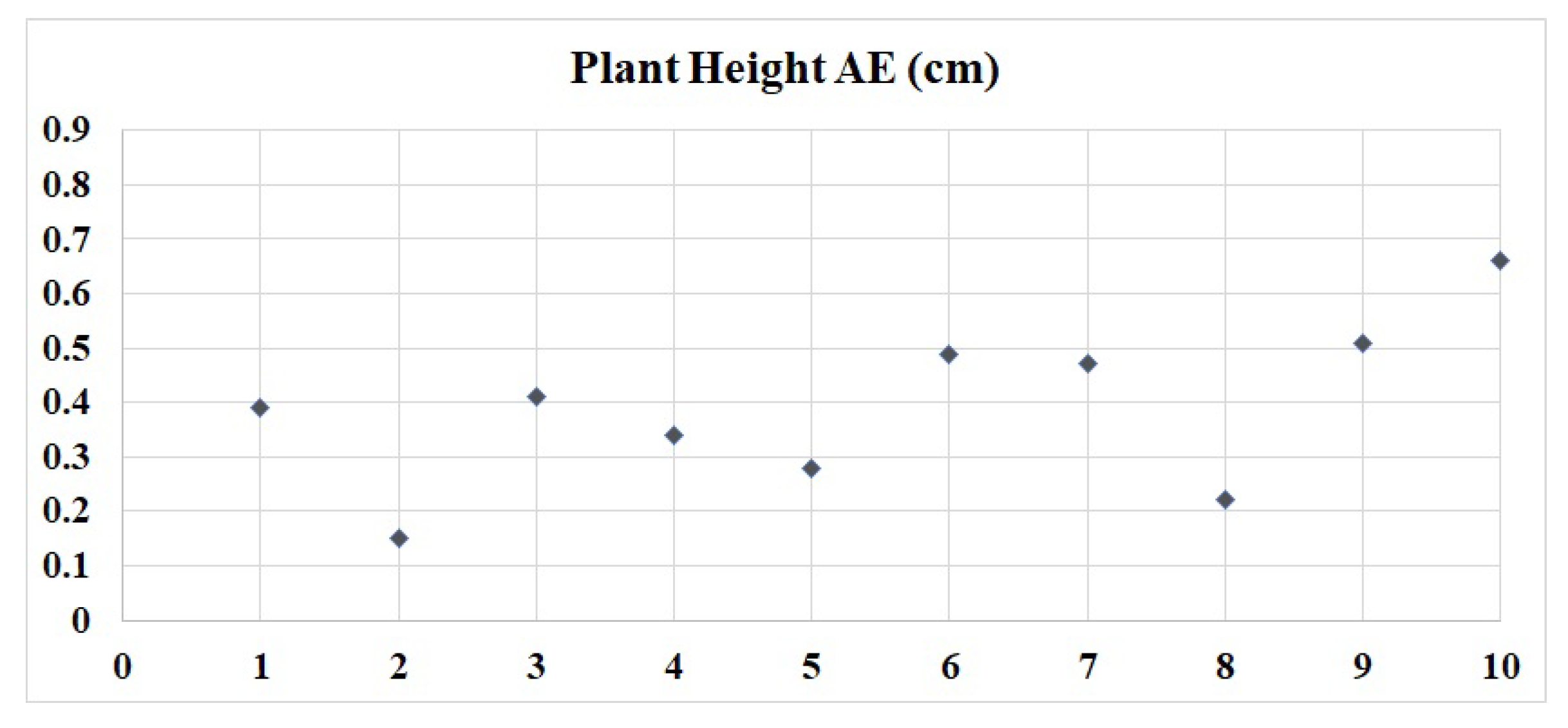
4. Experiment
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| 3D | Three-dimensional |
| RGB-D | Red, Green, Blue, and Depth |
| TOF | Time of Flight |
| ROI | Region of Interest |
| v2.0 | Version 2.0 |
| ICP | Iterative Closet Point |
| MLS | Moving Least Squares |
| Pos | Position |
| FLANN | Fast Library for Approximate Nearest Neighbors |
| LCCP | Locally Convex Connected Patches |
| min | minimum |
| max | maximum |
| AE | Absolute Error |
| RMSE | Root Mean Squared Error |
| MAE | Mean Absolute Error |
References
- Chaudhury, A.; Ward, C.; Talasaz, A.; Ivanov, A.G.; Huner, N.P.; Grodzinski, B.; Patel, R.V.; Barron, J.L. Computer Vision Based Autonomous Robotic System for 3D Plant Growth Measurement. In Proceedings of the 12th Conference on Computer and Robot Vision, Halifax, NS, Canada, 3–5 June 2015. [Google Scholar]
- Scharr, H.; Minervini, M.; French, A.P.; Klukas, C.; Kramer, D.M.; Liu, X.M.; Luengo, I.; Pape, J.M.; Polder, G.; Vukadinovic, D.; et al. Leaf segmentation in plant phenotyping: A collation study. Mach. Vis. Appl. 2016, 27, 585–606. [Google Scholar] [CrossRef]
- Kaiyan, L.; Lihong, X.; Junhui, W. Advances in the application of computer-vision to plant growth monitoring. Trans. Chin. Soc. Agric. Eng. 2004, 20, 279–283. [Google Scholar]
- Mishra, K.B.; Mishra, A.; Klem, K.; Govindjee, G. Plant phenotyping: A perspective. Ind. J. Plant Physiol. 2016, 21, 514–527. [Google Scholar] [CrossRef]
- Qiu, R.; Wei, S.; Zhang, M.; Li, H.; Li, M. Sensors for measuring plant phenotyping: A review. Int. J. Agric. Biol. Eng. 2018, 11, 1–17. [Google Scholar] [CrossRef]
- Sritarapipat, T.; Rakwatin, P.; Kasetkasem, T. Automatic Rice Crop Height Measurement Using a Field Server and Digital Image Processing. Sensors 2014, 14, 900–926. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kato, K.; Haga, S.; Misawa, T. Growth Estimation of Transplanted Paddy Rice by Digital Camera Image. Tohoku J. Crop Sci. 2001, 44, 77–78. [Google Scholar]
- Liping, G.; Aijun, X. Tree Height Measurement Method with Intelligent Terminal. J. Northeast Forest. Univ. 2018, 46, 28–34. [Google Scholar]
