Transcriptome Profiling of Ornithogalum dubium Leaves and Flowers to Identify Key Carotenoid Genes for CRISPR Gene Editing
Abstract
1. Introduction
2. Results and Discussion
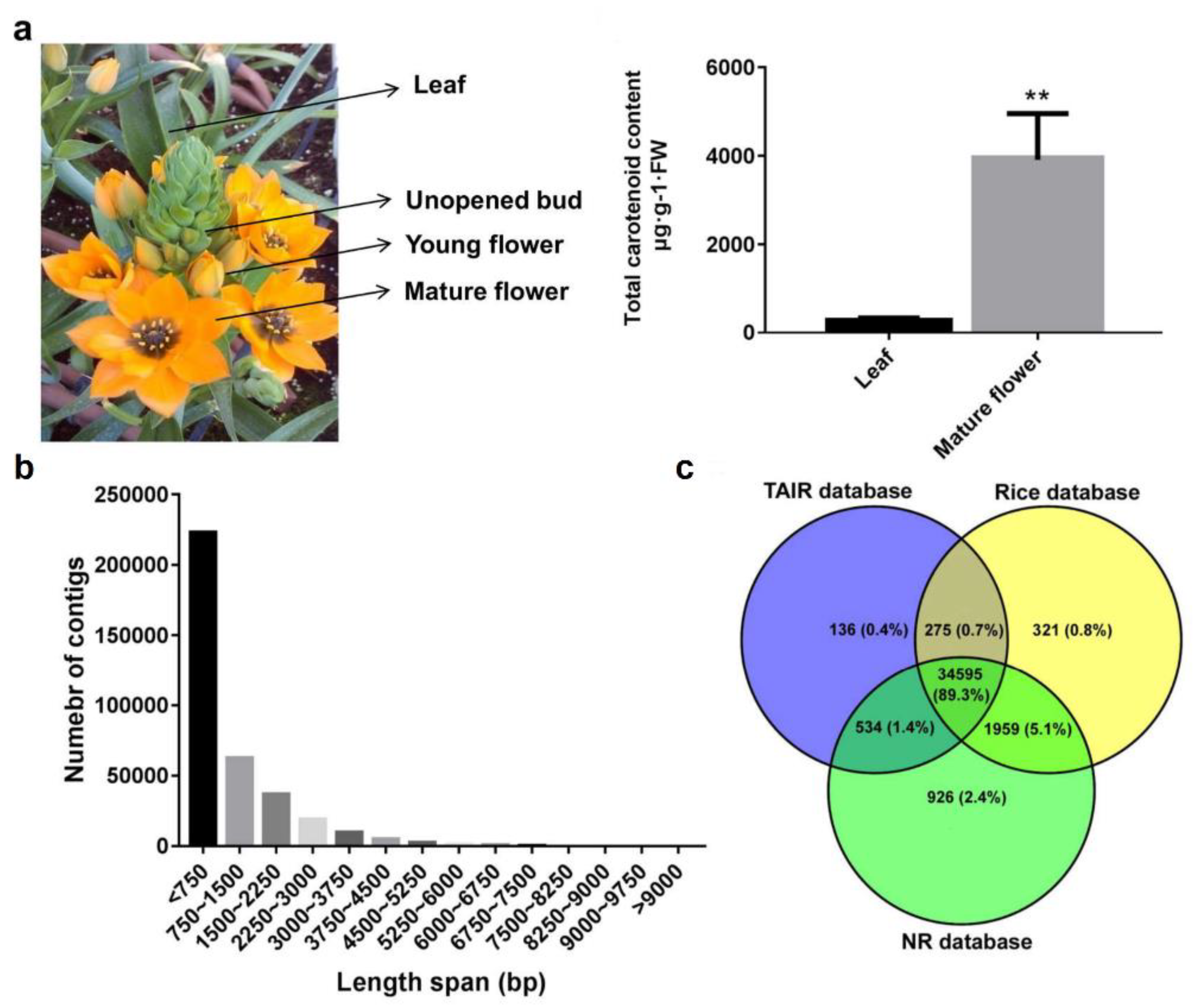
2.1. Transcriptome Sequencing, De Novo Assembly and Functional Annotation of O. dubium Leaves and Flowers
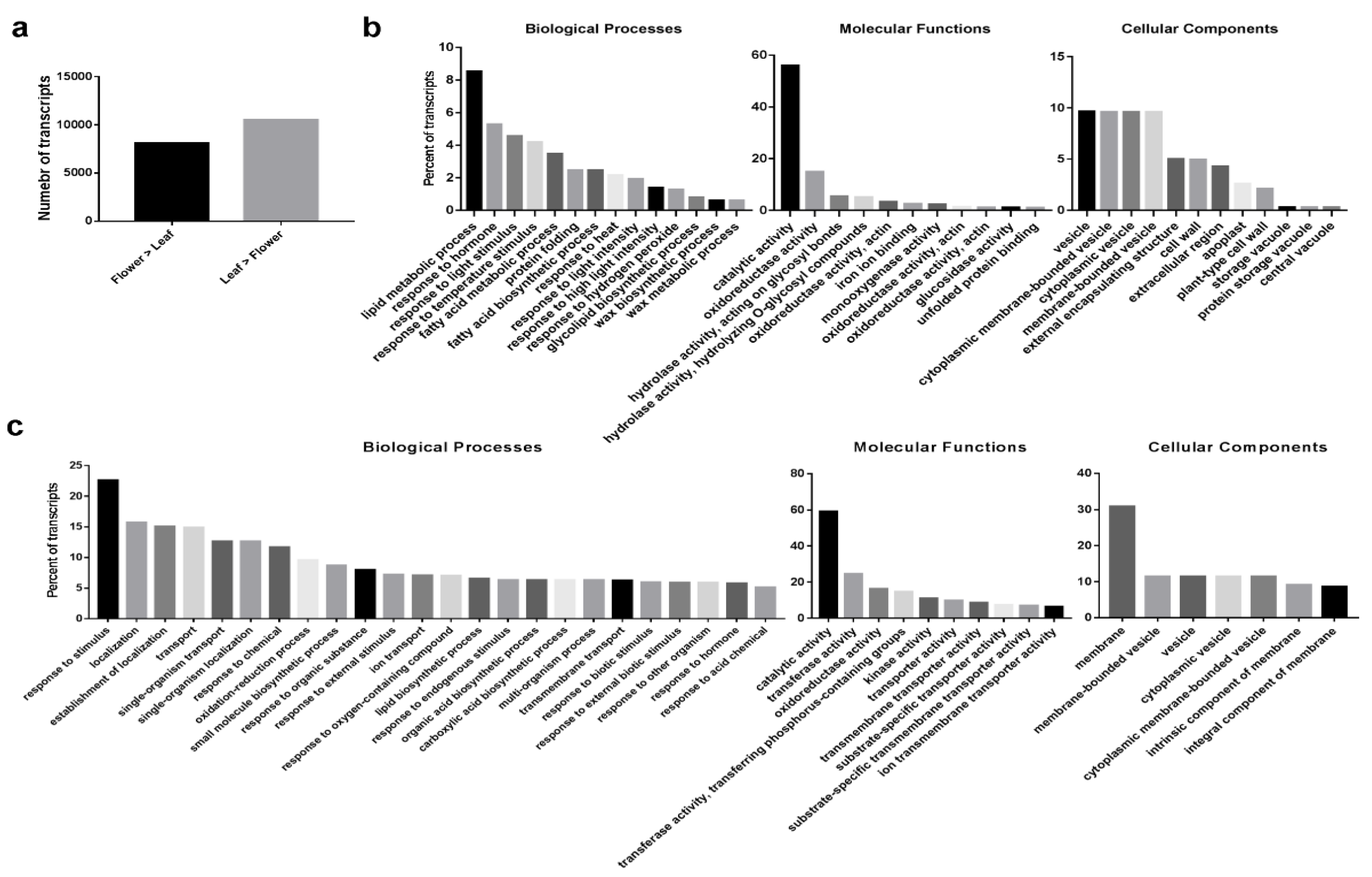
2.2. Differential Expression (DE) Transcripts between O. dubium Leaves and Flowers
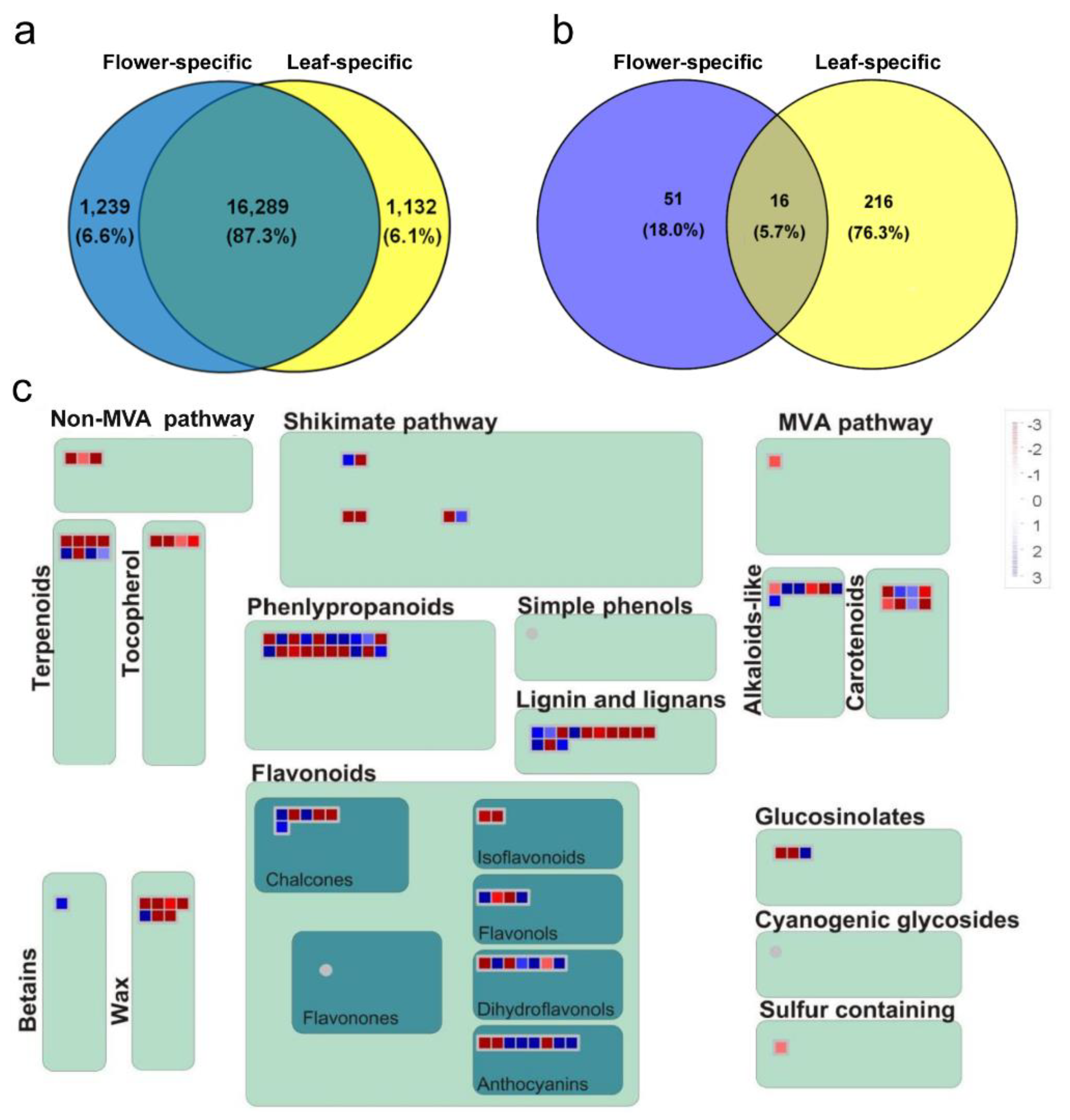
2.3. Common and Specific DE Transcripts between O. dubium Leaves and Flowers
2.4. Carotenoid Contents and Distribution between O. dubium Leaves and Flowers
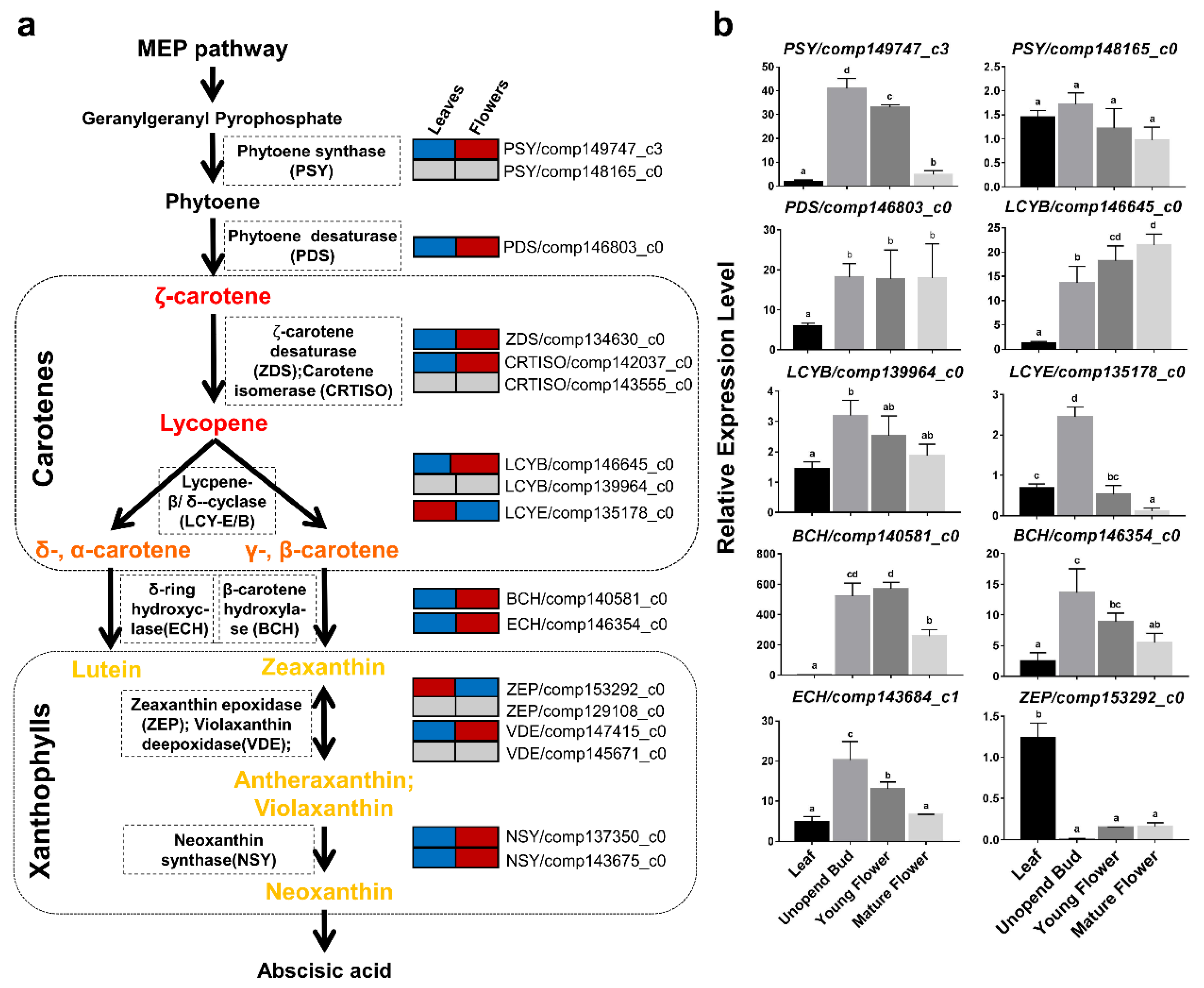
2.5. Identification of Key Transcripts in the Carotenoid Biosynthetic Pathway of O. dubium
3. Materials and Methods
3.1. Plant Materials
3.2. RNA Extraction, Pair-End Library Construction and Sequencing
3.3. Sequence Processing, Transcriptome Assembly and Functional Annotation
3.4. Differential Expression (DE) of Transcripts and GO-Enrichment Analysis
3.5. Carotenoid Extraction and HPLC Analysis
3.6. Quantitative PCR Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Noman, A.; Aqeel, M.; Deng, J.; Khalid, N.; Sanaullah, T.; He, S. Biotechnological advancements for improving floral attributes in ornamental plants. Front. Plant Sci. 2017, 8, 530. [Google Scholar] [CrossRef] [PubMed]
- Tun, O.M.; Lipsky, A.; Knaan, T.L.; Kerem, Z.; Yedidia, I. The plant activator BTH promotes Ornithogalum dubium and O. thyrsoides differentiation and regeneration in vitro. Biol. Plant. 2013, 57, 41–48. [Google Scholar] [CrossRef]
