Unraveling the Phylogenomic Relationships of the Most Diverse African Palm Genus Raphia (Calamoideae, Arecaceae)
Abstract
1. Introduction
2. Results
2.1. DNA Sequencing
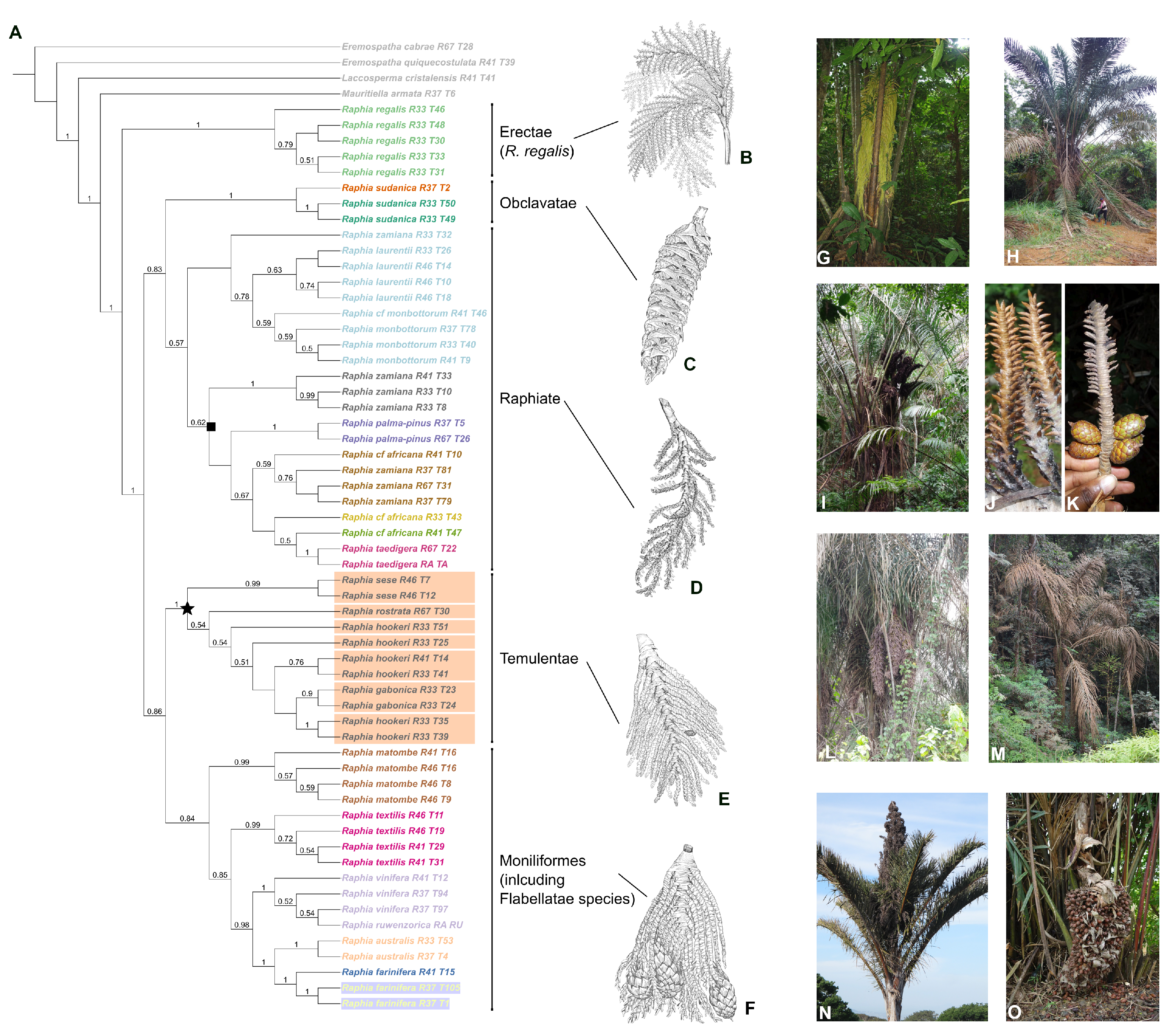
2.2. Evolutionary History of Raphia
2.3. Species Delimitation
2.4. Fine Scale Structure in Two Species-Complexes
3. Discussion
3.1. Synthesizing Morphology and Molecules: The Sections of Otedoh Reevaluated
3.2. Species Delimitation and Species Complexes
3.2.1. The Moniliformes (Including Flabellatae) Section
3.2.2. The Raphiate Section
3.2.3. The Temulentae Section
4. Materials and Methods
4.1. Species Sampling, Library Preparation, and DNA Sequencing
4.2. Contig Assembly and Multi-Sequence Alignment
Paralog Identification
4.3. Coalescent Phylogenetic Inference
4.4. Species Delimitation
4.5. Maximum-Likelihood Phylogenetic Inference
4.6. SNP Calling
4.7. Genetic Clustering
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CVL | Cameroon Volcanic Line |
| SNP | Single Nucleotide Polymorphism |
| PCA | Principal Components Analysis |
Appendix A
| Genus | Species Epithet | ID | Collector (Herbarium) | Country | Latitude | Longitude | Run | Tag | No. Mapped | Mean Depth | Stdev Depth |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eremospatha | cabrae | R227 | Couvreur 1165 (WAG) | Cameroon | 3.128984 | 9.973292 | RUN67 | TAG-28 | 441,790 | 315.48 | 469.205 |
| Eremospatha | quinquecostulata | R162 | Couvreur 1079 (WAG) | Gabon | −1.45585 | 12.5863 | RUN41 | TAG39 | 91,982 | 46.1243 | 68.317 |
| Laccosperma | cristalensis | R164 | Couvreur 1142 (WAG) | Gabon | 0.6059 | 10.4118 | RUN41 | TAG41 | 105,250 | 54.4592 | 86.7057 |
| Mauritiella | armata | R135 | Couvreur 257 (NY) | Bolivia | −13.496289 | −68.019923 | RUN37 | TAG6 | 406,142 | 213.159 | 1244.61 |
| Raphia | africana (cf) | R072 | Couvreur 971 (WAG) | Cameroon | 4.87036 | 9.26579 | RUN41 | TAG10 | 121,972 | 63.533 | 69.0331 |
| Raphia | africana (cf) | R077 | No voucher, close to Couvreur 971 (WAG) | Cameroon | 4.87064 | 9.26582 | RUN33 | TAG43 | 315,915 | 159.931 | 188.084 |
| Raphia | africana (cf) | R174 | No voucher, close to Couvreur 971 (WAG) | Cameroon | 4.12977 | 9.21399 | RUN41 | TAG47 | 50,272 | 26.0156 | 28.656 |
| Raphia | australis | R092 | MBC 99 874D (SANBI) | Cultivated, Kirstenbosch National Botanical Garden, South Africa | N/A | N/A | RUN33 | TAG53 | 25,5329 | 129.65 | 156.525 |
| Raphia | australis | R130 | MBC 99 874D (SANBI) | Cultivated, Kirstenbosch National Botanical Garden, South Africa | N/A | N/A | RUN37 | TAG4 | 913,167 | 473.13 | 627.945 |
