Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants
Abstract
1. Introduction
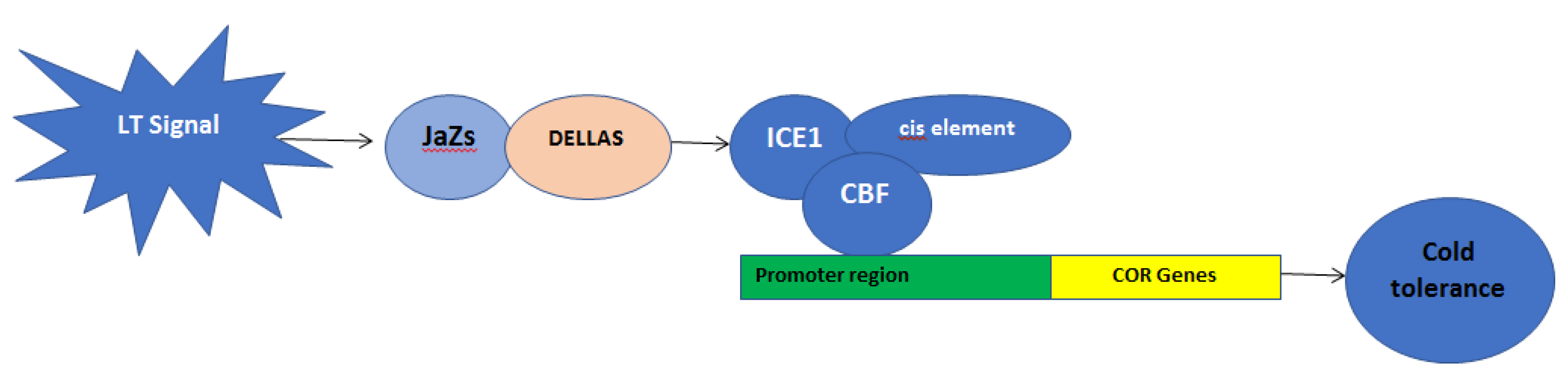
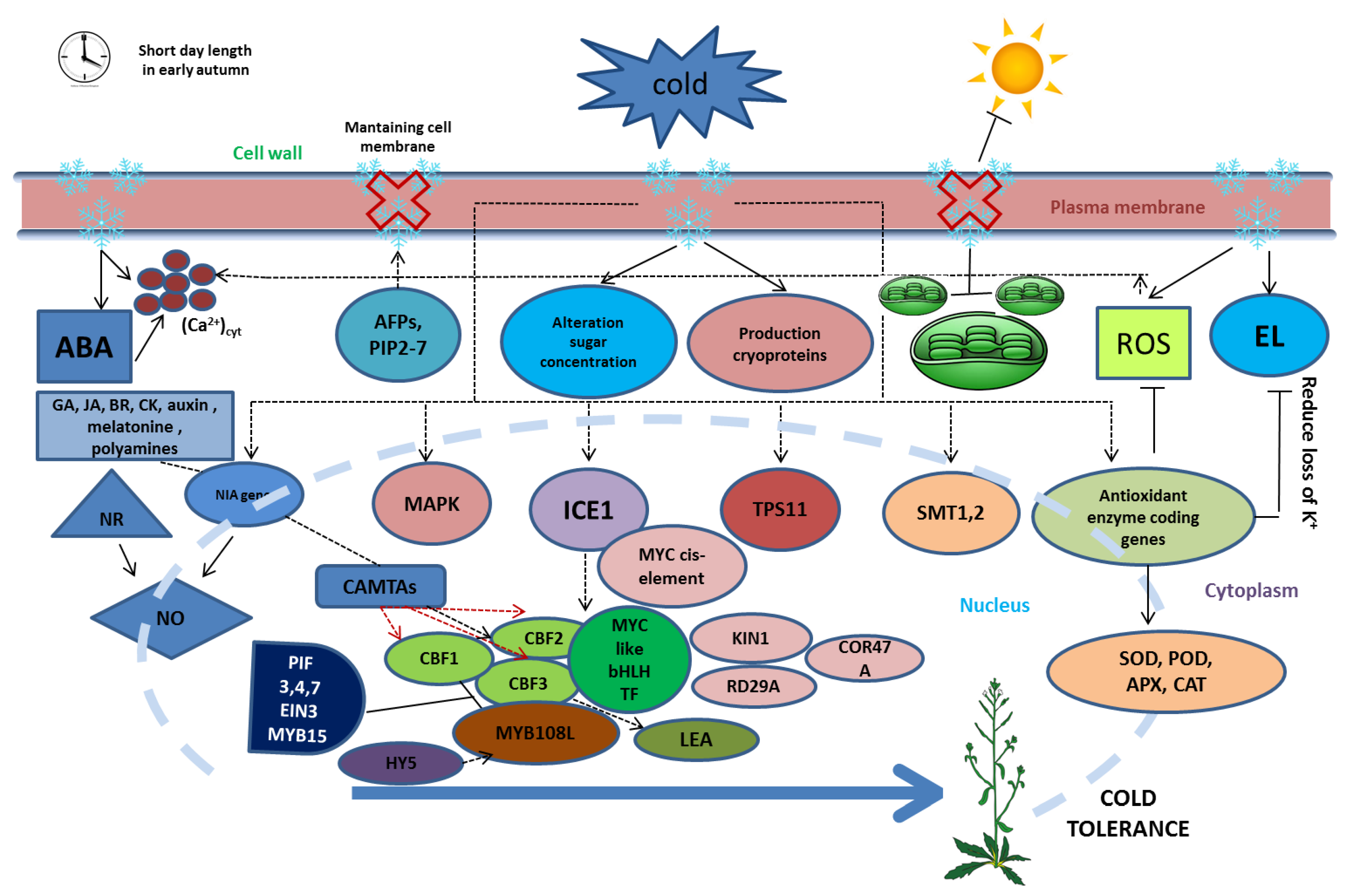
2. Genetical Changes during Cold Stress
3. Physiological Changes during Cold Stress
4. Influence Factors
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mboup, M.; Fischer, I.; Lainer, H.; Stephan, W. Trans-species polymorphism and allele-specific expression in the cbf gene family of wild tomatoes. Mol. Biol. Evol. 2012, 29, 3641–3652. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.; Clément, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Chen, H.; Wei, D.; Ma, H.; Lin, J. Arabidopsis I3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, L.; Zhu, J.; Liu, H.; Wang, A. Cloning and characterization of SiDHN, a novel dehydrin gene from Saussurea involucrata Kar. et Kir. that enhances cold and drought tolerance in tobacco. Plant Sci. 2017, 256, 160–169. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J. MAPping Kinase Regulation of ICE1 in Freezing Tolerance. Trends Plant Sci. 2018, 23, 91–93. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Zhang, F.; Jiang, Y.; Bai, L.-P.; Zhang, L.; Chen, L.-J.; Li, H.G.; Yin, Y.; Yan, W.-W.; Yi, Y.; Guo, Z.-F. The ICE-CBF-COR Pathway in Cold Acclimation and AFPs in Plants. Middle-East J. Sci. Res. 2011, 8, 493–498. [Google Scholar]
- Zhao, C.; Lang, Z.; Zhu, J.K. Cold responsive gene transcription becomes more complex. Trends Plant Sci. 2015, 20, 466–468. [Google Scholar] [CrossRef]
- Zhang, X.; Da Silva, J.A.T.; Niu, M.; Li, M.; He, C.; Zhao, J.; Zeng, S.; Duan, J.; Ma, G. Physiological and transcriptomic analyses reveal a response mechanism to cold stress in Santalum album L. Leaves. Sci. Rep. 2017, 7, 42165. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Carrer, M.; Castagneri, D.; Petit, G. Xylem anatomical responses to climate variability in Himalayan birch trees at one of the world’s highest forest limit. Perspect. Plant Ecol. Evol. Syst. 2018, 33, 34–41. [Google Scholar] [CrossRef]
- Hao, X.; Wang, B.; Wang, L.; Zeng, J.; Yang, Y.; Wang, X. Comprehensive transcriptome analysis reveals common and specific genes and pathways involved in cold acclimation and cold stress in tea plant leaves. Sci. Hortic. 2018, 240, 354–368. [Google Scholar] [CrossRef]
- Mata, C.I.; Hertog, M.L.; Van Raemdonck, G.; Baggerman, G.; Tran, D.; Nicolai, B.M. Omics analysis of the ethylene signal transduction in tomato as a function of storage temperature. Postharvest Biol. Technol. 2019, 155, 1–10. [Google Scholar] [CrossRef]
- Rubio, S.; Pérez, F.J. ABA and its signaling pathway are involved in the cold acclimation and deacclimation of grapevine buds. Sci. Hortic. 2019, 256, 108565. [Google Scholar] [CrossRef]
