The Allelic Diversity of the Gibberellin Signaling Pathway Genes in Aegilops tauschii Coss
Abstract
1. Introduction
2. Results
2.1. Rht-D1
2.2. Gid1-D
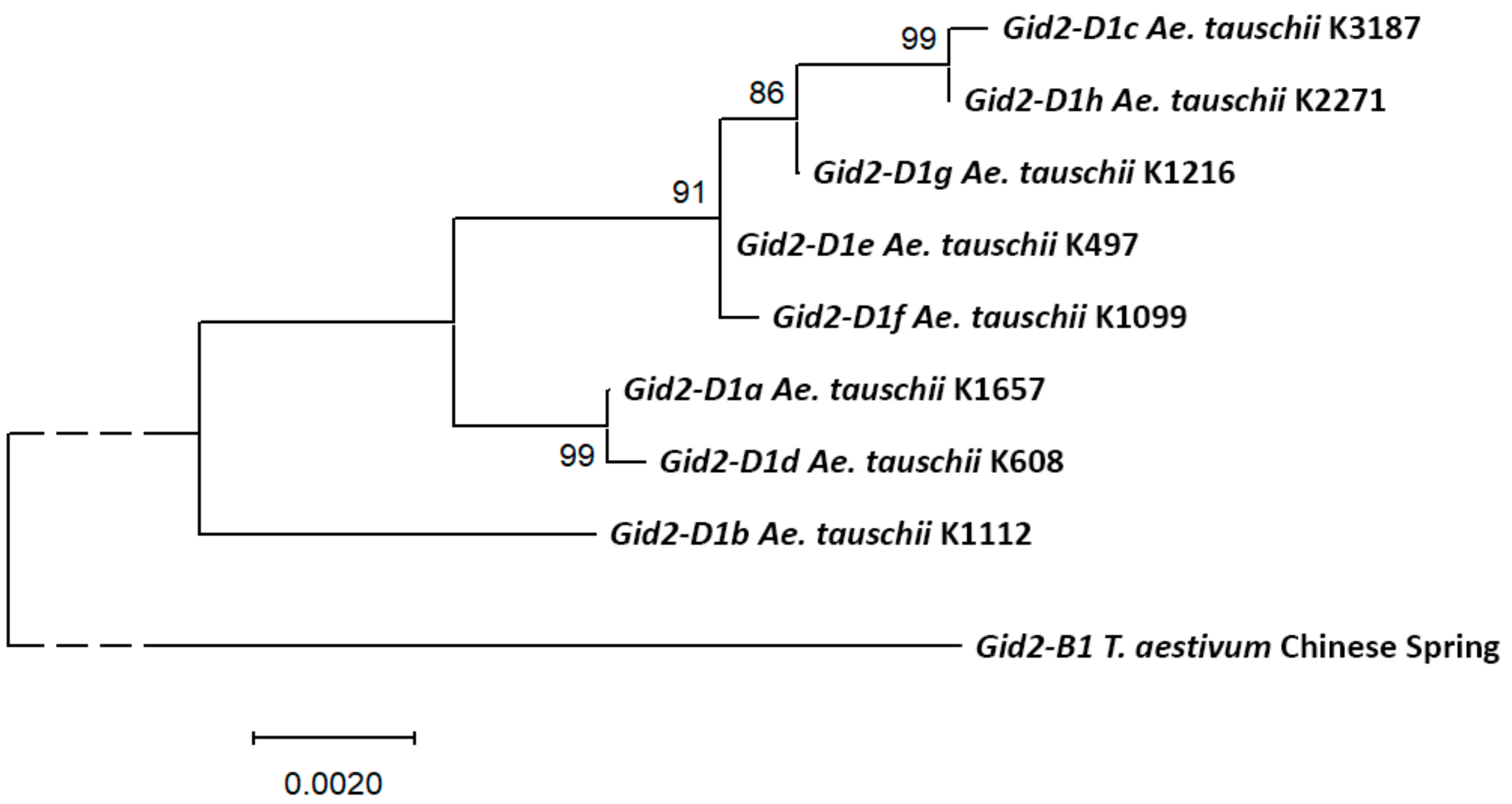
2.3. Gid2-D
2.4. Co-Occurrence Of Protein Isoforms
2.5. Data Availability
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA Extraction and PCR
4.3. Sequencing
4.4. Bioinformatic Treatment of the Sequencing Results
4.5. Statistical Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. “Green revolution” genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Gent, M.P.N.; Kiyomoto, R.K. Physiological and Agronomic Consequences of Rht Genes in Wheat. J. Crop Prod. 1997, 1, 27–46. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, W.J.; Morris, C.; Appels, R.; Xia, X.C. Catalogue of Gene Symbols for Wheat. In Proceedings of the 12th International Wheat Genetics Symposium, Yokohama, Japan, 8–14 September 2013. [Google Scholar]
- Borojevic, K.; Borojevic, K. The transfer and history of “reduced height genes” (Rht) in wheat from Japan to Europe. J. Hered. 2005, 96, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Divashuk, M.G.; Bespalova, L.A.; Vasilyev, A.V.; Fesenko, I.A.; Puzyrnaya, O.Y.; Karlov, G.I. Reduced height genes and their importance in winter wheat cultivars grown in southern Russia. Euphytica 2012, 190, 137–144. [Google Scholar] [CrossRef]
- Jatayev, S.; Sukhikh, I.; Vavilova, V.; Smolenskaya, S.E.; Goncharov, N.P.; Kurishbayev, A.; Zotova, L.; Absattarova, A.; Serikbay, D.; Hu, Y.-G.; et al. Green revolution “stumbles” in a dry environment: Dwarf wheat with Rht genes fails to produce higher grain yield than taller plants under drought. Plant Cell Environ. 2020, 43, 2355–2364. [Google Scholar] [CrossRef]
- Yoshida, H.; Hirano, K.; Sato, T.; Mitsuda, N.; Nomoto, M.; Maeo, K.; Koketsu, E.; Mitani, R.; Kawamura, M.; Ishiguro, S.; et al. DELLA protein functions as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN family proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 7861–7866. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Wu, K.; Ye, Y.; Yu, J.; Zhang, J.; Liu, Q.; Hu, M.; Li, H.; Tong, Y.; et al. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef]
- Hirsch, S.; Oldroyd, G.E. GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav. 2009, 4, 698–700. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.; Hsing, Y.C.; Kitano, H.; Yamaguchi, I.; et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef]