- Constantino, K.P.; Gonzales, E.J.; Lazaro, L.M.; Serrano, E.C.; Samson, B.P. Towards an Automated Plant Height Measurement and Tiller Segmentation of Rice Crops using Image Processing. In Mechatronics and Machine Vision in Practice 3; Billingsley, J., Brett, P., Eds.; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Qiu, R.; Miao, Y.; Ji, Y.; Zhang, M.; Li, H.; Liu, G. Measurement of Individual Maize Height Based on RGB-D Camera. Trans. Chin. Soc. Agric. Mach. 2017, 48, 211–219. [Google Scholar]
- Thi Phan, A.T.; Takahashi, K.; Rikimaru, A.; Higuchi, Y. Method for estimating rice plant height without ground surface detection using laser scanner measurement. J. Appl. Remote Sens. 2016, 10, 046018. [Google Scholar] [CrossRef]
- Li, H.; Wang, K.; Cao, Q.; Bian, H. Measurement of plant height based on stereoscopic vision under the condition of a single camera. In Proceedings of the Intelligent Control & Automation, Jinan, China, 6–9 July 2010. [Google Scholar]
- Young, K.C.; Zaman, Q.; Farooque, A.; Rehman, T.U.; Esau, T. An on-the-go ultrasonic plant height measurement system (UPHMS II) in the wild blueberry cropping system. Biosyst. Eng. 2017, 157, 35–44. [Google Scholar]
- Jiang, Y.; Li, C.; Paterson, A.H. High throughput phenotyping of cotton plant height using depth images under field conditions. Comp. Electron. Agric. 2016, 130, 57–68. [Google Scholar] [CrossRef]
- Yang, S.; Gao, W.; Mi, J.; Wu, M.; Wang, M.; Zheng, L. Method for Measurement of Vegetable Seedlings Height Based on RGB-D Camera. Trans. Chin. Soc. Agric. Mach. 2019, 50, 128–135. [Google Scholar]
- Vázquez-Arellano, M.; Paraforos, D.S.; Reiser, D.; Garrido-lzard, M.; Griepentrog, H.W. Determination of stem position and height of reconstructed maize plants using a time-of-flight camera. Comp. Electron. Agric. 2018, 154, 276–288. [Google Scholar] [CrossRef]
- Hämmerle, M.; Höfle, B. Direct derivation of maize plant and crop height from low-cost time-of-flight camera measurements. Plant Methods 2016, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Leemans, V.; Dumont, B.; Destain, M.F. Assessment of plant leaf area measurement by using stereo-vision. In Proceedings of the International Conference on 3D Imaging (IC3D), Liege, Belgium, 3–5 December 2013. [Google Scholar]