- Zheng, Y.; Luo, L.; Wei, J.; Chen, Q.; Yang, Y.; Hu, X.; Kong, X. The glutamate receptors AtGLR1. 2 and AtGLR1. 3 increase cold tolerance by regulating jasmonate signaling in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 506, 895–900. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Liu, J.; Song, S.; Hou, X.; Jia, C.; Li, J.; Miao, H.; Wang, Z.; Tie, W. An aquaporin gene MaPIP2-7 is involved in tolerance to drought, cold and salt stresses in transgenic banana (Musa acuminata L.). Plant Physiol. Biochem. 2020, 147, 66–76. [Google Scholar] [CrossRef]
- Tolosa, L.N.; Zhang, Z. The Role of Major Transcription Factors in Solanaceous Food Crops under Different Stress Conditions: Current and Future Perspectives. Plants 2020, 9, 56. [Google Scholar] [CrossRef]
- Puijalon, S.; Bouma, T.J.; Douady, C.J.; Groenendael, J.V.; Anten, N.P.R.; Martel, E.; Bornette, G. Plant resistance to mechanical stress: Evidence of an avoidance—Tolerance trade-off. New Phytol. 2011, 191, 1141–1149. [Google Scholar] [CrossRef]
- Jutsz, A.M.; Gnida, A. Mechanisms of stress avoidance and tolerance by plants used in phytoremediation of heavy metals. Arch. Environ. Prot. 2015, 41, 104–114. [Google Scholar] [CrossRef]
- Zarka, D.G.; Vogel, J.T.; Cook, D.; Thomashow, M.F. Cold Induction of Arabidopsis. Plant Physiol. 2003, 133, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, R.; Yin, Y.; Jiao, Z. Role of carbon ion beams irradiation in mitigating cold stress in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2018, 162, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Ungerer, M.C. Relaxed selection on the CBF/DREB1 regulatory genes and reduced freezing tolerance in the southern range of Arabidopsis thaliana. Mol. Biol. Evol. 2008, 25, 2547–2555. [Google Scholar] [CrossRef]
- Borba, A.R.; Serra, T.S.; Górska, A.; Gouveia, P.; Cordeiro, A.M.; Reyna-Llorens, I.; Kneřová, J.; Barros, P.M.; Abreu, I.A.; Oliveira, M.M.; et al. Synergistic binding of bHLH transcription factors to the promoter of the maize NADP-ME gene used in C4photosynthesis is based on an ancient code found in the ancestral C3state. Mol. Biol. Evol. 2018, 35, 1690–1705. [Google Scholar] [CrossRef]
- Londo, J.P.; Kovaleski, A.P.; Lillis, J.A. Divergence in the transcriptional landscape between low temperature and freeze shock in cultivated grapevine (Vitis vinifera). Hortic. Res. 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Rihan, H.Z.; Al-Issawi, M.; Fuller, M.P. Upregulation of CBF/DREB1 and cold tolerance in artificial seeds of cauliflower (Brassica oleracea var. botrytis). Sci. Hortic. 2017, 225, 299–309. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef]
- Yadav, S.K. Cold stress tolerance mechanisms in plants. Sustain. Agric. 2009, 2, 605–620. [Google Scholar] [CrossRef]
- Azzeme, A.M.; Abdullah, S.N.A.; Aziz, M.A.; Wahab, P.E.M. Oil palm drought inducible DREB1 induced expression of DRE/CRT- and non-DRE/CRT-containing genes in lowland transgenic tomato under cold and PEG treatments. Plant Physiol. Biochem. 2017, 112, 129–151. [Google Scholar] [CrossRef]