| Raphia | farinifera | R128 | Faye 36 (WAG) | Republic of Congo | −3.99056 | 11.30600 | RUN37 | TAG1 | 476,130 | 245.7 | 292.787 |
| Raphia | farinifera | R127 | Baker s.n. (K, DNA bank id: 14927) | Cultivated, Royal Botanic Gardens, Kew, U.K. | NA | NA | RUN37 | TAG105 | 486,723 | 250.42 | 309.47 |
| Raphia | farinifera | R132 | Dransfield 7516 (K) | Madagascar | −1.84988 | 13.85903 | RUN41 | TAG15 | 133,441 | 68.7643 | 85.4757 |
| Raphia | gabonica | R034 | Kamga Mogue 22 (WAG) | Gabon | −0.07916 | 11.00836 | RUN33 | TAG23 | 284,243 | 143.822 | 174.031 |
| Raphia | gabonica | R036 | Kamga Mogue 23 (WAG) | Gabon | −1.03044 | 10.51881 | RUN33 | TAG24 | 212,852 | 108.979 | 136.141 |
| Raphia | hookeri | R063 | Kamga Mogue 1 (WAG) | Cameroon | 3.07485 | 13.3663 | RUN33 | TAG35 | 319,096 | 162.68 | 210.004 |
| Raphia | hookeri | R069 | Kamga Mogue 12 (WAG) | Cameroon | 3.97037 | 13.2367 | RUN33 | TAG39 | 197,508 | 100.49 | 122.815 |
| Raphia | hookeri | R071 | Kamga Mogue 26 (WAG) | Cameroon | 4.11224 | 9.56915 | RUN33 | TAG41 | 292,201 | 149.509 | 181.385 |
| Raphia | hookeri | R124 | Couvreur 984 (WAG) | Cameroon | 3.52108 | 11.74376 | RUN41 | TAG14 | 78,554 | 40.8646 | 44.4911 |
| Raphia | hookeri | R037 | Kamga Mogue 25 (WAG) | Gabon | −0.82955 | 10.52294 | RUN33 | TAG25 | 278,678 | 142.219 | 175.028 |
| Raphia | hookeri | R089 | Michon 01 (G) | Togo | 6.39183 | 2.67703 | RUN33 | TAG51 | 201,880 | 103.103 | 124.193 |
| Raphia | laurentii | R208 | Lautenschläger 806 (JACQ) | Angola | −6.14997 | 15.40333 | RUN46 | TAG18 | 64,379 | 34.3542 | 37.7478 |
| Raphia | laurentii | R040 | Ayole 01 (YA) | Cameroon | 2.15329 | 15.7367 | RUN33 | TAG26 | 272,509 | 138.397 | 168.066 |
| Raphia | laurentii | R186 | Kamga Mogue 39 (WAG) | Democratic Republic of Congo | −0.8852 | 18.1337 | RUN46 | TAG10 | 87,838 | 47.1055 | 51.2042 |
| Raphia | laurentii | R198 | Kamga Mogue 42 (WAG) | Democratic Republic of Congo | −0.60673 | 18.2468 | RUN46 | TAG14 | 86,786 | 46.5246 | 51.1853 |
| Raphia | matombe | R206 | Lautenschläger 1095 (JACQ) | Angola | −7.94817 | 15.83894 | RUN46 | TAG16 | 59,006 | 31.6789 | 34.7279 |
| Raphia | matombe | R134 | 19392103 (BR) | Cultivated, Meise Botanical Garden, Belgium | NA | NA | RUN41 | TAG16 | 76,250 | 39.5975 | 44.2225 |
| Raphia | matombe | R181 | Kamga Mogue 37 (WAG) | Democratic Republic of Congo | −5.73485 | 14.2162 | RUN46 | TAG8 | 55,883 | 29.9284 | 31.68 |