- Xia, Y.; Xu, D.; Du, J.; Zhang, L.; Wang, A. On-line measurement of tobacco leaf area based on machine vision. Nongye Jixie Xuebao/Trans. Chin. Soc. Agric. Mach. 2012, 43, 167–173. [Google Scholar]
- Lin, K.Y.; Wu, J.H.; Chen, J.; Si, H.P. Measurement of Plant Leaf Area Based on Computer Vision. In Proceedings of the Sixth International Conference on Measuring Technology and Mechatronics Automation, Zhangjiajie, China, 10–11 January 2014. [Google Scholar]
- Zhong, Q.; Zhou, P.; Fu, B.; Liu, K. Measurement of pest-damaged area of leaf based on auto-matching of representative leaf. Trans. Chin. Soc. Agric. Eng. 2010, 26, 216–221. [Google Scholar]
- Liu, Z.C.; Xu, L.H.; Lin, C.F. An Improved Stereo Matching Algorithm Applied to 3D Visualization of Plant Leaf. In Proceedings of the 8th International Symposium on Computational Intelligence and Design (ISCID), Hangzhou, China, 12–13 December 2015. [Google Scholar]
- Zhao, L.; Yang, L.L.; Cui, S.G.; Wu, X.L.; Liang, F.; Tian, L.G. Method for Non-Destructive Measurement of Leaf Area Based on Binocular Vision. Appl. Mech. Mater. 2014, 577, 664–667. [Google Scholar] [CrossRef]
- Paproki, A.; Sirault, X.; Berry, S.; Furbank, R.; Fripp, J. Novel mesh processing based technique for 3D plant analysis. BMC Plant Biol. 2012, 12, 63. [Google Scholar] [CrossRef]
- Hui, F.; Zhu, J.Y.; Hu, P.C.; Meng, L.; Zhu, B.L.; Guo, Y.; Li, B.G.; Ma, Y.T. Image-based dynamic quantification and high-accuracy 3D evaluation of canopy structure of plant populations. Ann. Bot. 2018, 121, 1079–1088. [Google Scholar] [CrossRef]
- Paulus, S.; Dupuis, J.; Mahlein, A.-K.; Kuhlmann, H. Surface feature based classification of plant organs from 3D laserscanned point clouds for plant phenotyping. BMC Bioinform. 2013, 14, 238. [Google Scholar] [CrossRef]
- Liu, H.; Liu, J.L.; Shen, Y.; Pan, C.K. Segmentation Method of Supervoxel Clusterings and Salient Map. Trans. Chin. Soc. Agric. Mach. 2018, 12, 172–179. [Google Scholar]
- Gang, L.; Weijie, Z.; Cailing, G. Apple Leaf Point Cloud Clustering Based on Dynamic-K-threshold and Growth Parameters Extraction. Trans. Chin. Soc. Agric. Mach. 2019, 50, 163–169, 178. [Google Scholar]