- Kargiotidou, A.; Deli, D.; Galanopoulou, D.; Tsaftaris, A.; Farmaki, T. Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J. Exp. Bot. 2008, 59, 2043–2056. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Sun, Z.; Li, C.; Zhao, X.; Li, M.; Deng, R.; Huang, Y.; Zhao, H.; Chen, H.; Wu, Q. Overexpression of Fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis. Plant Physiol. Biochem. 2018, 125, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Kindgren. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. bioXriv 2018, 77058, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBF s in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, C.; Zhao, X.; Chen, S.; Qu, G.-z. Complete chloroplast genome sequence of Betula platyphylla: Gene organization, RNA editing, and comparative and phylogenetic analyses. BMC Genomics 2018, 19, 950. [Google Scholar] [CrossRef]
- Yang, T.; Huang, X.S. Deep sequencing-based characterization of transcriptome of Pyrus ussuriensis in response to cold stress. Gene 2018, 661, 109–118. [Google Scholar] [CrossRef]
- Barrero-Gil, J.; Salinas, J. CBFs at the Crossroads of Plant Hormone Signaling in Cold Stress Response. Mol. Plant 2017, 10, 542–544. [Google Scholar] [CrossRef]
- Wang, D.-Z.; Jin, Y.-N.; Ding, X.-H.; Wang, W.-J.; Zhai, S.-S.; Bai, L.-P.; Guo, Z.-F. Gene regulation and signal transduction in the ICE–CBF–COR signaling pathway during cold stress in plants. Biochem. (Mosc.) 2017, 82, 1103–1117. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Solano, R. Identification of plant transcription factor target sequences. Biochim. Biophys. Acta-Gene Regul. Mech. 2017, 1860, 21–30. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof Domain Proteins: Plant-Specific Transcription Factors Associated with Div. Library 2004, 45, 386–391. [Google Scholar]
- Matsukura, S.; Mizoi, J.; Yoshida, T.; Todaka, D.; Ito, Y.; Maruyama, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom. 2010, 283, 185–196. [Google Scholar] [CrossRef]
- Ying, M.; Kidou, S.i. Discovery of novel cold-induced CISP genes encoding small RNA-binding proteins related to cold adaptation in barley. Plant Sci. 2017, 260, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhu, Z.; Zhang, L.; Fang, L.; Zhang, J.; Wang, Q.; Li, S.; Liang, Z.; Xin, H. Overexpression of ethylene response factors VaERF080 and VaERF087 from Vitis amurensis enhances cold tolerance in Arabidopsis. Sci. Hortic. 2019, 243, 320–326. [Google Scholar] [CrossRef]
- Hao, J.; Yang, J.; Dong, J.; Fei, S.z. Characterization of BdCBF genes and genome-wide transcriptome profiling of BdCBF3-dependent and -independent cold stress responses in Brachypodium distachyon. Plant Sci. 2017, 262, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Wang, H.; Ni, X.; Gao, Z.; Iqbal, S. Integrating proteomic and transcriptomic analyses of loquat (Eriobotrya japonica Lindl.) in response to cold stress. Gene 2018, 677, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Chan, Z.; Wang, Y.; Cao, M.; Gong, Y.; Mu, Z.; Wang, H.; Hu, Y.; Deng, X.; He, X.J.; Zhu, J.K. RDM 4 modulates cold stress resistance in Arabidopsis partially through the CBF-mediated pathway. New Phytol. 