The Effect of Recombinant Tags on Citrus paradisi Flavonol-Specific 3-O Glucosyltransferase Activity
Abstract
1. Introduction
2. Results and Discussion
2.1. Effect of Tags on Optimal pH
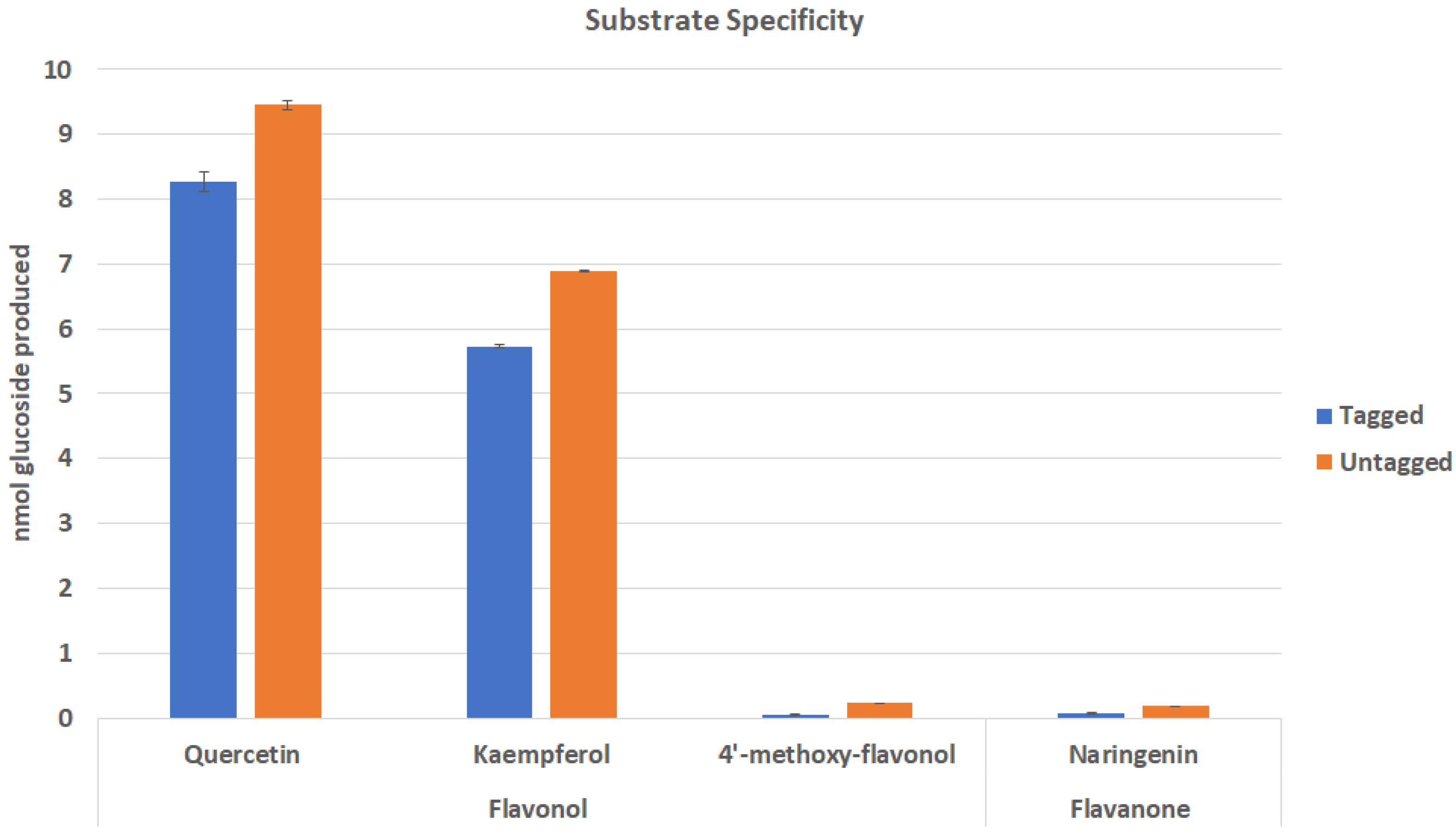
2.2. Effect of Tags on Substrate Specificity of Cp3GT
2.3. Effect of Tags on Kinetic Parameters
Effect of Tags and Metals on Cp3GT Activity
3. Conclusions and Directions for Future Research
4. Methods and Materials
4.1. Reagents and Materials
4.2. Insertion of Thrombin Cleavage Site and Transformation into Yeast
4.3. Removal of Recombinant Tags by Thrombin Digestion
4.4. Purification and GT Enzyme Assay
4.4.1. pH Optima
4.4.2. Substrate Specificity
4.4.3. Kinetics
4.4.4. Metals and UDP Inhibition
4.5. Statistial Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mathesius, U. Flavonoid functions in plants and their interactions with other organisms. Plants 2018, 7, 30. [Google Scholar] [CrossRef]
- Agati, G.; Tattini, M. Multiple functional roles of flavonoids in photoprotection. New Phytol. 2010, 186, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.W.; Murray, J.D. The role of flavonoids in nodulation host-range specificity: An update. Plants 2016, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, M.S.J. Importance of flavonoids in insect-plant interactions: Feeding and oviposition. Phytochemistry 2001, 56, 245–252. [Google Scholar] [CrossRef]
- Kozłowska, A.; Szostak-Wegierek, D. Flavonoids—Food sources and health benefits. Rocz. Panstw. Zakl. Hig. 2014, 65, 79–85. [Google Scholar] [PubMed]
- De Masi, L.; Bontempo, P.; Rigano, D.; Stiuso, P.; Carafa, V.; Nebbioso, A.; Piacente, S.; Montoro, P.; Aversano, R.; D’Amelia, V.; et al. Comparative phytochemical characterization, genetic profile, and antiproliferative activity of polyphenol-rich extracts from pigmented tubers of different Solanum tuberosum varieties. Molecules 2020, 25, 233. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Mumper, R.J. Plant phenolics: Extraction, analysis and their antioxidant and anticancer properties. Molecules 2010, 15, 7313–7352. [Google Scholar] [CrossRef]
- Ferreyra, M.L.F.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 3. [Google Scholar]
- Zhou, K.; Qiao, K.; Edgar, S.; Stephanopoulos, G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 2015, 33, 377–383. [Google Scholar] [CrossRef]
- Chen, W.; Yao, J.; Meng, J.; Han, W.; Tao, Y.; Chen, Y.; Guo, Y.; Shi, G.; He, Y.; Jin, J.M.; et al. Promiscuous enzymatic activity-aided multiple-pathway network design for metabolic flux rearrangement in hydroxytyrosol biosynthesis. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Terpe, K. Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.; Holliger, P.; Winter, G. Improved tumor targeting by disulphide stabilized diabodies expressed in Pichia pastoris. Protein Eng. 1997, 10, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Furqan, B.R.N. Heterologous expression and characterization of thermostable lipase (Lk1) in Pichia pastoris GS115. Biocatal. Agric. Biotechnol. 2020, 23, 101448. [Google Scholar] [CrossRef]