- Lipsky, A.; Cohen, A.; Ion, A.; Yedidia, I. Genetic transformation of Ornithogalum via particle bombardment and generation of Pectobacterium carotovorum-resistant plants. Plant Sci. 2014, 228, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for crop improvement: An update review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef] [PubMed]
- Francis, D.; Finer, J.J.; Grotewold, E. Challenges and opportunities for improving food quality and nutrition through plant biotechnology. Curr. Opin. Biotechnol. 2017, 44, 124–129. [Google Scholar] [CrossRef]
- Que, Z.Q.; Lu, Q.N.; Liu, X.L.; Li, S.Q.; Zeng, H.L.; Li, R.G.; Liu, T.; Yan, X.Y.; Shen, C.X. A method to create golden hull rice biological resources using CRISPR/Cas9 system. J. Biobased Mater. Bioenergy 2018, 12, 59–64. [Google Scholar] [CrossRef]
- Klimek-Chodacka, M.; Oleszkiewicz, T.; Lowder, L.G.; Baranski, R. Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 2018, 37, 575–586. [Google Scholar] [CrossRef]
- Watanabe, K.; Kobayashi, A.; Endo, M.; Sage-Ono, K.; Toki, S.; Ono, M. CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci. Rep. 2017, 7, 10028. [Google Scholar] [CrossRef]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef]
- D’Ambrosio, C.; Stigliani, A.L.; Giorio, G. CRISPR/Cas9 editing of carotenoid genes in tomato. Transgenic Res. 2018, 27, 367–378. [Google Scholar] [CrossRef]
- Griesbach, R.; Meyer, F.; Koopowitz, H. Creation of new flower colors in Ornithogalum via interspecific hybridization. J. Am. Soc. Hortic. Sci. 1993, 118, 409–414. [Google Scholar] [CrossRef]