- Wu, J.; Kong, X.; Wan, J.; Liu, X.; Zhang, X.; Guo, X.; Zhou, R.; Zhao, G.; Jing, R.; Fu, X.; et al. Dominant and Pleiotropic Effects of a GAI Gene in Wheat Results from a Lack of Interaction between DELLA and GID1. Plant Physiol. 2011, 157, 2120–2130. [Google Scholar] [CrossRef]
- Pearce, S.; Saville, R.; Vaughan, S.P.; Chandler, P.M.; Wilhelm, E.P.; Sparks, C.A.; Al-Kaff, N.; Korolev, A.; Boulton, M.I.; Phillips, A.L.; et al. Molecular Characterization of Rht-1 Dwarfing Genes in Hexaploid Wheat. Plant Physiol. 2011, 157, 1820–1831. [Google Scholar] [CrossRef] [PubMed]
- Bazhenov, M.S.; Divashuk, M.G.; Amagai, Y.; Watanabe, N.; Karlov, G.I. Isolation of the dwarfing Rht-B1p (Rht17) gene from wheat and the development of an allele-specific PCR marker. Mol. Breed. 2015, 35. [Google Scholar] [CrossRef]
- Van De Velde, K.; Chandler, P.M.; Van Der Straeten, D.; Rohde, A. Differential coupling of gibberellin responses by Rht-B1c suppressor alleles and Rht-B1b in wheat highlights a unique role for the DELLA N-terminus in dormancy. J. Exp. Bot. 2017, 68, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, J.; Wu, J.; Duan, J.; Liu, Y.; Ye, X.; Zhang, X.; Guo, X.; Gu, Y.; Zhang, L.; et al. A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 2012, 196, 282–291. [Google Scholar] [CrossRef]
- Wilhelm, E.P.; Mackay, I.J.; Saville, R.J.; Korolev, A.V.; Balfourier, F.; Greenland, A.J.; Boulton, M.I.; Powell, W. Haplotype dictionary for the Rht-1 loci in wheat. Theor. Appl. Genet. 2013, 126, 1733–1747. [Google Scholar] [CrossRef]
- Lou, X.; Li, X.; Li, A.; Pu, M.; Shoaib, M.; Liu, D.; Sun, J.; Zhang, A.; Yang, W. The 160 bp insertion in the promoter of Rht-B1i plays a vital role in increasing wheat height. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Li, A.; Yang, W.; Li, S.; Liu, D.; Guo, X.; Sun, J.; Zhang, A. Molecular characterization of three GIBBERELLIN-INSENSITIVE DWARF1 homologous genes in hexaploid wheat. J. Plant Physiol. 2013, 170, 432–443. [Google Scholar] [CrossRef]
- Lou, X.; Li, X.; Li, A.; Pu, M.; Shoaib, M.; Liu, D.; Sun, J.; Zhang, A.; Yang, W. Molecular Characterization of Three GIBBERELLIN-INSENSITIVE DWARF2 Homologous Genes in Common Wheat. PLoS ONE 2016, 11, e0157642. [Google Scholar] [CrossRef]
- Srinivasachary; Gosman, N.; Steed, A.; Hollins, T.W.; Bayles, R.; Jennings, P.; Nicholson, P. Semi-dwarfing Rht-B1 and Rht-D1 loci of wheat differ significantly in their influence on resistance to Fusarium head blight. Theor. Appl. Genet. 2008, 118, 695–702. [Google Scholar] [CrossRef]