2016, 209, 1527–1539. [Google Scholar] [CrossRef]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member, OsGH3-2, modulates auxin and methylation and chromatin patterning abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, Y.; Xu, S.; Zhang, Z.; Xu, Y.; Zhang, J.; Chong, K. Os MADS 57 together with Os TB 1 coordinates transcription of its target Os WRKY 94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231. [Google Scholar] [CrossRef]
- Shan, D.P.; Huang, J.G.; Yang, Y.T.; Guo, Y.H.; Wu, C.A.; Yang, G.D.; Gao, Z.; Zheng, C.C. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 2007, 176, 70–81. [Google Scholar] [CrossRef]
- Ortiz, D.; Hu, J.; Salas Fernandez, M.G. Genetic architecture of photosynthesis in Sorghum bicolor under non-stress and cold stress conditions. J. Exp. Bot. 2017, 68, 4545–4557. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, Q.; Sprague, S.A.; Park, J.; Oh, M.; Rajashekar, C.B.; Koiwa, H.; Nakata, P.A.; Cheng, N.; Hirschi, K.D.; et al. Tomato expressing Arabidopsis glutaredoxin gene AtGRXS17 confers tolerance to chilling stress via modulating cold responsive components. Hortic. Res. 2015, 2, 1–11. [Google Scholar] [CrossRef]
- Gang, H.; Liu, G.; Zhang, M.; Zhao, Y.; Jiang, J.; Chen, S. Comprehensive characterization of T-DNA integration induced chromosomal rearrangement in a birch T-DNA mutant. BMC Genom. 2019, 20, 311. [Google Scholar] [CrossRef] [PubMed]
- Gang, H.; Li, R.; Zhao, Y.; Liu, G.; Chen, S.; Jiang, J. Loss of GLK1 transcription factor function reveals new insights in chlorophyll biosynthesis and chloroplast development. J. Exp. Bot. 2019, 70, 3125–3138. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X.; Zhang, D.; Li, Q.; Zhao, X.; Chen, S. Genome-Wide Analysis of NAC Gene Family in Betula pendula. Forests 2019, 10, 741. [Google Scholar] [CrossRef]
- Wang, F.; Chen, S.; Liang, D.; Qu, G.-Z.; Chen, S.; Zhao, X. Transcriptomic analyses of Pinus koraiensis under different cold stresses. BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef]
- Wang, S.; Huang, H.; Han, R.; Liu, C.; Qiu, Z.; Liu, G.; Chen, S.; Jiang, J. Negative feedback loop between BpAP1 and BpPI/BpDEF heterodimer in Betula platyphylla× B. pendula. Plant Sci. 2019, 289, 110280. [Google Scholar] [CrossRef]
- Jyoti, A.; Kaushik, S.; Srivastava, V.K.; Datta, M.; Kumar, S.; Yugandhar, P.; Kothari, S.L.; Rai, V.; Jain, A. The potential application of genome editing by using CRISPR/Cas9, and its engineered and ortholog variants for studying the transcription factors involved in the maintenance of phosphate homeostasis in model plants. Semin. Cell Dev. Biol. 2019, 96, 77–90. [Google Scholar] [CrossRef]