- Smulders, M.J.M.; Arens, P. New developments in molecular techniques for breeding in ornamentals. In Ornamental Crops; Van Huylenbroeck, J., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 213–230. [Google Scholar]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A.; Kato, M.; Shimada, T.; Nashima, K.; Kishimoto, S.; Nagata, M. Molecular basis of carotenoid accumulation in horticultural crops. Hortic. J. 2019, 88, 135–139. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Ramakrishna, A.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Heiling, S.; Schuman, M.C.; Schoettner, M.; Mukerjee, P.; Berger, B.; Schneider, B.; Jassbi, A.R.; Baldwin, I.T. Jasmonate and ppHsystemin regulate key malonylation steps in the biosynthesis of 17-hydroxygeranyllinalool diterpene glycosides, an abundant and effective direct defence against herbivores in Nicotiana attenuata. Plant Cell 2010, 22, 273–292. [Google Scholar] [CrossRef]
- Behr, M.; Sergeant, K.; Leclercq, C.C.; Planchon, S.; Guignard, C.; Lenouvel, A.; Renaut, J.; Hausman, J.F.; Lutts, S.; Guerriero, G. Insights into the molecular regulation of monolignol-derived product biosynthesis in the growing hemp hypocotyl. BMC Plant Biol. 2018, 18, 1. [Google Scholar] [CrossRef]
- Park, C.S.; Go, Y.S.; Suh, M.C. Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 2016, 88, 257–270. [Google Scholar] [CrossRef]
- Scarano, A.; Chieppa, M.; Santino, A. Looking at flavonoid biodiversity in horticultural crops: A colored mine with nutritional benefits. Plants 2018, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Domonkos, I.; Kis, M.; Gombos, Z.; Ughy, B. Carotenoids, versatile components of oxygenic photosynthesis. Prog. Lipid Res. 2013, 52, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Bai, C.; Sanahuja, G.; Yuan, D.; Farré, G.; Naqvi, S.; Shi, L.; Capell, T.; Christou, P. The regulation of carotenoid pigmentation in flowers. Arch. Biochem. Biophys. 