Plant Ribonuclease J: An Essential Player in Maintaining Chloroplast RNA Quality Control for Gene Expression
Abstract
1. An Overview of Chloroplast Gene Expression
2. Relaxed Transcription in Chloroplasts
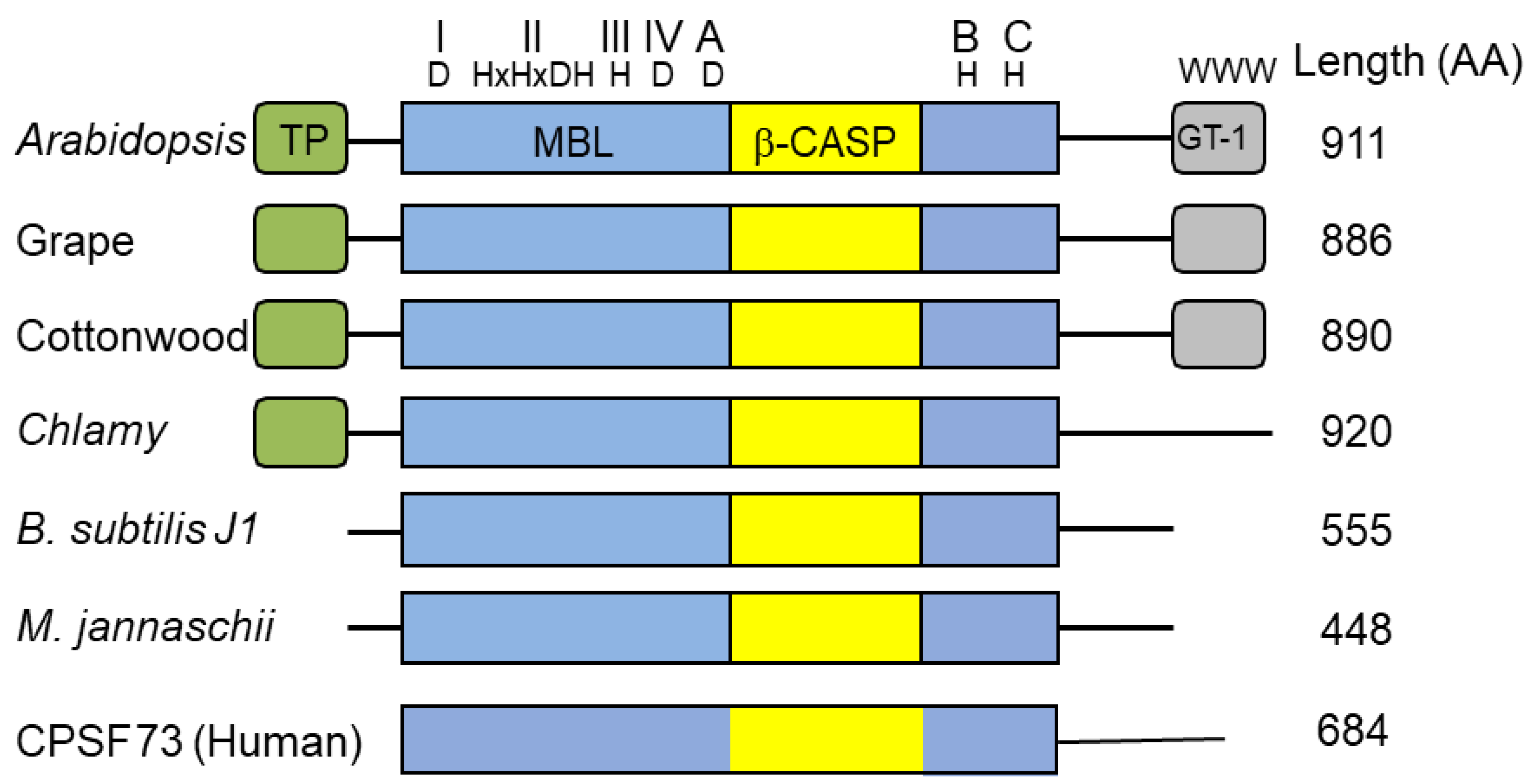
3. Ribonuclease J and β-CASP Proteins
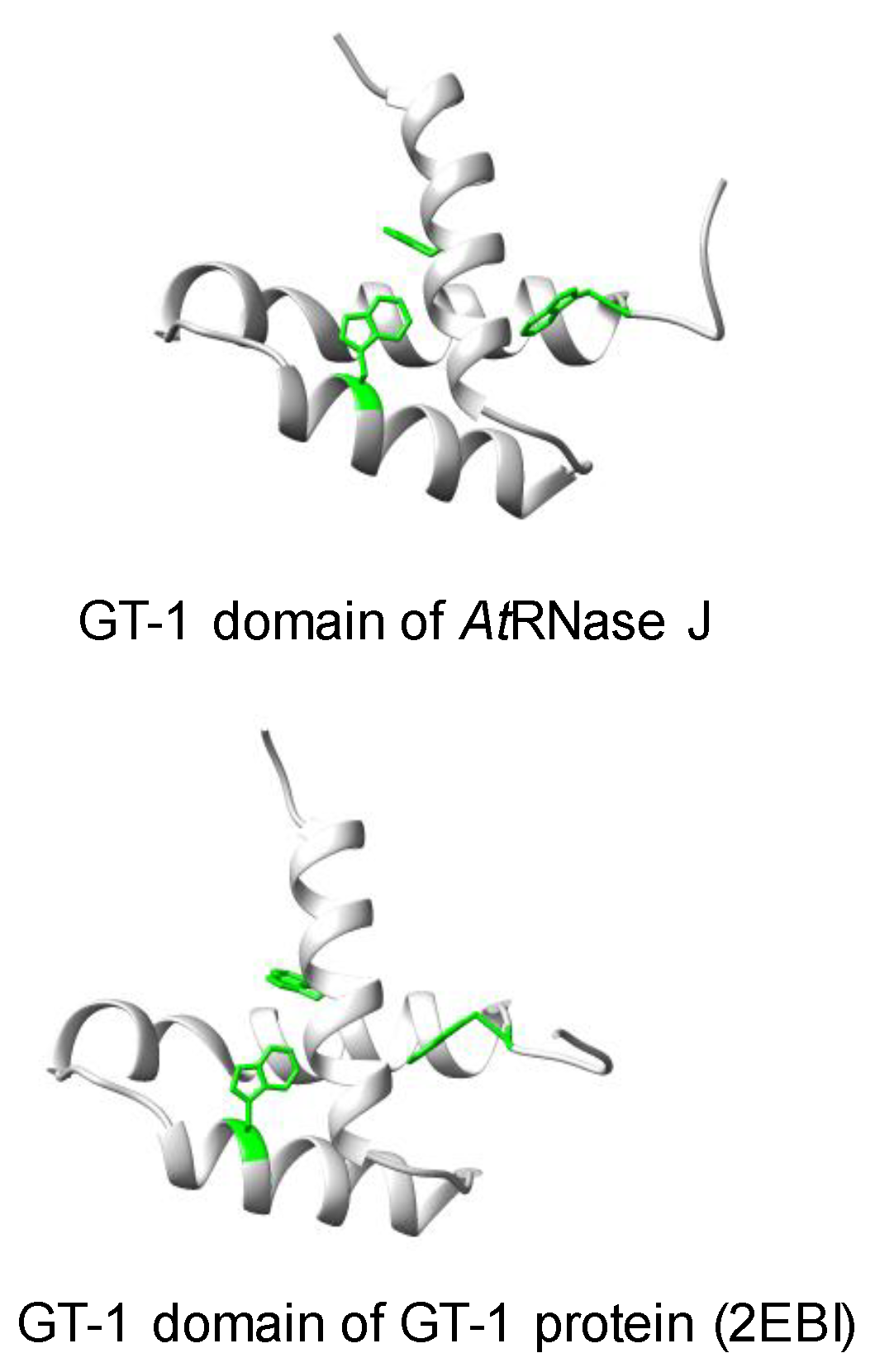
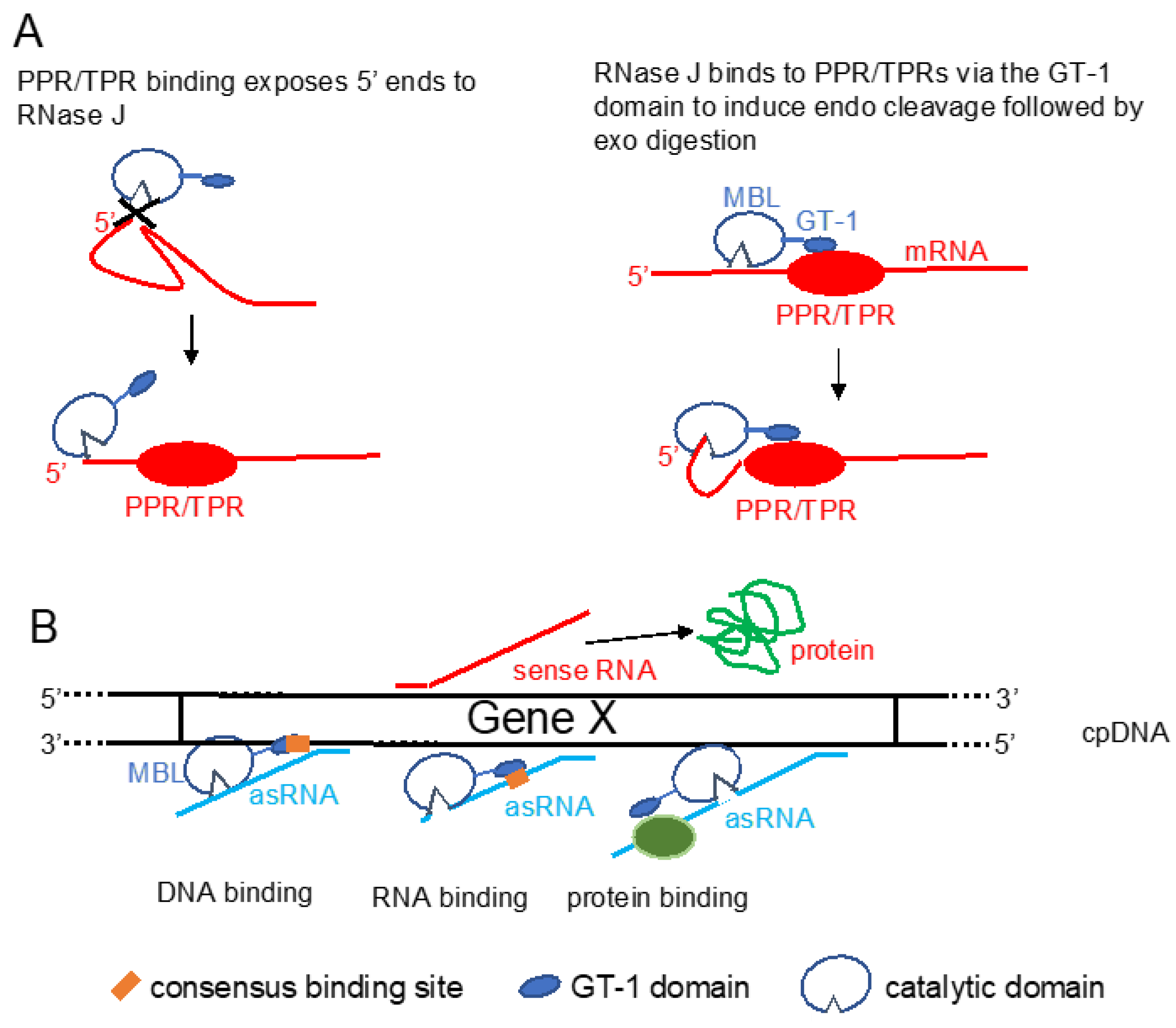
4. The Plant RNase J GT-1 Domain
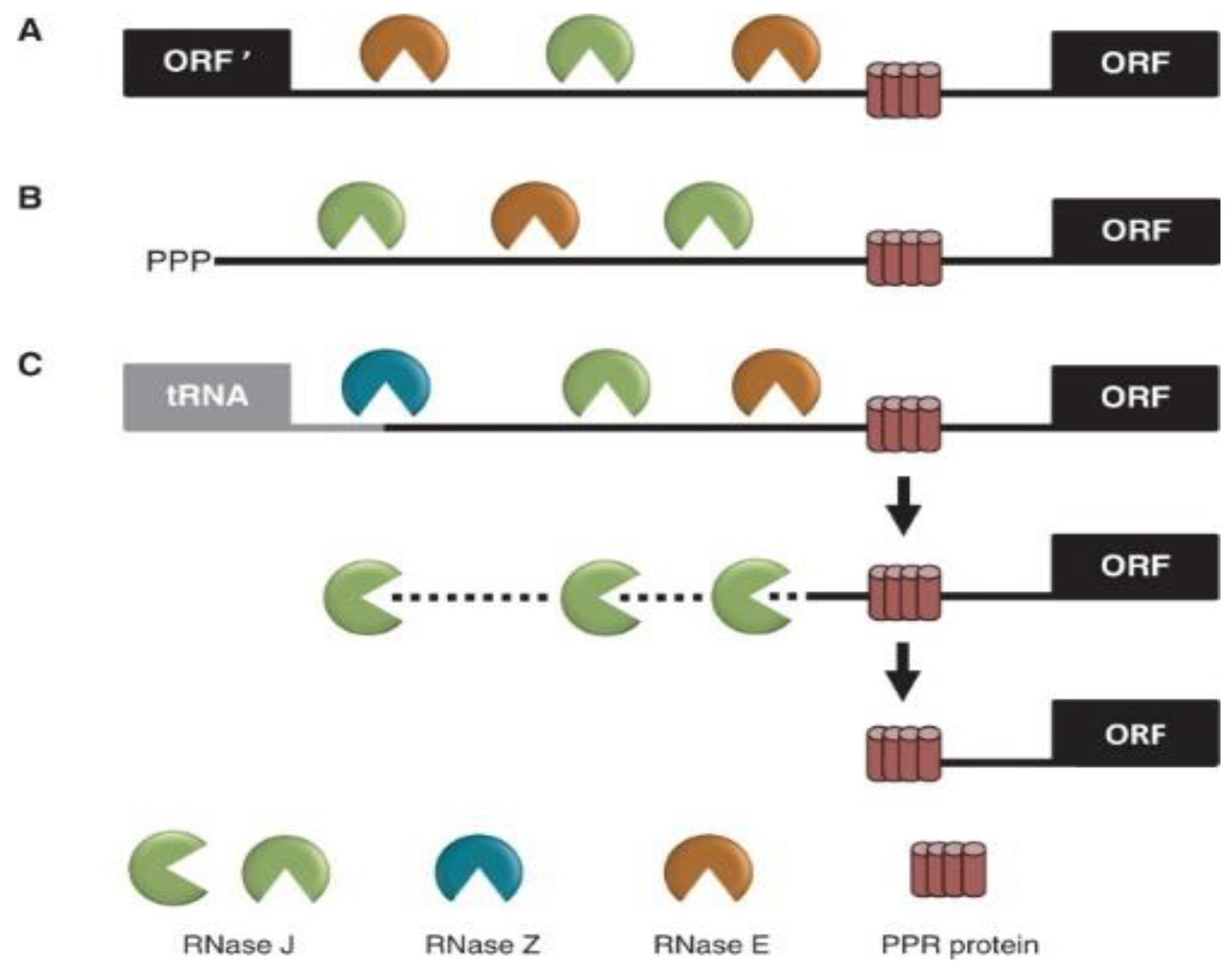
5. Consequences of Removing or Down-Regulating RNase J in Plants
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Doma, M.K.; Parker, R. RNA quality control in eukaryotes. Cell 2007, 131, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.L.; Zaher, H.S. How do cells cope with RNA damage and its consequences? J. Biol. Chem. 2019, 294, 15158–15171. [Google Scholar] [CrossRef] [PubMed]
- Peck, S.A.; Hughes, K.D.; Victorino, J.F.; Mosley, A.L. Writing a wrong: Coupled RNA polymerase II transcription and RNA quality control. Wiley Interdiscip. Rev. RNA 2019, 10, e1529. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, X. RNA Quality Control as a Key to Suppressing RNA Silencing of Endogenous Genes in Plants. Mol. Plant 2016, 9, 826–836. [Google Scholar] [CrossRef]
- Georg, J.; Hess, W.R. cis-Antisense RNA, Another Level of Gene Regulation in Bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 286–300. [Google Scholar] [CrossRef]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, J.; Zhang, W. Antisense RNA: The new favorite in genetic research. J. Zhejiang Univ. Sci. B 2018. [Google Scholar] [CrossRef]
- Sharwood, R.E.; Halpert, M.; Luro, S.; Schuster, G.; Stern, D.B. Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA 2011, 17, 2165–2176. [Google Scholar] [CrossRef]
- Sharwood, R.E.; Hotto, A.M.; Bollenbach, T.J.; Stern, D.B. Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA 2011, 17, 230–243. [Google Scholar] [CrossRef]
