Population Genetics of Odontarrhena (Brassicaceae) from Albania: The Effects of Anthropic Habitat Disturbance, Soil, and Altitude on a Ni-Hyperaccumulator Plant Group from a Major Serpentine Hotspot
Abstract
1. Introduction
2. Results
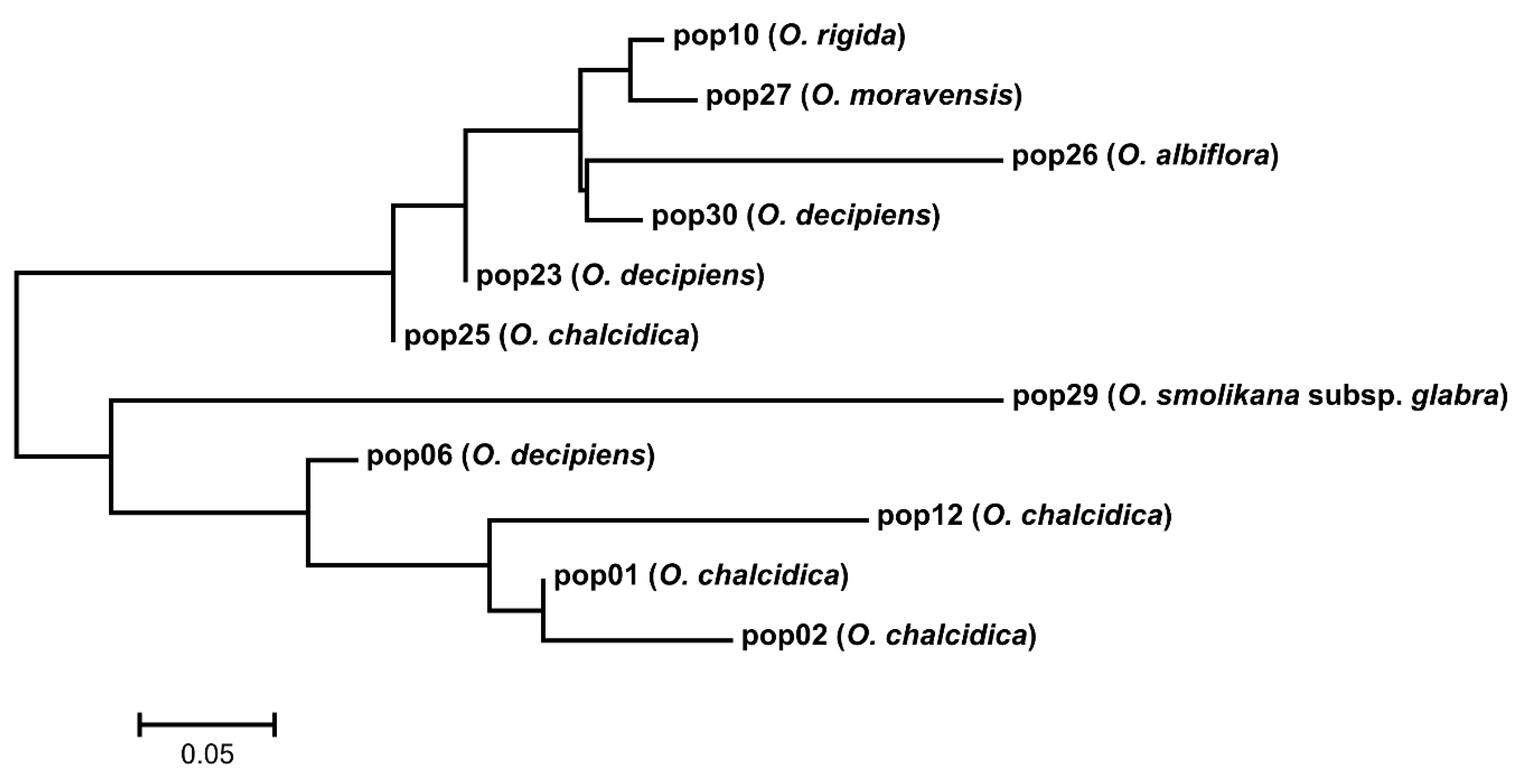
2.1. DNA Sequence Divergence and Phylogeny
2.2. Genetic Structure
2.3. Genetic Diversity
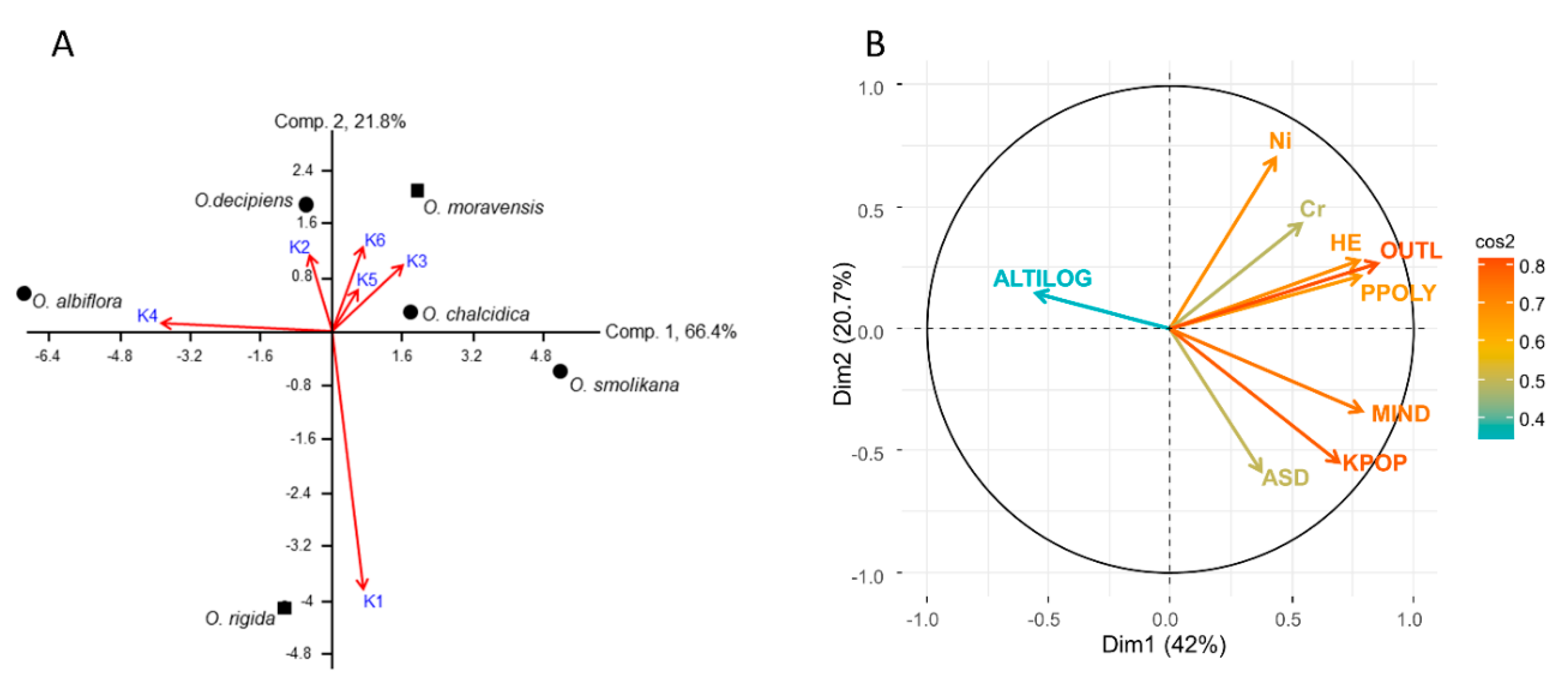
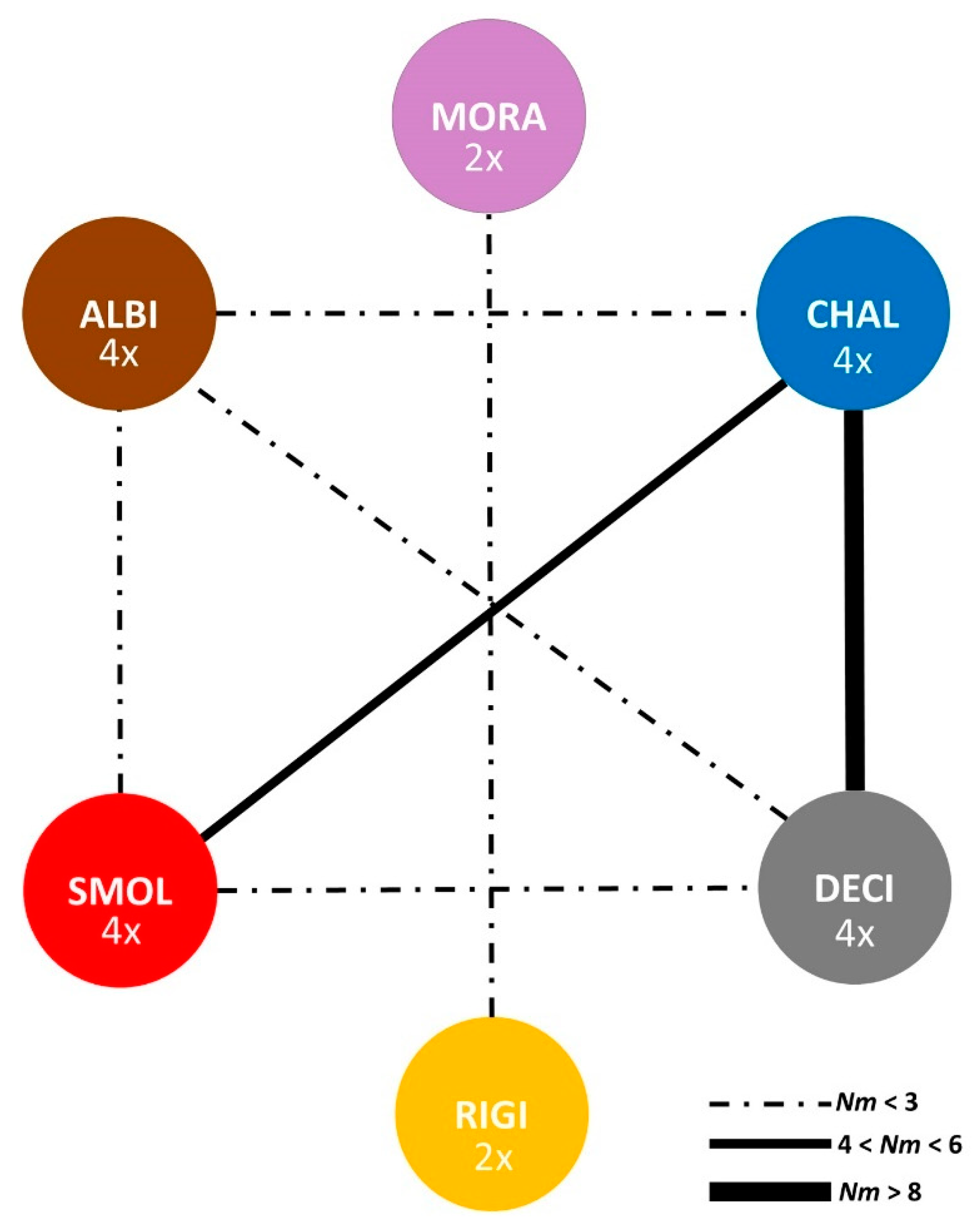
2.4. Genetic Distances and Gene Flow
3. Discussion
3.1. Phylogeny
3.2. Population Genetic Structure and Role of Anthropic Habitat Disturbance
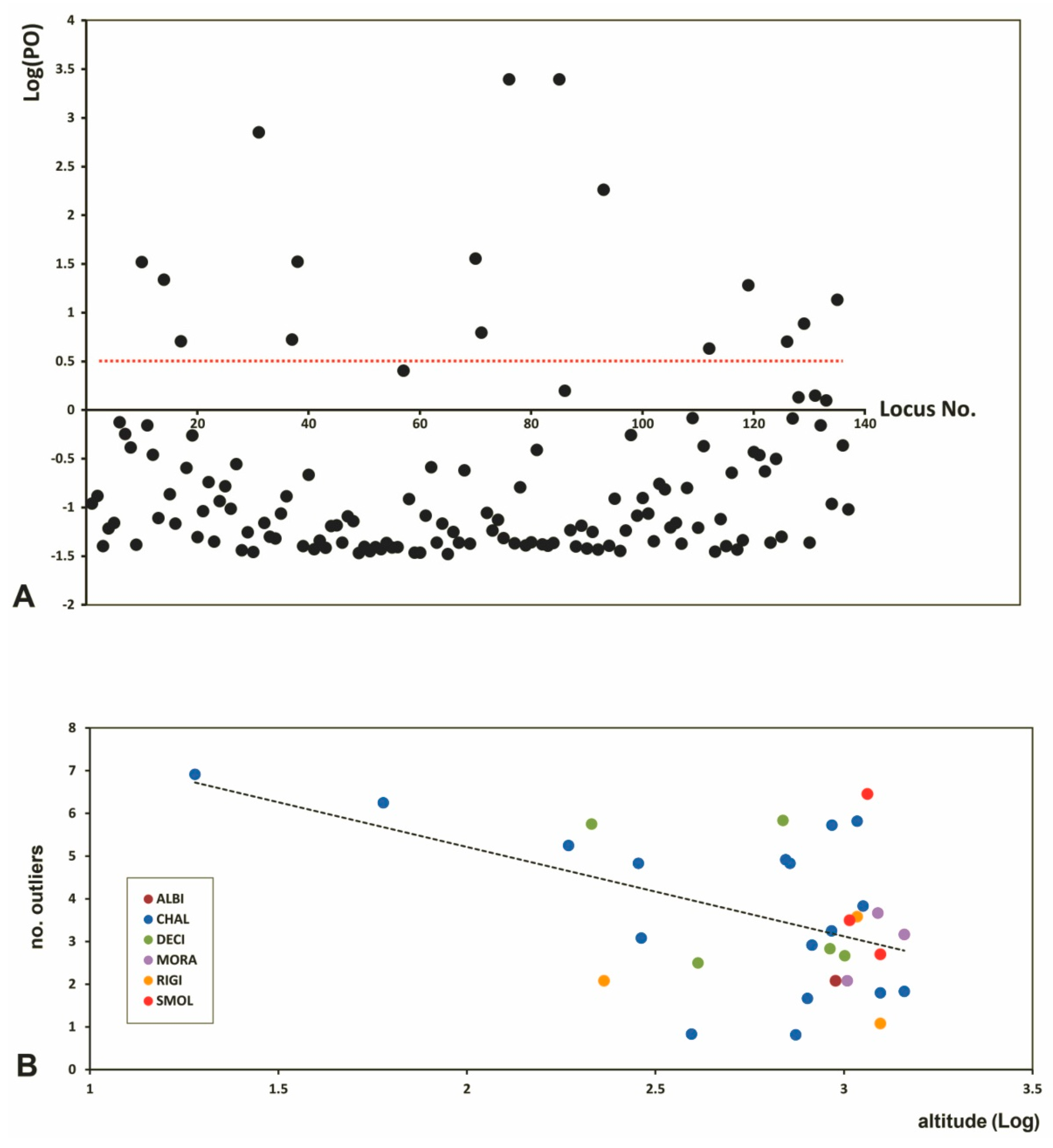
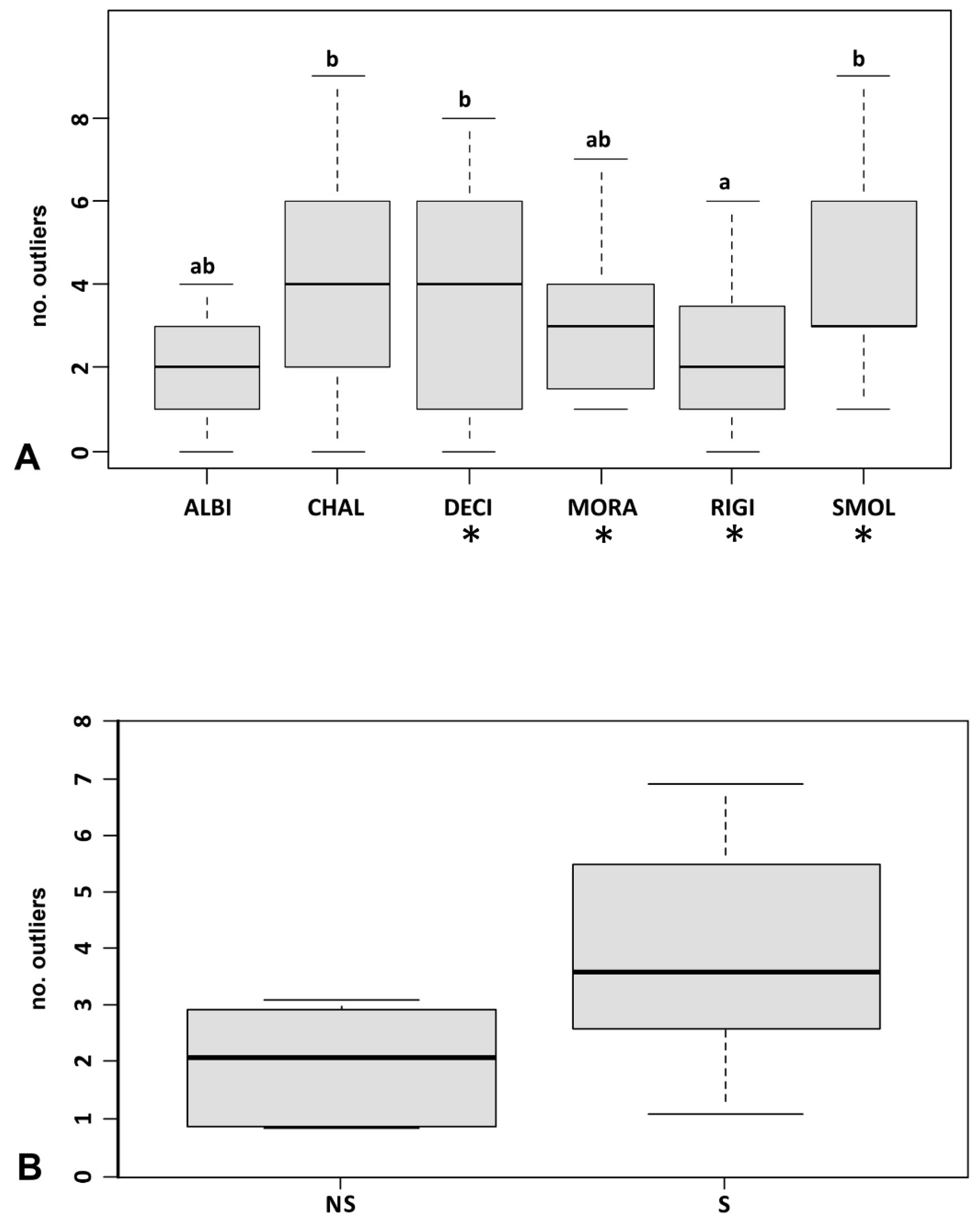
3.3. Genetic Diversity and Outlier Loci in Relation to Site Conditions
3.4. Implications for Conservation
4. Materials and Methods
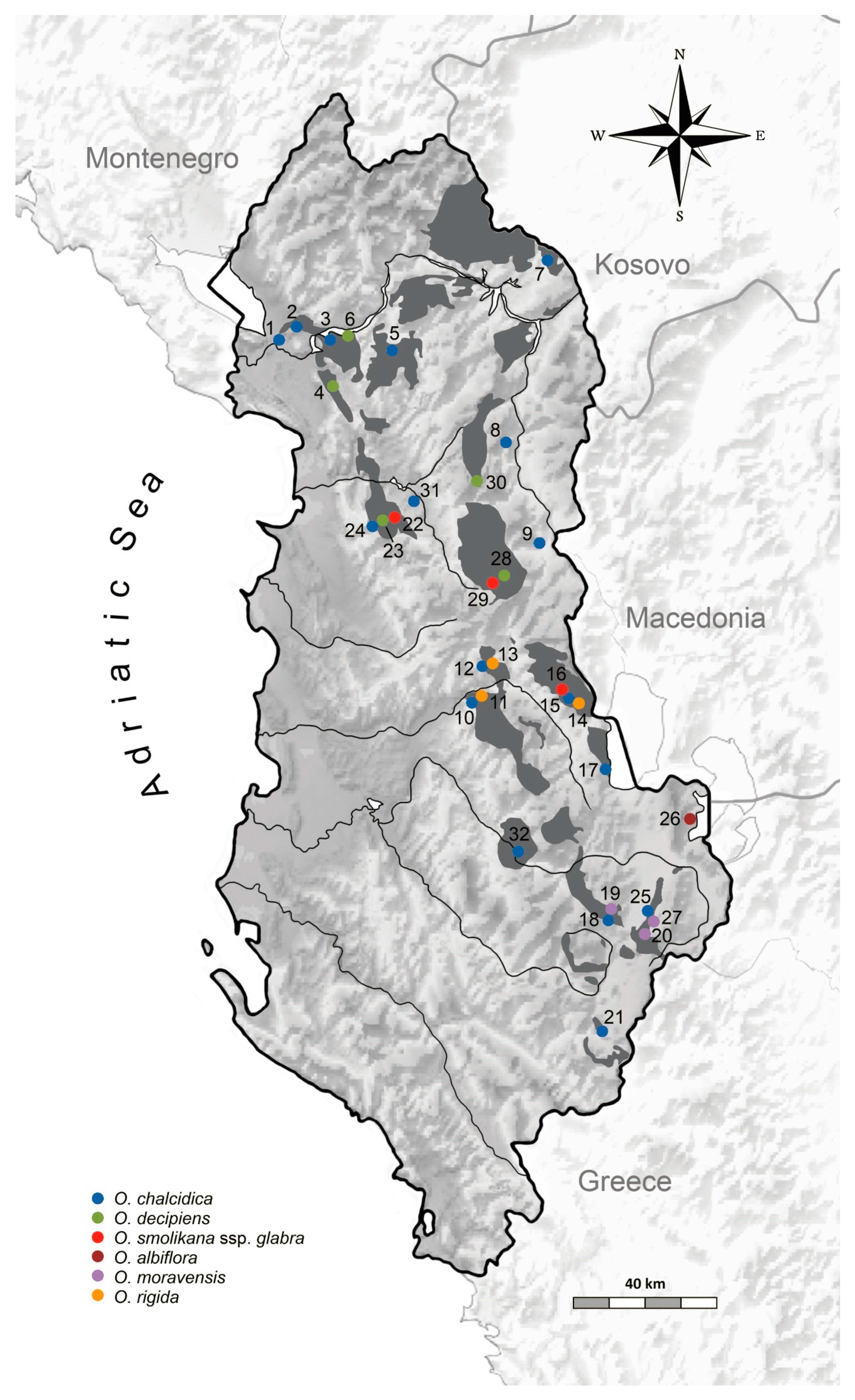
4.1. Plant Material and Sampling Design
4.2. DNA Isolation, Amplification, Sequencing, and Phylogenetic Analysis
4.3. Population Genetic Analyses