2010, 504, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Galpaz, N.; Ronen, G.; Khalfa, Z.; Zamir, D.; Hirschberg, J. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 2006, 18, 1947–1960. [Google Scholar] [CrossRef] [PubMed]
- Bartley, G.E.; Viitanen, P.V.; Bacot, K.O.; Scolnik, P.A. A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J. Biol. Chem. 1992, 267, 5036–5039. [Google Scholar]
- Bartley, G.E.; Scolnik, P.A. cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J. Biol. Chem. 1993, 268, 25718–25721. [Google Scholar]
- Fraser, P.D.; Truesdale, M.R.; Bird, C.R.; Schuch, W.; Bramley, P.M. Carotenoid biosynthesis during tomato fruit development. Plant Physiol. 1994, 105, 405–413. [Google Scholar] [CrossRef]
- Pecker, R.; Gabbay, F.X.; Cunningham, J.; Hirschberg, J. Cloning and characterization of the cDNA for lycopene beta-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol. Biol. 1996, 30, 807–819. [Google Scholar] [CrossRef]
- Ronen, G.; Carmel-Goren, L.; Zamir, D.; Hirschberg, J. An alternative pathway to beta -carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. USA 2000, 97, 11102–11107. [Google Scholar] [CrossRef]
- Corona, V.; Aracri, B.; Kosturkova, G.; Bartley, G.E.; Pitto, L.; Giorgetti, L.; Scolnik, P.A.; Giuliano, G. Regulation of a carotenoid biosynthesis gene promoter during plant development. Plant J. 1996, 9, 505–512. [Google Scholar] [CrossRef]
- Stanley, L.; Yuan, Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef] [PubMed]