- Yubing, H.; Min, Z.; Lihao, W.; Junhua, W.; Qiaoyan, W.; Rongchen, W.; Yunde, Z. Improvements of TKC technology accelerate isolation of transgene-free CRISPR/Cas9-edited rice plants. Rice Sci. 2019, 26, 109–117. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Al-Sadi, A.M.; Pour-Aboughadareh, A.; Burritt, D.J.; Tran, L.-S.P. Genome editing using CRISPR/Cas9–targeted mutagenesis: An opportunity for yield improvements of crop plants grown under environmental stresses. Plant Physiol. Biochem. 2018, 131, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y.-G. CRISPR/Cas9 platforms for genome editing in plants: Developments and applications. Mol. Plant 2016, 9, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, M.; Shao, W.; Wang, H.; Fan, R.; Chen, X.; Wang, X.; Zhan, Y.; Zeng, F. Molecular cloning and functional analysis of a UV-B photoreceptor gene, BpUVR8 (UV Resistance Locus 8), from birch and its role in ABA response. Plant Sci. 2018, 274, 294–308. [Google Scholar] [CrossRef]
- Shi, H.; Ye, T.; Zhong, B.; Liu, X.; Jin, R.; Chan, Z. At HAP 5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 2014, 203, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zeng, W.; Ding, Y.; Wang, Y.; Niu, L.; Yao, J.-l.; Pan, L.; Lu, Z.; Cui, G.; Li, G.; et al. Plant Science Peach ethylene response factor PpeERF2 represses the expression of ABA biosynthesis and cell wall degradation genes during fruit ripening. Plant Sci. 2019, 283, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Lei, L.; Yao, L.; Wang, L.; Hao, X.; Li, N.; Wang, Y.; Yin, P.; Guo, G.; Yang, Y. The involvements of calcium-dependent protein kinases and catechins in tea plant [Camellia sinensis (L.) O. Kuntze] cold responses. Plant Physiol. Biochem. 2019, 143, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, L.; Dong, G.; Xu, Z.; Li, G.; Liu, N.; Wang, A.; Zhu, J. A novel cold-regulated protein isolated from Saussurea involucrata confers cold and drought tolerance in transgenic tobacco (Nicotiana tabacum). Plant Sci. 2019, 289, 110246. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Khan, N.; Ma, X.; Hou, X. Identification, Evolution, and Expression Profiling of Histone Lysine Methylation Moderators in Brassica rapa. Plants 2019, 8, 526. [Google Scholar] [CrossRef]
- Liu, X.; Fu, L.; Qin, P.; Sun, Y.; Liu, J.; Wang, X. Overexpression of the wheat trehalose 6-phosphate synthase 11 gene enhances cold tolerance in Arabidopsis thaliana. Gene 2019, 710, 210–217. [Google Scholar] [CrossRef]
- Valitova, J.; Renkova, A.; Mukhitova, F.; Dmitrieva, S.; Beckett, R.P.; Minibayeva, F.V. Membrane sterols and genes of sterol biosynthesis are involved in the response of Triticum aestivum seedlings to cold stress. Plant Physiol. Biochem. 2019, 142, 452–459. [Google Scholar] [CrossRef]
- Visconti, S.; D’Ambrosio, C.; Fiorillo, A.; Arena, S.; Muzi, C.; Zottini, M.; Aducci, P.; Marra, M.; Scaloni, A.; Camoni, L. Overexpression of 14-3-3 proteins enhances cold tolerance and increases levels of stress-responsive proteins of Arabidopsis plants. Plant Sci. 2019, 289, 110215. [Google Scholar] [CrossRef]