| Raphia | matombe | R183 | Kamga Mogue 38 (WAG) | Democratic Republic of Congo | −5.65308 | 14.3181 | RUN46 | TAG9 | 66,676 | 35.6465 | 38.4587 |
| Raphia | monbuttorum | R059 | Kamdem 211 (WAG) | Cameroon | 3.885055556 | 14.39930556 | RUN41 | TAG9 | 104,568 | 54.2312 | 58.009 |
| Raphia | monbuttorum | R066 | Kamga Mogue 04 (WAG) | Cameroon | 3.07684 | 13.36761 | RUN37 | TAG78 | 407,023 | 208.475 | 245.939 |
| Raphia | monbuttorum | R070 | Kamga Mogue 13 (WAG) | Cameroon | 3.58237 | 13.14197 | RUN33 | TAG40 | 352731 | 180.143 | 231.782 |
| Raphia | monbuttorum | R173 | Kamga Mogue 31 (WAG) | Cameroon | 4.18332 | 13.10538 | RUN41 | TAG46 | 98,860 | 50.8202 | 56.6257 |
| Raphia | palma-pinus | R133 | Ouatara & Stauffer 14 (G) | Ghana | 5.39625 | −1.38277778 | RUN37 | TAG5 | 792,856 | 411.261 | 507.288 |
| Raphia | palma-pinus | TC-S1328 | Stauffer 857 (G) | Ivory Coast | 7.24598 | −-5.39625 | RUN67 | TAG-26 | 459,813 | 318.875 | 309.014 |
| Raphia | regalis | R055 | Couvreur 685 (WAG) | Cameroon | 2.49409 | 10.34844 | RUN33 | TAG30 | 184,893 | 94.4991 | 128.908 |
| Raphia | regalis | R056 | Couvreur 753 (WAG) | Cameroon | 3.4825 | 13.59469 | RUN33 | TAG31 | 210,853 | 107.898 | 137.475 |
| Raphia | regalis | R058 | Couvreur 398 (WAG) | Cameroon | 3.19962 | 10.51772 | RUN33 | TAG33 | 388,670 | 196.877 | 269.794 |
| Raphia | regalis | R081 | Wieringa 8539 (WAG) | Gabon | −2.37125 | 11.16443 | RUN33 | TAG46 | 161,079 | 81.8365 | 100.151 |
| Raphia | regalis | R083 | Wieringa 8547 (WAG) | Gabon | −2.3714 | 11.16399 | RUN33 | TAG48 | 150,544 | 76.7195 | 98.4529 |
| Raphia | rostrata | R229 | Kamga Mogue 43 (WAG) | Cameroon | 2.339 | 10.6025 | RUN67 | TAG-30 | 476,709 | 329.286 | 313.366 |
| Raphia | ruwenzorica | RA_RU | Robyns 4039 (BR) - herbarium sample | Democratic Republic of Congo | NA | NA | P6655_1076 | N/A | N/A | N/A | N/A |
| Raphia | sese | R179 | Kamga Mogue 36 (WAG) | Democratic Republic of Congo | −5.02068 | 15.1545 | RUN46 | TAG7 | 62,972 | 33.7432 | 37.0717 |
| Raphia | sese | R195 | Kamga Mogue 41 (WAG) | Democratic Republic of Congo | −0.8642 | 18.1458 | RUN46 | TAG12 | 88,659 | 47.5337 | 50.4421 |
| Raphia | sudanica | R086 | Michon 09 (G) | Benin | 7.55826 | 2.19247 | RUN33 | TAG49 | 379,755 | 193.881 | 231.648 |
| Raphia | sudanica | R129 | Bayton 70 (K) | Burkina Faso | 10.5938888 | −5.3069444 | RUN37 | TAG2 | 471,997 | 246.119 | 299.37 |
| Raphia | sudanica | R088 | Michon 56 (G) | Togo | 8.98317 | 1.49297 | RUN33 | TAG50 | 355,099 | 181.913 | 220.289 |
| Raphia | taedigera | RA_TA | Noblick 5015 (K) - (herbarium sample) | Brazil | −1.5666 | −48.73333 | AFNBJ_P2059_3047 | N/A | N/A | N/A | N/A |