- Ronen, G.; Cohen, M.; Zamir, D.; Hirschberg, J. Regulation of carotenoid biosynthesis during tomato fruit development: Expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 1999, 17, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Bang, H.; Kim, S.; Leskovar, D.; King, S. Development of a codominant CAPS marker for allelic selection between canary yellow and red watermelon based on SNP in lycopene β-cyclase (LCYB) gene. Mol. Breed. 2007, 20, 63–72. [Google Scholar] [CrossRef]
- Bang, H.; Yi, G.; Kim, S.; Leskovar, D.; Patil, B.S. Watermelon lycopene β-cyclase: Promoter characterization leads to the development of a PCR marker for allelic selection. Euphytica 2014, 200, 363–378. [Google Scholar] [CrossRef]
- Blas, A.L.; Ming, R.; Liu, Z.; Veatch, O.J.; Paull, R.E.; Moore, P.H.; Yu, Q. Cloning of the papaya chromoplast-specific lycopene β-cyclase, CpCYC-b, controlling fruit flesh color reveals conserved microsynteny and a recombination hot spot. Plant Physiol. 2010, 152, 2013–2022. [Google Scholar] [CrossRef]
- Alquezar, B.; Rodrigo, M.J.; Lado, J.; Zacarías, L. A comparative physiological and transcriptional study of carotenoid biosynthesis in white and red grapefruit (Citrus paradisi Macf.). Tree Genet. Genomes 2013, 9, 1257–1269. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zheng, X.; Zhu, K.; Xu, Q.; Deng, X. Molecular characterization, critical amino acid identification, and promoter analysis of a lycopene β-cyclase gene from citrus. Tree Genet. Genomes 2016, 12, 106. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Conesa, A.; Stefan, G.; Juan, M.G.G.; Javier, T.; Manuel, T.; Montserrat, R. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Bauer, S.; Grossmann, S.; Vingron, M.; Robinson, P.N. Ontologizer 2.0—A multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 2008, 24, 1650–1651. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef]