- Liebers, M.; Grubler, B.; Chevalier, F.; Lerbs-Mache, S.; Merendino, L.; Blanvillain, R.; Pfannschmidt, T. Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front. Plant Sci 2017, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Manavski, N.; Meurer, J.; Zhang, L.; Chi, W. Regulated chloroplast transcription termination. Biochim. Biophys. Acta BBA Bioenerg. 2019, 1860, 69–77. [Google Scholar] [CrossRef] [PubMed]
- De Longevialle, A.F.; Small, I.D.; Lurin, C. Nuclearly-encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant 2010, 3, 691–705. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Bentolila, S.; Hanson, M.R. The Unexpected Diversity of Plant Organelle RNA Editosomes. Trends Plant Sci. 2016, 21, 962–973. [Google Scholar] [CrossRef]
- Zoschke, R.; Bock, R. Chloroplast translation: Structural and functional organization, operational control and regulation. Plant Cell 2018, 30, 745–770. [Google Scholar] [CrossRef]
- Zhelyazkova, P.; Sharma, C.M.; Forstner, K.U.; Liere, K.; Vogel, J.; Börner, T. The primary transcriptome of barley chloroplasts: Numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 2012, 24, 123–136. [Google Scholar] [CrossRef]
- Castandet, B.; Germain, A.; Hotto, A.M.; Stern, D.B. Systematic sequencing of chloroplast transcript termini from Arabidopsis thaliana reveals >200 transcription initiation sites and the extensive imprints of RNA-binding proteins and secondary structures. Nucleic Acids Res. 2019, 47, 11889–11905. [Google Scholar] [CrossRef]
- Walter, M.; Piepenburg, K.; Schöttler, M.A.; Petersen, K.; Kahlau, S.; Tiller, N.; Drechsel, O.; Weingartner, M.; Kudla, J.; Bock, R. Knockout of the plastid RNase e leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 2010, 64, 851–863. [Google Scholar] [CrossRef]
- Schein, A.; Sheffy-Levin, S.; Glaser, F.; Schuster, G. The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA 2008, 14, 1057–1068. [Google Scholar] [CrossRef]