4.4. Relationships between Genetic Variables and Site Conditions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wójcik, M.; Gonnelli, C.; Selvi, F.; Dresler, S.; Rostański, A.; Vangronsveld, J. Metallophytes of serpentine and calamine soils–Their unique ecophysiology and potential for phytoremediation. In Phytoremediation. Advances in Botanical Research; Cuypers, A., Vangronsveld, J., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1–42. [Google Scholar]
- Reeves, R.D.; Baker, A.J.M.; Jaffré, T.; Erskine, P.D.; Echevarria, G.; van der Ent, A. A global database for plants that hyperaccumulate metal and metalloid trace elements. New Phytol. 2018, 218, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Van der Ent, A.; Echevarria, G.; Baker, A.J.M.; Morel, J.L. (Eds.) Agromining: Farming for Metals. Extracting Unconventional Resources Using Plants; Mineral Resource Reviews Series; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Broadley, M.R.; Willey, N.J.; Wilkins, J.C.; Baker, A.J.M.; Mead, A.; White, P.J. Phylogenetic variation in heavy metal accumulation in angiosperms. New Phytol. 2001, 152, 9–27. [Google Scholar] [CrossRef]
- Reeves, R.D.; Baker, A.J.M. Studies on metal uptake by plants from serpentine and non-serpentine populations of Thlaspi goesingense Hálácsy (Cruciferae). New Phytol. 1984, 98, 191–204. [Google Scholar]
- Krämer, U. Metal hyperaccumulation in plants. Annu. Rev. Plant Biol. 2010, 6, 517–534. [Google Scholar]
- Reeves, R.D.; Brooks, R.R.; Dudley, T.R. Uptake of Nickel by species of Alyssum, Bornmuellera, and other genera of old world tribus Alysseae. Taxon 1983, 32, 184–192. [Google Scholar] [CrossRef]
- Reeves, R.D. Nickel and zinc accumulation by species of Thlaspi L., Cochlearia L., and other genera of the Brassicaceae. Taxon 1988, 37, 309–318. [Google Scholar] [CrossRef]
- Peer, W.A.; Mahmoudian, M.; Freeman, J.L.; Lahner, B.; Richards, E.L.; Reeves, R.D.; Murphy, A.S.; Salt, D.E. Assessment of plants from the Brassicaceae family as genetic models for the study of nickel and zinc hyperaccumulation. New Phytol. 2006, 172, 248–260. [Google Scholar] [CrossRef]