- Hong, R.; Sun, Y.; Su, L.; Gu, L.; Wang, F.; Wu, J. High-level expression of Humicola insolens cutinase in Pichia pastoris without carbon starvation and its use in cotton fabric bioscouring. J. Biotechnol. 2019, 304, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, Y.; Yuan, Q. Study the effect of His-tag on chondroitinase ABC I based on characterization of enzyme. Int. J. Biol. Macromol. 2015, 78, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Dickson, J.M.J.; Lee, W.J.; Shepherd, P.R.; Buchanan, C.M. Enzyme activity effects of N-terminal His-tag attached to catalytic sub-unit of phosphoinositide-3-kinase. Biosci. Rep. 2013, 33, 857–863. [Google Scholar] [CrossRef]
- Sabaty, M.; Grosse, S.; Adryanczyk, G.; Boiry, S.; Biaso, F.; Arnoux, P.; Pignol, D. Detrimental effect of the 6 His C-terminal tag on YedY enzymatic activity and influence of the TAT signal sequence on YedY synthesis. BMC Biochem. 2013, 14, 28. [Google Scholar] [CrossRef]
- Yeon, Y.J.; Park, H.J.; Park, H.Y.; Yoo, Y.J. Effect of His-tag location on the catalytic activity of 3-hydroxybutyrate dehydrogenase. Biotechnol. Bioprocess Eng. 2014, 19, 798–802. [Google Scholar] [CrossRef]
- Smyth, D.R.; Mrozkiewicz, M.K.; McGrath, W.J.; Listwan, P.; Kobe, B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003, 12, 1313–1322. [Google Scholar] [CrossRef]
- Noirclerc-Savoye, M.; Flayhan, A.; Pereira, C.; Gallet, B.; Gans, P.; Ebel, C.; Breyton, C. Tail proteins of phage T5: Investigation of the effect of the His6-tag position, from expression to crystallisation. Protein Expr. Purif. 2015, 109, 70–78. [Google Scholar] [CrossRef]
- Woestenenk, E.A.; Hammarström, M.; Van Den Berg, S.; Härd, T.; Berglund, H. His tag effect on solubility of human proteins produced in Escherichia coli: A comparison between four expression vectors. J. Struct. Funct. Genom. 2004, 5, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Carson, M.; Johnson, D.H.; McDonald, H.; Brouillette, C.; DeLucas, L.J. His-tag impact on structure. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.K.; McIntosh, C.A. Identification, recombinant expression, and biochemical characterization of a flavonol 3-O-glucosyltransferase clone from Citrus paradisi. Phytochemistry 2009, 70, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, S.P.; Tolliver, B.M.; Zhang, C.; Owens, D.K.; McIntosh, C.A. Mutational analysis of substrate specificity in a Citrus paradisi flavonol 3-O-glucosyltransferase. J. Plant Biochem. Biotechnol. 2018, 27, 13–27. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Xu, Z.-S.; Xiong, A.-S. Identification and characterization of DcUSAGT1, a UDP-glucose: Sinapic acid glucosyltransferase from purple carrot taproots. PLoS ONE 2016, 11, e0154938. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, C.A.; Owens, D.K. Advances in flavonoid glycosyltransferase research: Integrating recent findings with long-term citrus studies. Phytochem. Rev. 2016, 15, 1075–1091. [Google Scholar] [CrossRef]