- Stoppel, R.; Manavski, N.; Schein, A.; Schuster, G.; Teubner, M.; Schmitz-Linneweber, C.; Meurer, J. RHON1 is a novel ribonucleic acid-binding protein that supports RNase e function in the Arabidopsis chloroplast. Nucleic Acids Res. 2012, 40, 8593–8606. [Google Scholar] [CrossRef]
- Yang, Z.; Li, M.; Sun, Q. RHON1 Co-transcriptionally Resolves R-Loops for Arabidopsis Chloroplast Genome Maintenance. Cell Rep. 2020, 30, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Germain, A.; Kim, S.H.; Gutierrez, R.; Stern, D.B. Ribonuclease II preserves chloroplast RNA homeostasis by increasing mRNA decay rates, and cooperates with polynucleotide phosphorylase in 3′ end maturation. Plant J. 2012, 72, 960–971. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Viola, S.; Cavaiuolo, M.; Drapier, D.; Eberhard, S.; Vallon, O.; Wollman, F.A.; Choquet, Y. MDA1, a nucleus-encoded factor involved in the stabilization and processing of the atpA transcript in the chloroplast of Chlamydomonas. Plant J. 2019, 98, 1033–1047. [Google Scholar]
- Manavski, N.; Schmid, L.-M.M.; Meurer, J. RNA-stabilization factors in chloroplasts of vascular plants. Essays Biochem. 2018, 62, 51–64. [Google Scholar]
- Cavaiuolo, M.; Kuras, R.; Wollman, F.A.; Choquet, Y.; Vallon, O. Small RNA profiling in Chlamydomonas: Insights into chloroplast RNA metabolism. Nucleic Acids Res. 2017, 45, 10783–10799. [Google Scholar] [CrossRef]
- Ruwe, H.; Wang, G.; Gusewski, S.; Schmitz-Linneweber, C. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 2016, 44, 7406–7417. [Google Scholar] [CrossRef]
- Sanitá Lima, M.; Smith, D.R. Pervasive transcription of mitochondrial, plastid, and nucleomorph genomes across diverse plastid-bearing species. Genome Biol. Evol. 2017, 9, 2650–2657. [Google Scholar] [CrossRef]
- Smith, D.R.R. Haematococcus lacustris: The makings of a giant-sized chloroplast genome. AoB Plants 2018, 10, ply058. [Google Scholar] [CrossRef]