- Nkrumah, P.N.; Baker, A.J.M.; Chaney, R.L.; Erskine, P.D.; Echevarria, G.; Morel, J.L.; van der Ent, A. Current status and challenges in developing nickel phytomining: An agronomic perspective. Plant Soil 2016, 406, 55–69. [Google Scholar] [CrossRef]
- Echevarria, G.; Bani, A.; Benizri, E.; Gonnelli, C.; Hazotte, C.; Kisser, J.; Konstantinou, M.; Kuppens, T.; Kyrkas, D.; Laubie, B.; et al. Developing sustainable agromining systems in agricultural ultramafic soils for nickel recovery. Front. Environ. Sci. 2018, 6. [Google Scholar] [CrossRef]
- Španiel, S.; Kempa, M.; Salmerón-Sánchez, E.; Fuertes-Aguilar, J.; Mota, J.F.; Al-Shehbaz, I.A.; German, D.A.; Olšavská, K.; Šingliarová, B.; Zozomová-Lihová, J.; et al. AlyBase: Database of names, chromosome numbers, and ploidy levels of Alysseae (Brassicaceae), with a new generic concept of the tribe. Plant Syst. Evol. 2015, 301, 2463–2491. [Google Scholar] [CrossRef]
- Brooks, R.R.; Morrison, R.S.; Reeves, R.D.; Dudley, T.R.; Akman, Y. Hyperaccumulation of nickel by Alyssum Linnaeus (Cruciferae). Proc. R. Soc. Lond. Ser. B Boil. Sci. 1979, 203, 387–403. [Google Scholar] [CrossRef]
- Brooks, R.R.; Shaw, S.; Marfil, A.A. Some observations on the ecology, metal uptake and nickel tolerance of Alyssum serpyllifolium subspecies from the Iberian peninsula. Vegetatio 1981, 45, 183–188. [Google Scholar] [CrossRef]
- Mengoni, A.; Gonnelli, C.; Brocchini, E.; Galardi, F.; Pucci, S.; Gabbrielli, R.; Bazzicalupo, M. Chloroplast genetic diversity and biogeography in the serpentine endemic Ni-hyperaccumulator Alyssum bertolonii. New Phytol. 2003, 157, 349–356. [Google Scholar] [CrossRef]
- Cecchi, L.; Gabbrielli, R.; Arnetoli, M.; Gonnelli, C.; Hasko, A.; Selvi, F. Evolutionary lineages of nickel hyperaccumulation and systematics in European Alysseae (Brassicaceae): Evidence from nrDNA sequence data. Ann. Bot. 2010, 106, 751–767. [Google Scholar] [CrossRef]