- Wahabzada, M.; Paulus, S.; Kersting, K.; Mahlein, A.K. Automated interpretation of 3D laserscanned point clouds for plant organ segmentation. BMC Bioinform. 2015, 16, 248. [Google Scholar] [CrossRef] [PubMed]
- Gélard, W.; Herbulot, A.; Devy, M.; Debaeke, P.; Mccormick, R.F.; Truong, S.K.; Mullet, J. Leaves Segmentation in 3D Point Cloud. In Proceedings of the International Conference on Advanced Concepts for Intelligent Vision Systems, Antwerp, Belgium, 18–21 September 2017. [Google Scholar]
- Gelard, W.; Devy, M.; Herbulot, A.; Burger, P. Model-based Segmentation of 3D Point Clouds for Phenotyping Sunflower Plants. In Proceedings of the International Joint Conference on Computer Vision Imaging and Computer Graphics Theory and Applications, Porto, Portugal, 27 February–1 March 2017; pp. 459–467. [Google Scholar]
- Vázquez-Arellano, M.; Reiser, D.; Paraforos, D.S.; Garrido-Izard, M.; Burce, M.E.C.; Griepentrog, H.W. 3-D reconstruction of maize plants using a time-of-flight camera. Comp. Electron. Agric. 2018, 145, 235–247. [Google Scholar] [CrossRef]
- Bao, Y.; Tang, L.; Shah, D. Robotic 3D Plant Perception and Leaf Probing with Collision-Free Motion Planning for Automated Indoor Plant Phenotyping. In Proceedings of the ASABE International Meeting, Spokane, WA, USA, 16–19 July 2017. [Google Scholar]
- Dupuis, J.; Paulus, S.; Behmann, J.; Plumer, L.; Kuhlmann, H. A Multi-Resolution Approach for an Automated Fusion of Different Low-Cost 3D Sensors. Sensors 2014, 14, 7563–7579. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Shi, L.; Ma, Y.; Deng, Q.; Li, B.; Guo, Y. Three-dimensional digitization in situ of rice canopies and virtual stratified-clipping method. Sci. Agric. Sin. 2009, 42, 1181–1189. [Google Scholar]
- Shah, D.; Tang, L.; Gai, J.; Putta-Venkata, R. Development of a Mobile Robotic Phenotyping System for Growth Chamber-based Studies of Genotype x Environment Interactions. IFAC Papers OnLine 2016, 49, 248–253. [Google Scholar] [CrossRef]
- Andújar, D.; Dorado, J.; Ribeiro, A. An Approach to the Use of Depth Cameras for Weed Volume Estimation. Sensors 2016, 16, 972. [Google Scholar] [CrossRef]