- Hotto, A.M.; Schmitz, R.J.; Fei, Z.; Ecker, J.R.; Stern, D.B. Unexpected Diversity of Chloroplast Noncoding RNAs as Revealed by Deep Sequencing of the Arabidopsis Transcriptome. G3 Genes Genomes Genet. 2011, 1, 559–570. [Google Scholar]
- Hotto, A.M.; Germain, A.; Stern, D.B. Plastid non-coding RNAs: Emerging candidates for gene regulation. Trends Plant Sci. 2012, 17, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Hotto, A.M.; Castandet, B.; Gilet, L.; Higdon, A.; Condon, C.; Stern, D.B. Arabidopsis chloroplast mini-ribonuclease III participates in rRNA maturation and intron recycling. Plant Cell 2015, 27, 724–740. [Google Scholar] [CrossRef] [PubMed]
- Malbert, B.; Rigaill, G.; Brunaud, V.; Lurin, C.; Delannoy, E. Bioinformatic analysis of chloroplast gene expression and RNA posttranscriptional maturations using RNA sequencing. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2018; Volume 1829, pp. 279–294. [Google Scholar]
- Michel, E.J.S.; Hotto, A.M.; Strickler, S.R.; Stern, D.B.; Castandet, B. A guide to the chloroplast transcriptome analysis using RNA-Seq. In Plastids; Springer: Berlin/Heidelberg, Germany, 2018; pp. 295–313. [Google Scholar]
- Castandet, B.; Hotto, A.M.; Strickler, S.R.; Stern, D.B. ChloroSeq, an Optimized Chloroplast RNA-Seq Bioinformatic Pipeline, Reveals Remodeling of the Organellar Transcriptome Under Heat Stress. G3 Genes Genomes Genet. 2016, 6, 2817–2827. [Google Scholar] [CrossRef] [PubMed]
- Even, S.; Pellegrini, O.; Zig, L.; Labas, V.; Vinh, J.; Brechemmier-Baey, D.; Putzer, H. Ribonucleases J1 and J2: Two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005, 33, 2141–2152. [Google Scholar] [CrossRef]
- Condon, C.; Gilet, L. The Metallo-β-Lactamase Family of Ribonucleases. In Ribonucleases; Nicholson, A.W., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 245–267. ISBN 978-3-642-21077-8. [Google Scholar]