- Isaacson, T.; Ronen, G.; Zamir, D.; Hirschberg, J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-Carotene and xanthophylls in plants. Plant Cell 2002, 14, 333–342. [Google Scholar] [CrossRef]
- Rodrigo, M.J.; Alquézar, B.; Alós, E.; Medina, V.; Carmona, L.; Bruno, M.; Al-Babili, S.; Zacarías, L. A novel carotenoid cleavage activity involved in the biosynthesis of citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 2013, 64, 4461–4478. [Google Scholar] [CrossRef]
- Misawa, N.; Nakagawa, M.; Kobayashi, K.; Yamano, S.; Nakamura, K.; Harashima, K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 1990, 172, 6704–6712. [Google Scholar] [CrossRef]
- Britton, G.; Liaaen-Jensen, S.; Pfander, H. Carotenoids Handbook; Birkhauser Verlag: Basel, Switzerland, 2004. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Sample | Leaves | Flowers |
|---|---|---|
| Raw reads | 83,956,119 | 97,041,800 |
| Clean reads (%) | 72,125,398 (85.9%) | 83,738,661(86.3%) |
| Total clean reads (%) | 155,864,059 (86.1%) | |
| Total assembled bases | 344,991,254 bp | |
| Sequence number | 360,689 | |
| Median contig length | 494 bp | |
| Length range | 201–16,643 bp | |
| Mean length | 956.5 bp | |
| N50 | 1759 bp | |
| GC content | 41.1% | |
| Carotenoid Composition | Leaves (Mean ± Std) | Flowers (Mean ± Std) |
|---|---|---|
| Phytoene | 0.0 | 20.7 ± 5.6 |
| Phytofluene | 0.0 | 16.0 ± 9.0 |