- Qin, L.; Zhao, J.; Li, T.; Hou, J.; Zhang, X.; Hao, C. TaGW2, a Good Reflection of Wheat Polyploidization and Evolution. Front. Plant Sci. 2017, 8, 318. [Google Scholar] [CrossRef]
- Schneider, A.; Molnár, I.; Molnár-Láng, M. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 2008, 163, 1–19. [Google Scholar] [CrossRef]
- McFadden, E.S.; Sears, E.R. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 1946, 37, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.-C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef]
- Tsunewaki, K. Plasmon analysis in the Triticum-Aegilops complex. Breed. Sci. 2009, 59, 455–470. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Nyine, M.; Adhikari, E.; Clinesmith, M.; Jordan, K.W.; Fritz, A.K.; Akhunov, E. Genomic Patterns of Introgression in Interspecific Populations Created by Crossing Wheat with Its Wild Relative. G3 2020, 10, 3651–3661. [Google Scholar] [CrossRef]
- Wang, J.; Luo, M.-C.; Chen, Z.; You, F.M.; Wei, Y.; Zheng, Y.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937. [Google Scholar] [CrossRef]
- Valkoun, J.; Kučerová, D.; Bartoš, P. Transfer of leaf rust resistance from Triticum monococcum L. to hexaploid wheat. Z. Pflanz. 1986, 96, 271–278. [Google Scholar]
- Ogbonnaya, F.C.; Abdalla, O.; Mujeeb-Kazi, A.; Kazi, A.G.; Xu, S.S.; Gosman, N.; Lagudah, E.S.; Bonnett, D.; Sorrells, M.E.; Tsujimoto, H. Synthetic Hexaploids: Harnessing Species of the Primary Gene Pool for Wheat Improvement. In Plant Breeding Reviews; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 35–122. ISBN 978-1-118-49786-9. [Google Scholar]
- Kishii, M. An Update of Recent Use of Aegilops Species in Wheat Breeding. Front. Plant Sci. 2019, 10, 585. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Kalia, B.; Wilson, D.L.; Bowden, R.L.; Singh, R.P.; Gill, B.S. Adult plant resistance to Puccinia triticina in a geographically diverse collection of Aegilops tauschii. Genet. Resour. Crop Evol. 2017, 64, 913–926. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Nishioka, E.; Kawahara, T.; Takumi, S. Genealogical analysis of subspecies divergence and spikelet-shape diversification in central Eurasian wild wheat Aegilops tauschii Coss. Plant Syst. Evol. 2009, 279, 233–244. [Google Scholar] [CrossRef]