- Dominski, Z.; Carpousis, A.J.; Clouet-d’Orval, B. Emergence of the beta-CASP ribonucleases: Highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta 2013, 1829, 532–551. [Google Scholar] [CrossRef]
- Clouet-d’Orval, B.; Batista, M.; Bouvier, M.; Quentin, Y.; Fichant, G.; Marchfelder, A.; Maier, L.K. Insights into RNA processing pathways and associated RNA-degrading enzymes in Archaea. FEMS Microbiol. Rev. 2018, 42, 579–613. [Google Scholar] [CrossRef]
- Phung, D.K.; Rinaldi, D.; Langendijk-Genevaux, P.S.; Quentin, Y.; Carpousis, A.J.; Clouet-d’Orval, B. Archaeal β-CASP ribonucleases of the aCPSF1 family are orthologs of the eukaryal CPSF-73 factor. Nucleic Acids Res. 2013, 41, 1091–1103. [Google Scholar] [CrossRef]
- Callebaut, I.; Moshous, D.; Mornon, J.P.; de Villartay, J.P. Metallo-beta-lactamase fold within nucleic acids processing enzymes: The beta-CASP family. Nucleic Acids Res 2002, 30, 3592–3601. [Google Scholar] [CrossRef]
- Dominski, Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 67–93. [Google Scholar] [CrossRef]
- Clouet-d’Orval, B.; Phung, D.K.; Langendijk-Genevaux, P.S.; Quentin, Y. Universal RNA-degrading enzymes in Archaea: Prevalence, activities and functions of β-CASP ribonucleases. Biochimie 2015, 118, 278–285. [Google Scholar] [CrossRef]
- Zheng, X.; Feng, N.; Li, D.; Dong, X.; Li, J. New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family. Mol. Microbiol. 2017, 106, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.-Y.Y.; Bralley, P.; Jones, G.H.; Luisi, B.F. Linkage of catalysis and 5′ end recognition in ribonuclease RNase J. Nucleic Acids Res. 2015, 43, 8066–8076. [Google Scholar] [CrossRef] [PubMed]