- Haroth, S.; Feussner, K.; Kelly, A.A.; Zienkiewicz, K.; Shaikhqasem, A.; Herrfurth, C.; Feussner, I. The glycosyltransferase UGT76E1 significantly contributes to 12-O-glucopyranosyl-jasmonic acid formation in wounded Arabidopsis thaliana leaves. J. Biol. Chem. 2019, 294, 9858–9872. [Google Scholar] [CrossRef]
- Cui, L.; Yao, S.; Dai, X.; Yin, Q.; Liu, Y.; Jiang, X.; Wu, Y.; Qian, Y.; Pang, Y.; Gao, L.; et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 2016, 67, 2285–2297. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.L.; Lai, C.J.S.; Wang, R.S.; Kang, L.P.; Ma, T.; Zhao, Z.H.; Gao, W.; Huang, L.Q. Functional characterization of three flavonoid glycosyltransferases from Andrographis paniculata. R. Soc. Open Sci. 2019, 6, 190150. [Google Scholar] [CrossRef]
- Ono, E.; Homma, Y.; Horikawa, M.; Kunikane-Doi, S.; Imai, H.; Takahashi, S.; Kawai, Y.; Ishiguro, M.; Fukui, Y.; Nakayama, T. Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 2010, 22, 2856–2871. [Google Scholar] [CrossRef]
- Gasser, B.; Saloheimo, M.; Rinas, U.; Dragosits, M.; Rodríguez-Carmona, E.; Baumann, K.; Giuliani, M.; Parrilli, E.; Branduardi, P.; Lang, C.; et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: A host comparative overview. Microb. Cell Fact. 2008, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Corsini, L.; Hothorn, M.; Scheffzek, K.; Sattler, M.; Stier, G. Thioredoxin as a fusion tag for carrier-driven crystallization. Protein Sci. 2008, 17, 2070–2079. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; Masullo, M.; Ruocco, M.R.; Grimaldi, P.; Lanzotti, M.A.; Arcari, P.; Zagari, A.; Vitagliano, L. Structure and stability of a thioredoxin reductase from Sulfolobus solfataricus: A thermostable protein with two functions. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Cura, V.; Gangloff, M.; Eiler, S.; Moras, D.; Ruff, M. Cleaved thioredoxin fusion protein enables the crystallization of poorly soluble ERα in complex with synthetic ligands. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, S.P.; Owens, D.K.; Sibhatu, M.B.; Sarkar, T.R.; Strong, C.L.; Mallampalli, V.K.P.S.; Asiago, J.; Cooke, J.; Kiser, S.; Lin, Z.; et al. Identification, recombinant expression, and biochemical analysis of putative secondary product glucosyltransferases from Citrus paradisi. J. Agric. Food Chem. 2016, 64, 1957–1969. [Google Scholar] [CrossRef] [PubMed]
- Kogawa, K.; Kato, N.; Kazuma, K.; Noda, N.; Suzuki, M. Purification and characterization of UDP-glucose: Anthocyanin 3′,5′-O-glucosyltransferase from Clitoria ternatea. Planta 2007, 226, 1501–1509. [Google Scholar] [CrossRef]