- Wang, W.; Gao, T.; Chen, J.; Yang, J.; Huang, H.; Yu, Y. The late embryogenesis abundant gene family in tea plant (Camellia sinensis): Genome-wide characterization and expression analysis in response to cold and dehydration stress. Plant Physiol. Biochem. 2019, 135, 277–286. [Google Scholar] [CrossRef]
- Wang, Y.; Mao, Z.; Jiang, H.; Zhang, Z.; Chen, X. A feedback loop involving MdMYB108L and MdHY5 controls apple cold tolerance. Biochem. Biophys. Res. Commun. 2019, 512, 381–386. [Google Scholar] [CrossRef]
- Yang, X.; Wang, R.; Hu, Q.; Li, S.; Mao, X.; Jing, H.; Zhao, J.; Hu, G.; Fu, J.; Liu, C. DlICE1, a stress-responsive gene from Dimocarpus longan, enhances cold tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2019, 142, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.-F.; Kang, H.-G.; Park, M.-Y.; Jeong, H.; Sun, H.-J.; Song, P.-S.; Lee, H.-Y. Zoysia japonica MYC type transcription factor ZjICE1 regulates cold tolerance in transgenic Arabidopsis. Plant Sci. 2019, 289, 110254. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kong, X.; Huang, H.; Wu, W.; Park, J.; Yun, D.-J.; Lee, B.-h.; Shi, H.; Zhu, J.-K. STCH4/REIL2 Confers Cold Stress Tolerance in Arabidopsis by Promoting rRNA Processing and CBF Protein Translation. Cell Rep. 2020, 30, 229–242.e225. [Google Scholar] [CrossRef] [PubMed]
- Puhakainen, T. Short-Day Potentiation of Low Temperature-Induced Gene Expression of a C-Repeat-Binding Factor-Controlled Gene during Cold Acclimation in Silver Birch. Plant Physiol. 2004, 136, 4299–4307. [Google Scholar] [CrossRef]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-induced electrolyte leakage: The role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef]
- Khan, H.; Shah, S.H.; Uddin, N.; Azhar, N.; Asim, M.; Syed, S.; Ullah, F.; Tawab, F.; Inayat, J. Biochemical and Physiological Changes of Different Plants Species in Response To Heat and Cold Stress. ARPN J. Agric. Biol. Sci. 2015, 10, 213–216. [Google Scholar]
- Janská, A.; Maršík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Abiri, R.; Azmi, N.; Maziah, M.; Norhana, Z.; Yusof, B.; Atabaki, N.; Sahebi, M.; Valdiani, A.; Kalhori, N.; Azizi, P.; et al. Role of ethylene and the APETALA 2/ethylene response factor superfamily in rice under various abiotic and biotic stress conditions. Environ. Exp. Bot. 2017, 134, 33–44. [Google Scholar] [CrossRef]
- Gupta, K.J.; Hincha, D.K.; Mur, L.A. NO way to treat a cold. New Phytol. 2011, 189, 360–363. [Google Scholar] [CrossRef]
- Beck, E.H.; Heim, R.; Hansen, J. Plant resistance to cold stress: Mechanisms and environmental signals triggering frost hardening and dehardening. J. Biosci. 2004, 29, 449–459. [Google Scholar] [CrossRef]
- Longo, V.; Kamran, R.V.; Michaletti, A.; Toorchi, M.; Zolla, L.; Rinalducci, S. Proteomic and Physiological Response of Spring Barley Leaves to Cold Stress. Int. J. Plant Biol. Res. 2017, 5, 1–10. [Google Scholar]