| Raphia | taedigera | Env0563 | MBC 94803 A (MBC) | Cultivated, Montgomery Botanical Garden | N/A | N/A | RUN67 | TAG-22 | 666,580 | 446.883 | 465.14 |
| Raphia | textilis | R209 | Lautenschläger 1086 (JACQ) | Angola | −6.74019 | 16.20117 | RUN46 | TAG19 | 50,163 | 26.882 | 30.0834 |
| Raphia | textilis | R192 | Kamga Mogue 40 (WAG) | Democratic Republic of Congo | −0.88513 | 18.1337 | RUN46 | TAG11 | 78,738 | 42.0848 | 46.2868 |
| Raphia | textilis | R149 | Couvreur 743 (WAG) | Gabon | −1.84988 | 13.85903 | RUN41 | TAG29 | 132,014 | 68.4236 | 80.0799 |
| Raphia | textilis | R151 | Couvreur 1075 (WAG) | Gabon | −1.40695 | 12.5712 | RUN41 | TAG31 | 80,524 | 41.722 | 45.5424 |
| Raphia | vinifera | R105 | Couvreur 638 (WAG) | Cameroon | 6.366476985 | 10.894598 | RUN41 | TAG12 | 87,308 | 45.4189 | 49.1511 |
| Raphia | vinifera | R113 | No voucher, Couvreur 638 (WAG) | Cameroon | 6.27413 | 10.51091 | RUN37 | TAG94 | 286,322 | 145.81 | 177.287 |
| Raphia | vinifera | R116 | No voucher | Cameroon | 5.48034 | 10.05056 | RUN37 | TAG97 | 367,120 | 189.172 | 220.664 |
| Raphia | zamiana | R057 | Couvreur 427 (WAG) | Cameroon | 3.59972 | 11.2877 | RUN33 | TAG32 | 411,897 | 209.629 | 268.899 |
| Raphia | zamiana | R093 | Ayole 20 (YA) | Cameroon | 3.23685 | 10.02514 | RUN37 | TAG79 | 300,001 | 153.527 | 177.289 |
| Raphia | zamiana | R095 | Ayole 32 (YA) | Cameroon | 2.80897 | 10.52734 | RUN37 | TAG81 | 420,757 | 215.428 | 250.534 |
| Raphia | zamiana | R230 | Kamga Mogue 45 (WAG) | Cameroon | 3.137672 | 9.971397 | RUN67 | TAG-31 | 468,825 | 318.682 | 376.272 |
| Raphia | zamiana | R007 | Kamga Mogue 17 (WAG) | Gabon | 1.59848 | 11.62294 | RUN33 | TAG8 | 217,692 | 110.68 | 136.444 |
| Raphia | zamiana | R009 | Kamga Mogue 17 (WAG) | Gabon | −2.25428 | 11.14284 | RUN33 | TAG10 | 228,828 | 116.278 | 144.853 |
| Raphia | zamiana | R154 | Couvreur 1122 (WAG) | Gabon | −0.1473 | 11.726 | RUN41 | TAG33 | 56,712 | 29.1983 | 31.1171 |
Appendix B


References
- Dransfield, J.; Uhl, N.W.; Asmussen, C.B.; Baker, W.J.; Harley, M.M.; Lewis, C.E. Genera Palmarum: The Evolution and Classification of Palms; Kew Publishing: Kew, UK, 2008. [Google Scholar]
- Stauffer, F.W.; Ouattara, D.N.; Roguet, D.; da Giau, S.; Michon, L.; Bakayoko, A.; Ekpe, P. An update to the African palms (Arecaceae) floristic and taxonomic knowledge, with emphasis on the West African region. Webbia 2017, 72, 1–14. [Google Scholar] [CrossRef]