- De la Sierra-Gallay, I.L.; Zig, L.; Jamalli, A.; Putzer, H. Structural insights into the dual activity of RNase J. Nat. Struct. Mol. Biol. 2008, 15, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Dorléans, A.; Li de la Sierra-Gallay, I.; Piton, J.; Zig, L.; Gilet, L.; Putzer, H.; Condon, C. Molecular basis for the recognition and cleavage of RNA by the bifunctional 5′-3′ exo/endoribonuclease RNase J. Structure 2011, 19, 1252–1261. [Google Scholar] [CrossRef]
- Condon, C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010, 7, 316–321. [Google Scholar] [CrossRef]
- Levy, S.; Portnoy, V.; Admon, J.; Schuster, G. Distinct activities of several RNase J proteins in methanogenic archaea. RNA Biol. 2011, 8, 1073–1083. [Google Scholar] [CrossRef]
- Liponska, A.; Jamalli, A.; Kuras, R.; Suay, L.; Garbe, E.; Wollman, F.-A.; Laalami, S.; Putzer, H. Tracking the elusive 5 ′ exonuclease activity of Chlamydomonas reinhardtii RNase J. Plant Mol. Biol. 2018, 96, 641–653. [Google Scholar] [CrossRef]
- Mathy, N.; Benard, L.; Pellegrini, O.; Daou, R.; Wen, T.; Condon, C. 5′-to-3′ exoribonuclease activity in bacteria: Role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 2007, 129, 681–692. [Google Scholar] [CrossRef]
- Halpert, M.; Liveanu, V.; Glaser, F.; Schuster, G. The Arabidopsis chloroplast RNase J displays both exo- and robust endonucleolytic activities. Plant Mol. Biol. 2019, 99, 17–29. [Google Scholar] [CrossRef]
- Mandel, C.R.; Kaneko, S.; Zhang, H.; Gebauer, D.; Vethantham, V.; Manley, J.L.; Tong, L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 2006, 444, 953–956. [Google Scholar] [CrossRef]
- Dominski, Z. The hunt for the 3′ endonuclease. Wiley Interdiscip. Rev. RNA 2010, 1, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.; Choi, E.A.; Shi, Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip. Rev. RNA 2011, 2, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.C.; Sullivan, K.D.; Marzluff, W.F.; Dominski, Z. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol. Cell. Biol. 2009, 29, 31–42. [Google Scholar] [CrossRef]