- Savitch, L.V.; Ivanov, A.G.; Gudynaite-Savitch, L.; Huner, N.P.A.; Simmonds, J. Cold stress effects on PSI photochemistry in Zea mays: Differential increase of FQR-dependent cyclic electron flow and functional implications. Plant Cell Physiol. 2011, 52, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Cantrel, C.; Vazquez, T.; Puyaubert, J.; Rezé, N.; Lesch, M.; Kaiser, W.M.; Dutilleul, C.; Guillas, I.; Zachowski, A.; Baudouin, E. Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol. 2011, 189, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Laine, P.; Bigot, J.; Ourry, A.; Boucaud, J. Effects of low temperature on nitrate uptake, and xylem and phloem flows of nitrogen, in Secale cereale L. and Brassica napus L. New Phytol. 1994, 127, 675–683. [Google Scholar] [CrossRef]
- Sun, J.; Zheng, T.; Yu, J.; Wu, T.; Wang, X.; Chen, G.; Tian, Y.; Zhang, H.; Wang, Y.; Terzaghi, W. TSV, a putative plastidic oxidoreductase, protects rice chloroplasts from cold stress during development by interacting with plastidic thioredoxin Z. New Phytol. 2017, 215, 240–255. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Y.; Wu, J.; Han, X.; Gu, X.; Lu, T.; Zhang, Z. The RNA editing factor DUA 1 is crucial to chloroplast development at low temperature in rice. New Phytol. 2019, 221, 834–849. [Google Scholar] [CrossRef]
- Garsed, S.; Davey, H.; Galley, D. The Effects of Light and Temperature on the Growth of and Balances of Carbon, Nitrogen and Potassium between Vicia faba L. and Aphis fabae Scop. New Phytol. 1987, 107, 77–102. [Google Scholar] [CrossRef]
- Maibam, P.; Nawkar, G.M.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. The influence of light quality, circadian rhythm, and photoperiod on the CBF-mediated freezing tolerance. Int. J. Mol. Sci. 2013, 14, 11527–11543. [Google Scholar] [CrossRef]
- Shinozaki, K.; Kazuko, Y. Molecular responses to drought and cold stress. Biotechnology 1996, 7, 161–167. [Google Scholar] [CrossRef]
- Bigot, J.; Boucaud, J. Effects of synthetic plant growth retardants and abscisic acid on root functions of Brassica rapa plants exposed to low root-zone temperature. New Phytol. 1998, 139, 255–265. [Google Scholar] [CrossRef]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Munné-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chen, S.; Jiang, J. Transcriptome analysis reveals the role of BpGH3.5 in root elongation of Betula platyphylla × B. pendula. Plant CellTissue Organ Cult. 2015, 121, 605–617. [Google Scholar] [CrossRef]
- Yang, G.; Chen, S.; Wang, S.; Liu, G.; Li, H.; Huang, H.; Jiang, J. BpGH3.5, an early auxin-response gene, regulates root elongation in Betula platyphylla × Betula pendula. Plant CellTissue Organ Cult. 2015, 120, 239–250. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, J.; Peng, W.; Peng, D.; Zhuo, Y.; Zhu, D.; Huang, X.; Tang, D.; Guo, M.; He, R.; et al. Dwarfism in Brassica napus L. induced by the over-expression of a gibberellin 2-oxidase gene from Arabidopsis thaliana. Mol. Breed. 2012, 29, 115–127. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef]