- Ciesla, W.P.; Bobak, D.A. Clostridium difficile toxins A and B are cation-dependent UDP-glucose hydrolases with differing catalytic activities. J. Biol. Chem. 1998, 273, 16021–16026. [Google Scholar] [CrossRef]
- Ford, C.M.; Boss, P.K.; Hæj, P.B. Cloning and characterization of Vitis vinifera UDP-glucose. Flavonoid 3-O-Glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J. Biol. Chem. 1998, 273, 9224–9233. [Google Scholar] [CrossRef]
- Yamazaki, M.; Gong, Z.; Fukuchi-Mizutani, M.; Fukui, Y.; Tanaka, Y.; Kusumi, T.; Saito, K. Molecular cloning and biochemical characterization of a novel anthocyanin 5-O-glucosyltransferase by mRNA differential display for plant forms regarding anthocyanin. J. Biol. Chem. 1999, 274, 7405–7411. [Google Scholar] [CrossRef]
- Taguchi, G.; Ubukata, T.; Hayashida, N.; Yamamoto, H.; Okazaki, M. Cloning and characterization of a glucosyltransferase that reacts on 7-hydroxyl group of flavonol and 3-hydroxyl group of coumarin from tobacco cells. Arch. Biochem. Biophys. 2003, 420, 95–102. [Google Scholar] [CrossRef]
- Noguchi, A.; Saito, A.; Homma, Y.; Nakao, M.; Sasaki, N.; Nishino, T.; Takahashi, S.; Nakayama, T. A UDP-glucose: Isoflavone 7-O-glucosyltransferase from the roots of soybean (Glycine max) seedlings: Purification, gene cloning, phylogenetics, and an implication for an alternative strategy of enzyme catalysis. J. Biol. Chem. 2007, 282, 23581–23590. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, C.A.; Latchinian, L.; Mansell, R.L. Flavanone-specific 7-O-glucosyltransferase activity in Citrus paradisi seedlings: Purification and characterization. Arch. Biochem. Biophys. 1990, 282, 50–57. [Google Scholar] [CrossRef]
- Owens, D.K.; Mcintosh, C.A.; Flavonoids, C.; Glycosides, F. The Biological Activity of Phytochemicals; Springer: New York, NY, USA, 2011. [Google Scholar]
- Mclntosh, C.A.; Mansell, R.L. Biosynthesis of naringin in Citrus Paradisi: UDP-glucosyl-transferase activity in grapefruit seedlings. Phytochemistry 1990, 29, 1533–1538. [Google Scholar] [CrossRef]
- Zea, C.J.; Camci-Unal, G.; Pohl, N.L. Thermodynamics of binding of divalent magnesium and manganese to uridine phosphates: Implications for diabetes-related hypomagnesaemia and carbohydrate biocatalysis. Chem. Cent. J. 2008, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; He, X.; Achnine, L.; Blount, J.W.; Dixon, R.A.; Wang, X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 2005, 17, 3141–3154. [Google Scholar] [CrossRef] [PubMed]
- Offen, W.; Martinez-Fleites, C.; Yang, M.; Kiat-Lim, E.; Davis, B.G.; Tarling, C.A.; Ford, C.M.; Bowles, D.J.; Davies, G.J. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006, 25, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Brazier-Hicks, M.; Offen, W.A.; Gershater, M.C.; Revett, T.J.; Lim, E.K.; Bowles, D.J.; Davies, G.J.; Edwards, R. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 20238–20243. [Google Scholar] [CrossRef]