- Cosiaux, A.; Gardiner, L.M.; Stauffer, F.W.; Bachman, S.P.; Sonké, B.; Baker, W.J.; Couvreur, T.L.P. Low extinction risk for an important plant resource: Conservation assessments of continental African palms (Arecaceae/Palmae). Biol. Conserv. 2018, 221, 323–333. [Google Scholar] [CrossRef]
- Moore, H.E. Palms in the tropical forest ecosystems of Africa and South America. In Tropical Forest Ecosystems of Africa and South America: A Comparative Review; Meggers, B.J., Ayensu, E., Duckworth, W.D., Eds.; Smithsonian Institution Press: Washintong, DC, USA, 1973; pp. 63–88. [Google Scholar]
- Couvreur, T.L.P. Odd man out: Why are there fewer plant species in African rain forests? Plant Syst. Evol. 2015, 301, 1299–1313. [Google Scholar] [CrossRef]
- Dransfield, J. The palms of Africa and their relationships. In Modern Systematic Studies in African Botany; Goldblatt, P., Lowry, P.P., Eds.; Missouri Botanical Garden Press: St. Louis, MO, USA, 1988; pp. 95–103. [Google Scholar]
- Mogue Kamga, S.; Niangadouma, R.; Stauffer, F.W.; Sonké, B.; Couvreur, T.L.P. Two new species of Raphia (Palmae/Arecaceae) from Cameroon and Gabon. PhytoKeys 2018, 111, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, G.R. Long-term Persistence of Raphia taedigera Mart. Swamps in Nicaragua1. Biotropica 1999, 31, 565–569. [Google Scholar] [CrossRef]
- Urquhart, G.R. Paleoecological evidence of Raphia in the Pre-Columbian Neotropics. J. Trop. Ecol. 1997, 13, 783–792. [Google Scholar] [CrossRef]
- Otedoh, M.O. Systematic Studies in Raphia Palms. Ph.D. Thesis, University of Reading, Reading, UK, 1976. [Google Scholar]
- Otedoh, M.O. The African origin of Raphia taedigera—Palmae. J. Niger. Inst. Oil Palm Res 1977, 42, 11–16. [Google Scholar]
- Mogue Kamga, S.; Brokamp, G.; Cosiaux, A.; Awono, A.; Fürniss, S.; Barfod, A.S.; Muafor, F.J.; Le Gall, P.; Sonké, B.; Couvreur, T.L.P. Use and cultural significance of Raphia palms. Econ. Botany 2020, 74. in press. [Google Scholar] [CrossRef]
- Lautenschläger, T.; Neinhuis, C. (Eds.) Riquezas Naturais de Uíge—Uma Breve Introdução sobre o Estado Atual, a Utilização, a Ameaça e a Preservação da Biodiversidade; Technische Universität Dresden: Dresden, Germany, 2014. [Google Scholar]
- Obahiagbon, F.I. A review of the origin, morphology, cultivation, economic products, health and physiological implications of raphia palm. Afr. J. Food Sci. 2009, 3, 447–453. [Google Scholar]
- Couvreur, T.L.P.; Fumtim, J. A l’Echelle du Raphia/On Raphia and Man. 2017. Available online: https://www.youtube.com/watch?v=avSoLIusCCs&t=1167s (accessed on 8 April 2020).