| Gene Name | Family | Species | Type of Stress Condition | References |
|---|---|---|---|---|
| FtbHLH2 | bHLH | Fagopyrum tataricum | Cold stress | [31] |
| BpUVR8 | UVR | Betula platyphylla | ABA response and cold stress | [60] |
| FDA2-3 | FDA | Gossypium hirsutum | Cold stress | [30] |
| FDA2-4 | ||||
| FDA8 | Arabidopsis thaliana | Cold stress | [30] | |
| Sb08g007310 | GST | Sorghum bicolor | Cold stress | [49] |
| Sb06g018220 | ZEP | Sorghum bicolor | Epoxidation of zeaxanthin in the xanthophyll cycle | [49] |
| AtGRXS17 | Trx | Solanum lycopersicum | Chilling stress | [50] |
| AtCBF3 | AP2/ERF | Arabidopsis | Cold Stress | [50] |
| VaERF080 | AP2/ERF | Vitis amurensis | Cold stress | [31] |
| VaERF087 | ||||
| SiDHN | DHN | Saussurea involucrata | Freezing stress and drought stress | [5] |
| OsGH3-2 | GH3 | Oryza sativa | Drought and cold stress | [46] |
| MYBS3 | MYB | Oryza sativa | Cold stress | [4] |
| RDM4 | Arabidopsis | Cold stress and freezing stress | [45] | |
| OsMADS57 | Oryza sativa | Chilling stress | [47] | |
| GHDREB1 | DREB | Gossypium hirsutum | Chilling stress | [48] |
| AtHAP5A, AtXTH21 | Arabidopsis thaliana | Freezing stress | [61] | |
| PUB25/26 | Arabidpsis thaliana | Freezing stress | [62] | |
| MaPIP2-7 | AQP | Musa acuminata | Drought, cold and salt stress | |
| MaPIP2-7 | AQP | Musa acuminata | Drought, cold and salt stress | [17] |
| CsCPKs | CPK | Camellia sinensis | Cold tolerance | [63] |
| COR413 | COR | Saussurea involucrata | Cold and drought tolerance | [64] |
| SET, JmJC | Brassica rapa | Heat and cold stress | [65] | |
| TaTPS11 | Triticum aestivum | Cold stress | [66] | |
| TaSMT1, TaSMT2 | Triticum aestivum | Cold stress | [67] | |
| 14-3-3ε, 14-3-3ω | Arabidopsis thaliana | Cold and oxidative stress | [68] | |
| CsLEA | LEA | Camellia sinensis | Cold and dehydration stress | [69] |
| MdMYB108L | MYB | Malus domestica | Cold stress | [70] |
| MdHY5 | bZIP | Malus domestica | Cold stress | [70] |
| DlICE1 | bHLH | Dimocarpus longan | Cold stress | [71] |
| ZjICE1 | bHLH | Zoysia japonica | Cold, dehydration and salt stress | [72] |
| VvCBF | DREB | Vitis vinifera | Cold stress | [15] |
| AtGLR1.2AtGLR1.3 | Arabidopsis thaliana | Cold stress | [16] | |
| STCH4 | Arabidopsis thaliana | Cold stress | [73] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ritonga, F.N.; Chen, S. Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants 2020, 9, 560. https://doi.org/10.3390/plants9050560
Ritonga FN, Chen S. Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants. 2020; 9(5):560. https://doi.org/10.3390/plants9050560
Chicago/Turabian StyleRitonga, Faujiah Nurhasanah, and Su Chen. 2020. "Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants" Plants 9, no. 5: 560. https://doi.org/10.3390/plants9050560
APA StyleRitonga, F. N., & Chen, S. (2020). Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants, 9(5), 560. https://doi.org/10.3390/plants9050560