| β-carotene | 54.8 ± 22.0 | 80.2 ± 22.9 ** b |
| Lutein | 149.7 ± 35.4 | 880.7 ± 192.8 ** |
| β-cryptoxanthin | 0.0 | 117.6 ± 39.1 |
| Zeaxanthin | 9.0 ± 3.3 | 1,561.5 ± 304.5 *** |
| Violaxanthin | 12.5 ± 5.8 | 128.8 ± 59.0 * |
| Neoxanthin | 2.7 ± 0.7 | 147.2 ± 94.1 * |
| a UK2 | 24.2 ± 9.1 | 9.4 ± 8.9 |
| UK3 | 15.0 ± 5.8 | 21.3 ± 4.9 |
| UK5 | 0.0 | 528.6 ± 226.9 |
| UK11 | 0.0 | 159.0 ± 44.0 |
| others | 27.0 ± 11.5 | 241.8 ± 53.9 ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Z.; Arazi, T.; Hod, N.; Zohar, M.; Isaacson, T.; Doron-Faigenboim, A.; Reznik, N.; Yedidia, I. Transcriptome Profiling of Ornithogalum dubium Leaves and Flowers to Identify Key Carotenoid Genes for CRISPR Gene Editing. Plants 2020, 9, 540. https://doi.org/10.3390/plants9040540
Wei Z, Arazi T, Hod N, Zohar M, Isaacson T, Doron-Faigenboim A, Reznik N, Yedidia I. Transcriptome Profiling of Ornithogalum dubium Leaves and Flowers to Identify Key Carotenoid Genes for CRISPR Gene Editing. Plants. 2020; 9(4):540. https://doi.org/10.3390/plants9040540
Chicago/Turabian StyleWei, Zunzheng, Tzahi Arazi, Nofar Hod, Matat Zohar, Tal Isaacson, Adi Doron-Faigenboim, Noam Reznik, and Iris Yedidia. 2020. "Transcriptome Profiling of Ornithogalum dubium Leaves and Flowers to Identify Key Carotenoid Genes for CRISPR Gene Editing" Plants 9, no. 4: 540. https://doi.org/10.3390/plants9040540
APA StyleWei, Z., Arazi, T., Hod, N., Zohar, M., Isaacson, T., Doron-Faigenboim, A., Reznik, N., & Yedidia, I. (2020). Transcriptome Profiling of Ornithogalum dubium Leaves and Flowers to Identify Key Carotenoid Genes for CRISPR Gene Editing. Plants, 9(4), 540. https://doi.org/10.3390/plants9040540