- Profizi, J.P. Swampy Area Transformations by Exploitation of Raphia hookeri (Arecaceae) in Southern Benin (West Africa). Hum. Ecol. 1988, 16, 87–94. [Google Scholar] [CrossRef]
- Dargie, G.C.; Lewis, S.L.; Lawson, I.T.; Mitchard, E.T.A.; Page, S.E.; Bocko, Y.E.; Ifo, S.A. Age, extent and carbon storage of the central Congo Basin peatland complex. Nature 2017, 542, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Rainey, H.J.; Iyenguet, F.C.; Malanda, G.A.F.; Madzoké, B.; Santos, D.D.; Stokes, E.J.; Maisels, F.; Strindberg, S. Survey of Raphia swamp forest, Republic of Congo, indicates high densities of Critically Endangered western lowland gorillas Gorilla gorilla gorilla. Oryx 2010, 44, 124–132. [Google Scholar] [CrossRef]
- Couvreur, T.L.P.; Helmstetter, A.J.; Koenen, E.J.M.; Bethune, K.; Brandão, R.D.; Little, S.A.; Sauquet, H.; Erkens, R.H.J. Phylogenomics of the Major Tropical Plant Family Annonaceae Using Targeted Enrichment of Nuclear Genes. Front. Plant Sci. 2019, 9, 1941. [Google Scholar] [CrossRef] [PubMed]
- Tuley, P. The Palms of Africa; The Tendrine Press: Zennor, UK, 1995. [Google Scholar]
- Otedoh, M.O. A revision of the genus Raphia Beauv. (Palmae). J. Niger. Inst. Oil Palm Res 1982, 6, 145–189. [Google Scholar]
- Beccari, O. Studio monografico del genere “Raphia“. Webbia 1910, 3, 37–130. [Google Scholar] [CrossRef]
- Russell, T.A. The Raphia Palms of West Africa. Kew Bulletin 1965, 19, 173–196. [Google Scholar] [CrossRef]
- Tuley, P.; Russell, T.A. The Raphia palms reviewed. Nigerian Field 1966, 31, 54–65. [Google Scholar]
- Baker, W.J.; Dransfield, J.; Hedderson, T.A. Phylogeny, character evolution, and a new classification of the calamoid palms. Syst. Botany 2000, 25, 297–322. [Google Scholar] [CrossRef]
- Heyduk, K.; Trapnell, D.W.; Barrett, C.F.; Leebens-Mack, J. Phylogenomic analyses of species relationships in the genus Sabal (Arecaceae) using targeted sequence capture. Biol. J. Linn. Soc. 2016, 117, 106–120. [Google Scholar] [CrossRef]
- Ouattara, D.N.; Stauffer, F.W.; Bakayoko, A. Lectotypification de Raphia sudanica A. Chev. (Arecaceae, Calamoideae), et commentaires sur la biologie et la conservation de l’espèce. Adansonia 2014, 36, 53–61. [Google Scholar] [CrossRef]
- Liu, X.; Fu, Y.X. Exploring population size changes using SNP frequency spectra. Nat. Gen. 2015, 47, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Tonini, J.; Moore, A.; Stern, D.; Shcheglovitova, M.; Ortí, G. Concatenation and Species Tree Methods Exhibit Statistically Indistinguishable Accuracy under a Range of Simulated Conditions. PLoS Curr. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; Gatesy, J. The gene tree delusion. Mol. Phylogen. Evol. 2016, 94, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sayyari, E.; Mirarab, S. ASTRAL-III: Increased scalability and impacts of contracting low support branches. In Lecture Notes in Computer Science; including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics; Springer: Berlin, Germany, 2017; Volume 10562, pp. 53–75. [Google Scholar] [CrossRef]
- Mogue Kamga, S.; Sonké, B.; Couvreur, T.L.P. Raphia vinifera (Arecaceae; Calamoideae): Misidentified for far too long. Biodiver. Data J. 2019, 7, e37757. [Google Scholar] [CrossRef]
- Robyns, W.; Tournay, R. Monocotylées nouvelles ou critiques de la région du Parc National Albert (Congo belge). Bulletin du Jardin botanique de l’État a Bruxelles 1955, 25, 239–260. [Google Scholar] [CrossRef]
- Stauffer, F.W.; Ouattara, D.; Stork, A.L. Palmae. In Tropical African Flowering Plants: Monocotyledons 2; Lebrun, J.P., Stork, A.L., Eds.; Conservatoire et Jardin botaniques de la Ville de Genève: Geneva, Switzerland, 2014; Volume 8, pp. 326–354. [Google Scholar]
- Couvreur, T.L.P. Monograph of the syncarpous African genera Isolona and Monodora (Annonaceae). Syst. Botany Monogr. 2009, 87, 1–150. [Google Scholar]