- Levy, S.; Allerston, C.K.; Liveanu, V.; Habib, M.R.; Gileadi, O.; Schuster, G. Identification of LACTB2, a metallo-beta-lactamase protein, as a human mitochondrial endoribonuclease. Nucleic Acids Res. 2016, 44, 1813–1832. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Q.; Wang, H.; Zhang, H.; Xu, X.; Li, C.; Yang, C. Comprehensive analysis of trihelix genes and their expression under biotic and abiotic stresses in Populus trichocarpa. Sci. Rep. 2016, 6, 36274. [Google Scholar] [CrossRef]
- Wang, W.; Wu, P.; Liu, T.; Ren, H.; Li, Y.; Hou, X. Genome-wide Analysis and Expression Divergence of the Trihelix family in Brassica Rapa: Insight into the Evolutionary Patterns in Plants. Sci. Rep. 2017, 7, 6463. [Google Scholar] [CrossRef]
- Xiao, J.; Hu, R.; Gu, T.; Han, J.; Qiu, D.; Su, P.; Feng, J.; Chang, J.; Yang, G.; He, G. Genome-wide identification and expression profiling of trihelix gene family under abiotic stresses in wheat. BMC Genom. 2019, 20. [Google Scholar] [CrossRef]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors—Light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef]
- Nagata, T.; Niyada, E.; Fujimoto, N.; Nagasaki, Y.; Noto, K.; Miyanoiri, Y.; Murata, J.; Hiratsuka, K.; Katahira, M. Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: Mechanism for an increase in DNA-binding affinity through phosphorylation. Proteins Struct. Funct. Bioinform. 2010, 78, 3033–3047. [Google Scholar] [CrossRef]
- Tzafrir, I.; Pena-Muralla, R.; Dickerman, A.; Berg, M.; Rogers, R.; Hutchens, S.; Sweeney, T.C.; McElver, J.; Aux, G.; Patton, D.; et al. Identification of genes required for embryo development in arabidopsis. Plant Physiol. 2004, 135, 1206–1220. [Google Scholar] [CrossRef]