- Yamamoto, S.; Hayashi, S.; Saito, S.; Ochiai, Y. Measurement of growth information of a strawberry plant using a natural interaction device. In Proceedings of the American Society of Agricultural and Biological Engineers Annual International Meeting, Dallas, TX, USA, 29 July–1 August 2012; pp. 5547–5556. [Google Scholar]
- Li, X.; Wang, X.; Wei, H.; Zhu, X.; Huang, H. A technique system for the measurement, reconstruction and character extraction of rice plant architecture. PLoS ONE 2017, 12, e0177205. [Google Scholar] [CrossRef]
- Sun, G.; Wang, X. Three-Dimensional Point Cloud Reconstruction and Morphology Measurement Method for Greenhouse Plants Based on the Kinect Sensor Self-Calibration. Agronomy 2019, 9, 596. [Google Scholar] [CrossRef]
- Heming, J.; Yujia, M.; Zhikai, X.; Baizhuo, Z.; Xiaoxu, P.; Jinduo, L.I. Reconstruction of three dimensional model of plant based on point cloud stitching. Appl. Sci. Technol. 2019, 46, 19–24. [Google Scholar]
- Yang, M.; Cui, J.; Jeong, E.S.; Cho, S.I. Development of High-resolution 3D Phenotyping System Using Depth Camera and RGB Camera. In Proceedings of the ASABE International Meeting, Spokane, WA, USA, 16–19 July 2017. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Xiang, L.; Wu, Q.; Jiang, H. Automatic Non-Destructive Growth Measurement of Leafy Vegetables Based on Kinect. Sensors 2018, 18, 806. [Google Scholar] [CrossRef] [PubMed]
- Rusu, R.B.; Marton, Z.C.; Blodow, N.; Dollha, M. Towards 3D Point Cloud Based Object Maps for Household Environments. Robot. Auton. Syst. 2008, 56, 927–941. [Google Scholar] [CrossRef]
- Besl, P.J.; Mckay, H.D. A method for registration of 3-D shapes. IEEE Trans. Pattern Anal. Mach. Intell. 1992, 14, 239–256. [Google Scholar] [CrossRef]
- Yawei, W.; Yifei, C. Fruit Morphological Measurement Based on Three-Dimensional Reconstruction. Agronomy 2020, 10, 455. [Google Scholar]
- Gelfand, N.; Ikemoto, L.; Rusinkiewicz, S.; Levoy, M. Geometrically Stable Sampling for the ICP Algorithm. In Proceedings of the Fourth International Conference on 3-D Digital Imaging and Modeling, Banff, AB, Canada, 6–10 October 2003; pp. 260–267. [Google Scholar]