- Dudnikov, A.J. Geographic patterns of low-polymorphic enzyme-encoding genes allelic variation in Aegilops tauschii. J. Syst. Evol. 2013, 51, 715–721. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Takumi, S.; Kawahara, T. Intraspecific lineage divergence and its association with reproductive trait change during species range expansion in central Eurasian wild wheat Aegilops tauschii Coss. (Poaceae). BMC Evol. Biol. 2015, 15, 213. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Wu, S.; Tiwari, V.; Sehgal, S.; Raupp, J.; Wilson, D.; Abbasov, M.; Gill, B.; Poland, J. Genomic Analysis Confirms Population Structure and Identifies Inter-Lineage Hybrids in Aegilops tauschii. Front. Plant Sci. 2019, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Aghaei, M.J.; Mozafari, J.; Taleei, A.R.; Naghavi, M.R.; Omidi, M. Distribution and diversity of Aegilops tauschii in Iran. Genet. Resour. Crop Evol. 2008, 55, 341–349. [Google Scholar] [CrossRef]
- Dvorak, J.; Luo, M.-C.; Yang, Z.-L.; Zhang, H.-B. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 1998, 97, 657–670. [Google Scholar] [CrossRef]
- Naghavi, M.R.; Amirian, R. Morphological Characterization of Accessions of Aegilops tauschii. Int. J. Agric. Biol. 2005, 7, 392–394. [Google Scholar]
- Takumi, S.; Nishioka, E.; Morihiro, H.; Kawahara, T.; Matsuoka, Y. Natural variation of morphological traits in wild wheat progenitor Aegilops tauschii Coss. Breed. Sci. 2009, 59, 579–588. [Google Scholar] [CrossRef][Green Version]
- Mizuno, N.; Yamasaki, M.; Matsuoka, Y.; Kawahara, T.; Takumi, S. Population structure of wild wheat D-genome progenitor Aegilops tauschii Coss.: Implications for intraspecific lineage diversification and evolution of common wheat. Mol. Ecol. 2010, 19, 999–1013. [Google Scholar] [CrossRef]
- Mirzaghaderi, G.; Mason, A.S. Broadening the bread wheat D genome. Theor. Appl. Genet. 2019, 132, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- Doyle, P.J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; NATO ASI Series; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. ISBN 978-3-642-83964-1. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Lischer, H.E.L.; Shimizu, K.K. Reference-guided de novo assembly approach improves genome reconstruction for related species. BMC Bioinform. 2017, 18, 474. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Zaharia, M.; Bolosky, W.J.; Curtis, K.; Fox, A.; Patterson, D.; Shenker, S.; Stoica, I.; Karp, R.M.; Sittler, T. Faster and More Accurate Sequence Alignment with SNAP. arXiv 2011, arXiv:1111.5572. [Google Scholar]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- GitHub-Samtools/Bcftools. Available online: https://github.com/samtools/bcftools (accessed on 19 December 2019).