- Cecchi, L.; Colzi, I.; Coppi, A.; Gonnelli, C.; Selvi, F. Diversity and biogeography of Ni-hyperaccumulators of Alyssum section Odontarrhena (Brassicaceae) in the central western Mediterranean: Evidence from karyology, morphology and DNA sequence data. Bot. J. Linn. Soc. 2013, 173, 269–289. [Google Scholar] [CrossRef]
- Sobczyk, M.K.; Smith, J.A.C.; Pollard, A.J.; Filatov, D.A. Evolution of nickel hyperaccumulation and serpentine adaptation in the Alyssum serpyllifolium species complex. Heredity 2016, 118, 31–41. [Google Scholar] [CrossRef]
- Mengoni, A.; Chianni, E.; Galardi, F.; Gonnelli, C.; Baker, A.J.M.; Bazzicalupo, M.; Reeves, R.D.; Adiguzel, N.; Gabbrielli, R. Evolutionary dynamics of nickel hyperaccumulation in Alyssum revealed by its nrDNA analysis. New Phytol. 2003, 159, 691–699. [Google Scholar] [CrossRef]
- Adamidis, G.C.; Dimitrakopoulos, P.G.; Manolis, A.; Papageorgiou, A.C. Genetic diversity and population structure of the serpentine endemic Ni hyperaccumulator Alyssum lesbiacum. Plant Syst. Evol. 2014, 300, 2051–2060. [Google Scholar] [CrossRef]