- Chen, H.; Zou, W.; Zhao, J. Ribonuclease J is required for chloroplast and embryo development in Arabidopsis. J. Exp. Bot. 2015, 66, 2079–2091. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Wang, J.; Zhou, Y.; An, D.; Xiao, Q.; Wang, W.; Wu, Y. EMB-7L is required for embryogenesis and plant development in maize involved in RNA splicing of multiple chloroplast genes. Plant Sci. 2019, 287, 110203. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-C.; Belmonte, F.M.; Harada, J.; Inoue, K. Indispensable Roles of Plastids in Arabidopsis thaliana Embryogenesis. Curr. Genom. 2010, 11, 338–349. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luro, S.; Germain, A.; Sharwood, R.E.E.; Stern, D.B. RNase J participates in a pentatricopeptide repeat protein-mediated 5′ end maturation of chloroplast mRNAs. Nucleic Acids Res. 2013, 41, 9141–9151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Germain, A.; Hotto, A.M.; Barkan, A.; Stern, D.B. RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA 2013, 4, 295–316. [Google Scholar] [CrossRef]
- Pfalz, J.; Bayraktar, O.A.; Prikryl, J.; Barkan, A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009, 28, 2042–2052. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hotto, A.M.; Stern, D.B.; Schuster, G. Plant Ribonuclease J: An Essential Player in Maintaining Chloroplast RNA Quality Control for Gene Expression. Plants 2020, 9, 334. https://doi.org/10.3390/plants9030334
Hotto AM, Stern DB, Schuster G. Plant Ribonuclease J: An Essential Player in Maintaining Chloroplast RNA Quality Control for Gene Expression. Plants. 2020; 9(3):334. https://doi.org/10.3390/plants9030334
Chicago/Turabian StyleHotto, Amber M., David B. Stern, and Gadi Schuster. 2020. "Plant Ribonuclease J: An Essential Player in Maintaining Chloroplast RNA Quality Control for Gene Expression" Plants 9, no. 3: 334. https://doi.org/10.3390/plants9030334
APA StyleHotto, A. M., Stern, D. B., & Schuster, G. (2020). Plant Ribonuclease J: An Essential Player in Maintaining Chloroplast RNA Quality Control for Gene Expression. Plants, 9(3), 334. https://doi.org/10.3390/plants9030334