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Goncharov, N.P.; Chikida, N.N. Genetics of growth habit in Aegilops squarrosa L. Genetika 1995, 31, 396–399. [Google Scholar]


| Allele | Frequency | Protein Isoform |
|---|---|---|
| Rht-D1a_7 | 0.42 | B |
| Rht-D1a_5 | 0.33 | A |
| Rht-D1a_8 | 0.08 | A |
| Rht-D1a_9 | 0.04 | A |
| Rht-D1a_10 | 0.04 | C |
| Rht-D1a_11 | 0.04 | B |
| Rht-D1a_12 | 0.04 | D |
| Amino Acid Variations | Protein Isoforms | PROVEAN Score | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| T162V | − | + | − | + | 0.294 |
| G334S | + | − | − | − | −3.761 * |
| G622A | − | + | − | − | 0.674 |
| Allele | Frequency | Protein Isoform |
|---|---|---|
| Gid1-D1a | 0.25 | A |
| Gid1-D1b | 0.15 | B |
| Gid1-D1c | 0.13 | A |
| Gid1-D1d | 0.08 | A |
| Gid1-D1e | 0.08 | B |
| Gid1-D1f | 0.06 | A |
| Gid1-D1g | 0.04 | A |
| Gid1-D1h | 0.04 | A |
| Gid1-D1i | 0.04 | A |
| Gid1-D1j | 0.04 | A |
| Gid1-D1k | 0.04 | A |
| Gid1-D1l | 0.02 | B |
| Gid1-D1m | 0.02 | A |
| Allele | Frequency | Protein Isoform |
|---|---|---|
| Gid2-D1a | 0.33 | A |
| Gid2-D1b | 0.17 | B |
| Gid2-D1c | 0.13 | A |
| Gid2-D1d | 0.13 | A |
| Gid2-D1e | 0.13 | A |
| Gid2-D1f | 0.04 | A |
| Gid2-D1g | 0.04 | A |
| Gid2-D1h | 0.04 | A |
| RHT-1 | The p Value of Fischer’s Exact Test * | |||||
|---|---|---|---|---|---|---|
| A | B | C | D | |||
| GID1 | A | 10 | 22 | 2 | 2 | 0.0001 |
| B | 12 | 0 | 0 | 0 | ||
| GID2 | A | 14 | 22 | 2 | 2 | 0.0036 |
| B | 8 | 0 | 0 | 0 | ||
| GID1 | The p Value of Fischer’s Exact Test | |||
|---|---|---|---|---|
| A | B | |||
| GID2 | A | 33 | 7 | 0.0166 |
| B | 3 | 5 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bazhenov, M.S.; Chernook, A.G.; Goncharov, N.P.; Chikida, N.N.; Belousova, M.K.; Karlov, G.I.; Divashuk, M.G. The Allelic Diversity of the Gibberellin Signaling Pathway Genes in Aegilops tauschii Coss. Plants 2020, 9, 1696. https://doi.org/10.3390/plants9121696
Bazhenov MS, Chernook AG, Goncharov NP, Chikida NN, Belousova MK, Karlov GI, Divashuk MG. The Allelic Diversity of the Gibberellin Signaling Pathway Genes in Aegilops tauschii Coss. Plants. 2020; 9(12):1696. https://doi.org/10.3390/plants9121696
Chicago/Turabian StyleBazhenov, Mikhail S., Anastasiya G. Chernook, Nikolay P. Goncharov, Nadezhda N. Chikida, Mariya Kh. Belousova, Gennady I. Karlov, and Mikhail G. Divashuk. 2020. "The Allelic Diversity of the Gibberellin Signaling Pathway Genes in Aegilops tauschii Coss" Plants 9, no. 12: 1696. https://doi.org/10.3390/plants9121696
APA StyleBazhenov, M. S., Chernook, A. G., Goncharov, N. P., Chikida, N. N., Belousova, M. K., Karlov, G. I., & Divashuk, M. G. (2020). The Allelic Diversity of the Gibberellin Signaling Pathway Genes in Aegilops tauschii Coss. Plants, 9(12), 1696. https://doi.org/10.3390/plants9121696