- Kadu, C.A.C.; Schueler, S.; Konrad, H.; Muluvi, G.M.M.; Eyog-Matig, O.; Muchugi, A.; Williams, V.L.; Ramamonjisoa, L.; Kapinga, C.; Foahom, B.; et al. Phylogeography of the Afromontane Prunus africana reveals a former migration corridor between East and West African highlands. Mol. Ecol. 2011, 20, 165–178. [Google Scholar] [CrossRef]
- Dale, I.R. Palms of Kenya. J. East Afr. Uganda Nat. Hist. Society 1938, 23, 183–186. [Google Scholar]
- Russell, T.A. Palmae. In Flora of West Tropical Africa, 2nd ed.; Hutchinson, J., Dalziel, J.M., Hepper, F., Eds.; Crown Agents for Oversea Governments and Administrations: London, UK, 1968; Volume 3, pp. 159–169. [Google Scholar]
- Dransfield, J. Palmae. In Flora of Tropical East Africa; Polhill, R., Ed.; A.A. Balkema: Rotterdam, The Netherlands, 1986; pp. 1–58. [Google Scholar]
- Dransfield, J.; Beentje, H.J. The Palms of Madagascar; Royal Botanic Gardens and International Palm Society: Kew, UK, 1995. [Google Scholar]
- Baker, W.J.; Smith, G.F. Palmae. In Plants of Angola; Number 23 in Strelitzia; Figueiredo, E., Ed.; South African National Biodiversity Institute (SANBI): Pretoria, South Africa, 2008; pp. 172–173. [Google Scholar]
- Faye, A.; Moutsamboté, J.; Couvreur, T.L.P. Palms in southern Republic of Congo. PALMS 2016, 59, 181–190. [Google Scholar]
- Dransfield, J.; Rakotoarinivo, M. The biogeography of Madagascar palms. In The Biology of Island Floras; Bramwell, D., Caujapé-Castells, J., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 179–196. [Google Scholar]
- Jumelle, H.; Perrier de la Bathié, M. Palmiers. In Flore de Madagascar et des Comores (Plantes Vasculaires); Humbert, H., Ed.; Impr. Officielle. Tananarive: Tananarive, Madagascar, 1945; Volume 30, pp. 1–185. [Google Scholar]
- Mann, G.; Wendland, H.A. On the Palms of Western Tropical Africa. Trans. Linn. Soc. Lond. 1864, 24, 421–440. [Google Scholar] [CrossRef]
- Hardy, O.J.; Born, C.; Budde, K.; Daïnou, K.; Dauby, G.; Duminil, J.Ô.; Ewédjé, E.E.B.; Gomez, C.; Heuertz, M.; Koffi, G.K.; et al. Comparative phylogeography of African rain forest trees: A review of genetic signatures of vegetation history in the Guineo-Congolian region. Comptes Rendus Geosci. 2013, 345, 284–296. [Google Scholar] [CrossRef]
- Helmstetter, A.J.; Amoussou, B.E.N.; Bethune, K.; Kamdem, N.G.; Kakaï, R.G.; Sonké, B.; Couvreur, T.L.P. Phylogenomic data reveal how a climatic inversion and glacial refugia shape patterns of diversity in an African rain forest tree species. bioRxiv 2019. [Google Scholar] [CrossRef]
- Baker, W.J.; Savolainen, V.; Asmussen-Lange, C.B.; Chase, M.W.; Dransfield, J.; Forest, F.; Harley, M.M.; Uhl, N.W.; Wilkinson, M. Complete generic-level phylogenetic analyses of palms (Arecaceae) with comparisons of supertree and supermatrix approaches. Syst. Biol. 2009, 58, 240–256. [Google Scholar] [CrossRef] [PubMed]
- Rohland, N.; Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Gen. Res. 2012, 22, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.G.; Gardner, E.M.; Liu, Y.; Medina, R.; Goffinet, B.; Shaw, A.J.; Zerega, N.J.C.; Wickett, N.J. HybPiper: Extracting Coding Sequence and Introns for Phylogenetics from High-Throughput Sequencing Reads Using Target Enrichment. Appl. Plant Sci. 2016, 4, 1600016. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Sayyari, E.; Mirarab, S. Fast Coalescent-Based Computation of Local Branch Support from Quartet Frequencies. Mol. Biol. Evol. 2016, 33, 1654–1668. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Junier, T.; Zdobnov, E.M. The Newick utilities: High-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 2010, 26, 1669–1670. [Google Scholar] [CrossRef]
- Rabiee, M.; Mirarab, S. SODA: Multi-locus species delimitation using quartet frequencies. bioRxiv 2019. [Google Scholar] [CrossRef]