- Lancaster, P.; Salkauskas, K. Surfaces generated by moving least squares methods. Math. Comput. 1981, 37, 141–158. [Google Scholar] [CrossRef]
- Available online: http://www.pointclouds.org/documentation/tutorials/resampling.php (accessed on 5 January 2020).
- Available online: http://docs.pointclouds.org/trunk/kdtree__flann_8hpp_source.html (accessed on 24 January 2020).
- Stein, S.C.; Wörgötter, F.; Schoeler, M. Convexity based object partitioning for robot applications. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA), Hong Kong, China, 31 May–7 June 2014. [Google Scholar]
- Hu, Z. Research on Boundary Detection and Hole Repairing Based on 3D Laser Scanning Point Cloud; China University of Mining and Technology: Xuzhou, China, 2016. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |


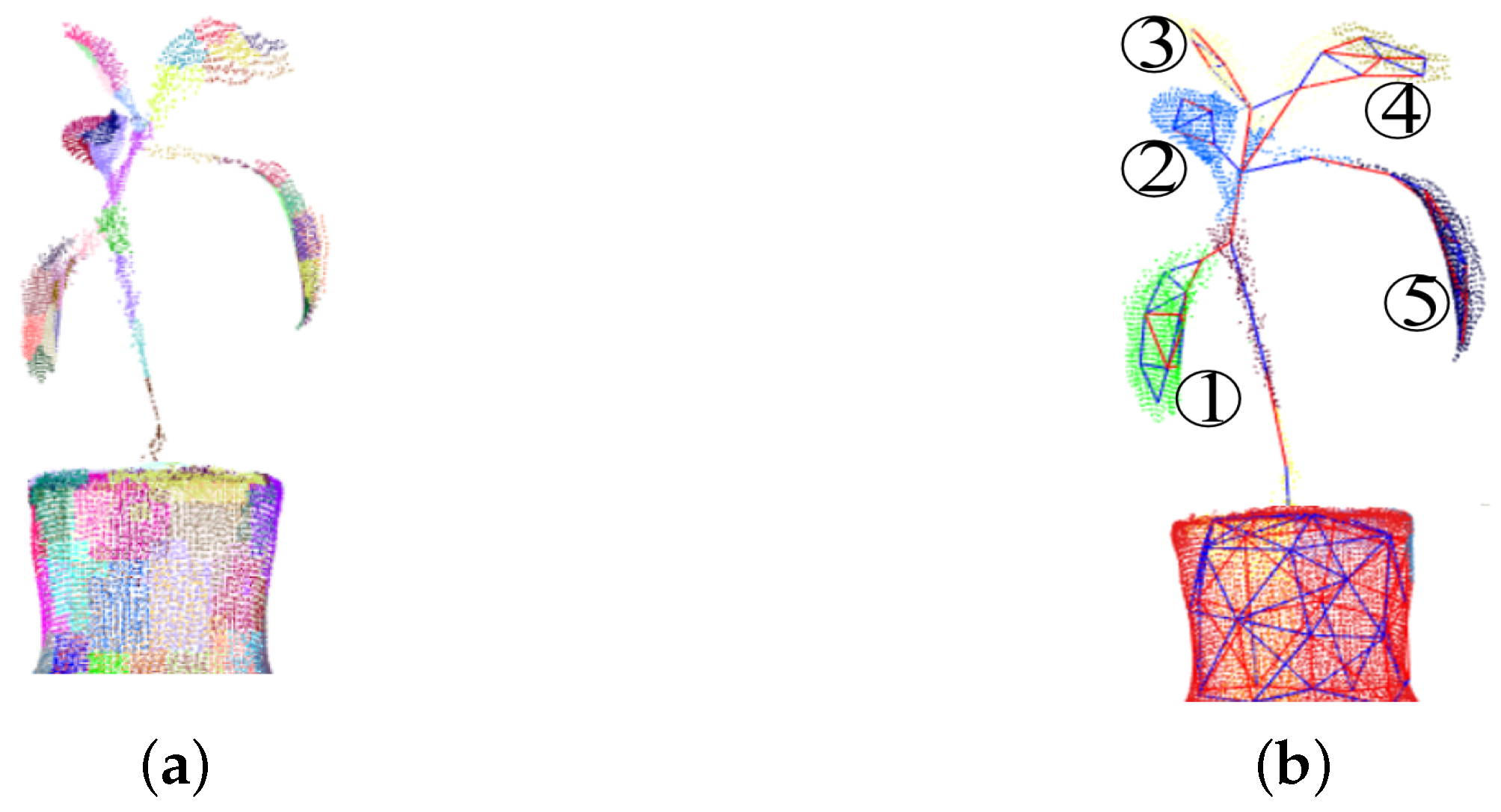




| Expriment | Object | Index | Length (cm) | Width (cm) | Area (cm2) | Plant Height (cm) |
|---|---|---|---|---|---|---|
| This Paper | Pepper | RMSE | 0.2639 | 0.2735 | 0.964 | 0.417 |
| Fang [25] | Cucumber | 0.28 | 0.32 | 4.34 | - | |
| Eggplant | 0.16 | 0.23 | 3.89 | - | ||
| Pepper | 0.33 | 0.15 | 1.33 | - | ||
| Paproki [24] | Gossypium hirsutum | 0.97 | 0.728 | - | 1.9 | |
| Liu [28] | apple tree | 0.59 | 0.538 | - | - | |
| This Paper | Pepper | MAE | 0.2537 | 0.2676 | 0.957 | 0.392 |
| Kaiyan [3] | Pepper | 0.183 | 0.124 | - | 0.344 | |
| Yang [15] | Cucumber | - | - | - | 0.23 | |
| Liu [28] | apple tree | 0.55 | 0.51 | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Chen, Y. Non-Destructive Measurement of Three-Dimensional Plants Based on Point Cloud. Plants 2020, 9, 571. https://doi.org/10.3390/plants9050571
Wang Y, Chen Y. Non-Destructive Measurement of Three-Dimensional Plants Based on Point Cloud. Plants. 2020; 9(5):571. https://doi.org/10.3390/plants9050571
Chicago/Turabian StyleWang, Yawei, and Yifei Chen. 2020. "Non-Destructive Measurement of Three-Dimensional Plants Based on Point Cloud" Plants 9, no. 5: 571. https://doi.org/10.3390/plants9050571
APA StyleWang, Y., & Chen, Y. (2020). Non-Destructive Measurement of Three-Dimensional Plants Based on Point Cloud. Plants, 9(5), 571. https://doi.org/10.3390/plants9050571