- Li, L.; Modolo, L.V.; Escamilla-Trevino, L.L.; Achnine, L.; Dixon, R.A.; Wang, X. Crystal structure of Medicago truncatula UGT85H2—Insights into the structural basis of a multifunctional (Iso)flavonoid glycosyltransferase. J. Mol. Biol. 2007, 370, 951–963. [Google Scholar] [CrossRef]
- Modolo, L.V.; Li, L.; Pan, H.; Blount, J.W.; Dixon, R.A.; Wang, X. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (Iso)flavonoids. J. Mol. Biol. 2009, 392, 1292–1302. [Google Scholar] [CrossRef]
- Hiromoto, T.; Honjo, E.; Noda, N.; Tamada, T.; Kazuma, K.; Suzuki, M.; Blaber, M.; Kuroki, R. Structural basis for acceptor-substrate recognition of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. Protein Sci. 2015, 24, 395–407. [Google Scholar] [CrossRef]
- Zong, G.; Li, J.; Gao, Y.; Fei, S.; Liu, X.; Wang, X.; Shen, Y. Overexpression, purification, biochemical and structural characterization of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana. Protein Expr. Purif. 2019, 156, 44–49. [Google Scholar] [CrossRef] [PubMed]

| Substrate | Kmapp (µ) | Vmax (pKat/µg) | Kcat (s−1) | Kcat/Kmapp (µM/s) | |
|---|---|---|---|---|---|
| Quercetin* | Tagged | 51.9 ± 8.29 | 31.7 ± 3.39 | 1.84 ± 0.19 | 0.035 ± 0.001 |
| Untagged | 42.4 ± 3.48 | 26.5 ± 1.06 | 1.54 ± 0.06 | 0.036 ± 0.001 | |
| Kaempferol* | Tagged | 44.1 ± 3.41 | 22.0 ± 0.56 | 1.28 ± 0.02 | 0.029 ± 0.001 |
| Untagged | 53.7 ± 10.6 | 23.1 ± 3.00 | 1.34 ± 0.17 | 0.025 ± 0.001 | |
| UDPG** | Tagged | 49.7 ± 5.32 | 10.9 ± 1.59 | 0.637 ± 0.092 | 0.012 ± 0.002 |
| Untagged | 55.0 ± 4.97 | 12.7 ± 2.15 | 0.738 ± 0.125 | 0.012 ± 0.0004 |
| % Relative Activity | ||||
|---|---|---|---|---|
| Metal | Tagged | Untagged | ||
| 1 mM | 10 mM | 1 mM | 10 mM | |
| Untreated | 100 | 100 | 100 | 100 |
| ZnCl2 | 44 | 28 | 25 | 23 |
| KCl | 117 | 119 | 114 | 112 |
| FeSO4 | 42 | 16 | 42 | 5 |
| NaCl | 118 | 121 | 100 | 95 |
| Na2SO4 | 92 | 88 | 92 | 87 |
| CaCl2 | 89 | 69 | 84 | 72 |
| CuSO4 | 79 | 3 | 70 | 3 |
| MnCl2 | 87 | 87 | 60 | 56 |
| MgCl2 | 104 | 123 | 103 | 140 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Birchfield, A.S.; McIntosh, C.A. The Effect of Recombinant Tags on Citrus paradisi Flavonol-Specific 3-O Glucosyltransferase Activity. Plants 2020, 9, 402. https://doi.org/10.3390/plants9030402
Birchfield AS, McIntosh CA. The Effect of Recombinant Tags on Citrus paradisi Flavonol-Specific 3-O Glucosyltransferase Activity. Plants. 2020; 9(3):402. https://doi.org/10.3390/plants9030402
Chicago/Turabian StyleBirchfield, Aaron S., and Cecilia A. McIntosh. 2020. "The Effect of Recombinant Tags on Citrus paradisi Flavonol-Specific 3-O Glucosyltransferase Activity" Plants 9, no. 3: 402. https://doi.org/10.3390/plants9030402
APA StyleBirchfield, A. S., & McIntosh, C. A. (2020). The Effect of Recombinant Tags on Citrus paradisi Flavonol-Specific 3-O Glucosyltransferase Activity. Plants, 9(3), 402. https://doi.org/10.3390/plants9030402