- Yang, Z. The BPP program for species tree estimation and species delimitation. Curr. Zool. 2015, 61, 854–865. [Google Scholar] [CrossRef]
- Brown, J.W.; Walker, J.F.; Smith, S.A. Phyx: Phylogenetic tools for unix. Bioinformatics 2017, 33, 1886–1888. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Meth. 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Kainer, D.; Mayer, C.; Stamatakis, A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014, 14, 82. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Andermann, T.; Cano, Á.; Zizka, A.; Bacon, C.; Antonelli, A. SECAPR—A bioinformatics pipeline for the rapid and user-friendly processing of targeted enriched Illumina sequences, from raw reads to alignments. PeerJ 2018, 6, e5175. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Gen. Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Li, H.; Barrett, J. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Gen. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Lissambou, B.J.; Couvreur, T.L.P.; Atteke, C.; Stévart, T.; Piñeiro, R.; Dauby, G.; Monthe, F.K.; Ikabanga, D.U.; Sonké, B.; M’batchi, B.; et al. Species delimitation in the genus Greenwayodendron based on morphological and genetic markers reveals new species. TAXON 2019, 68, 442–454. [Google Scholar] [CrossRef]
- Monthe, F.K.; Duminil, J.; Kasongo Yakusu, E.; Beeckman, H.; Bourland, N.; Doucet, J.L.; Sosef, M.S.M.; Hardy, O.J. The African timber tree Entandrophragma congoense (Pierre ex De Wild.) A. Chev. is morphologically and genetically distinct from Entandrophragma angolense (Welw.) C.DC. Tree Gen. Gen. 2018, 14, 66. [Google Scholar] [CrossRef]
- Daïnou, K.; Blanc-Jolivet, C.; Degen, B.; Kimani, P.; Ndiade-Bourobou, D.; Donkpegan, A.S.L.; Tosso, F.; Kaymak, E.; Bourland, N.; Doucet, J.L.; et al. Revealing hidden species diversity in closely related species using nuclear SNPs, SSRs and DNA sequences – a case study in the tree genus Milicia. BMC Evol. Biol. 2016, 16, 259. [Google Scholar] [CrossRef]
- Melo, W.A.; Freitas, C.G.; Bacon, C.D.; Collevatti, R.G. The road to evolutionary success: Insights from the demographic history of an Amazonian palm. Heredity 2018, 121, 183–195. [Google Scholar] [CrossRef]
- Loiseau, O.; Olivares, I.; Paris, M.; de La Harpe, M.; Weigand, A.; Koubínová, D.; Rolland, J.; Bacon, C.D.; Balslev, H.; Borchsenius, F.; et al. Targeted Capture of Hundreds of Nuclear Genes Unravels Phylogenetic Relationships of the Diverse Neotropical Palm Tribe Geonomateae. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helmstetter, A.J.; Kamga, S.M.; Bethune, K.; Lautenschläger, T.; Zizka, A.; Bacon, C.D.; Wieringa, J.J.; Stauffer, F.; Antonelli, A.; Sonké, B.; et al. Unraveling the Phylogenomic Relationships of the Most Diverse African Palm Genus Raphia (Calamoideae, Arecaceae). Plants 2020, 9, 549. https://doi.org/10.3390/plants9040549
Helmstetter AJ, Kamga SM, Bethune K, Lautenschläger T, Zizka A, Bacon CD, Wieringa JJ, Stauffer F, Antonelli A, Sonké B, et al. Unraveling the Phylogenomic Relationships of the Most Diverse African Palm Genus Raphia (Calamoideae, Arecaceae). Plants. 2020; 9(4):549. https://doi.org/10.3390/plants9040549
Chicago/Turabian StyleHelmstetter, Andrew J., Suzanne Mogue Kamga, Kevin Bethune, Thea Lautenschläger, Alexander Zizka, Christine D. Bacon, Jan J. Wieringa, Fred Stauffer, Alexandre Antonelli, Bonaventure Sonké, and et al. 2020. "Unraveling the Phylogenomic Relationships of the Most Diverse African Palm Genus Raphia (Calamoideae, Arecaceae)" Plants 9, no. 4: 549. https://doi.org/10.3390/plants9040549
APA StyleHelmstetter, A. J., Kamga, S. M., Bethune, K., Lautenschläger, T., Zizka, A., Bacon, C. D., Wieringa, J. J., Stauffer, F., Antonelli, A., Sonké, B., & Couvreur, T. L. P. (2020). Unraveling the Phylogenomic Relationships of the Most Diverse African Palm Genus Raphia (Calamoideae, Arecaceae). Plants, 9(4), 549. https://doi.org/10.3390/plants9040549