- Estrade, N.; Cloquet, C.; Echevarria, G.; Sterckeman, T.; Deng, T.; Tang, Y.; Morel, J.-L. Weathering and vegetation controls on nickel isotope fractionation in surface ultramafic environments (Albania). Earth Planet. Sci. Lett. 2015, 423, 24–35. [Google Scholar] [CrossRef]
- Cecchi, L.; Bettarini, I.; Colzi, I.; Coppi, A.; Echevarria, G.; Pazzagli, L.; Bani, A.; Gonnelli, C.; Selvi, F. The genus Odontarrhena (Brassicaceae) in Albania: taxonomy and nickel accumulation in a critical group of metallophytes from a major serpentine hot-spot. Phytotaxa 2018, 351, 1–28. [Google Scholar] [CrossRef]
- Nyárády, E.J. Synopsis specierum, variationum et formatum sectionis Odontarrhenae generis Alyssum. Analele Academiei Republicii Populare Române, Secţia de Ştiinţe Geologice, Geografice şi Biologice, Seria A 1949, 1, 67–194. [Google Scholar]
- Meyer, F.K. Beiträge zur Flora von Albanien. Haussknechtia Beiheft 2011, 15, 1–220. [Google Scholar]
- Wolf, A.T.; Howe, R.W.; Hamrick, J.L. Genetic diversity and population structure of the serpentine endemic Calystegia collina (Convolvulaceae) in northern California. Am. J. Bot. 2000, 87, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Mengoni, A.; Selvi, F.; Cusimano, N.; Galardi, F.; Gonnelli, C. Genetic diversity inferred from AFLP fingerprinting in populations of Onosma echioides (Boraginaceae) from serpentine and nrDNA sequence data. Plant Biosyst. 2006, 140, 211–219. [Google Scholar] [CrossRef]
- Coppi, A.; Cecchi, L.; Mengoni, A.; Pustahija, F.; Tomović, G.; Selvi, F. Low genetic diversity and contrasting patterns of differentiation in the two monotypic genera Halacsya and Paramoltkia (Boraginaceae) endemic to the Balkan serpentines. Flora -Morphol. Distrib. Funct. Ecol. Plants 2014, 209, 5–14. [Google Scholar] [CrossRef]
- Anderson, E.; Stebbins, G.L., Jr. Hybridization as an evolutionary stimulus. Evolution 1954, 8, 378–388. [Google Scholar] [CrossRef]
- Pierce, A.A.; Gutierrez, R.; Rice, A.M.; Pfennig, K.S. Genetic variation during range expansion: Effects of habitat novelty and hybridization. Proc. R. Soc. B Boil. Sci. 2017, 284, 20170007. [Google Scholar] [CrossRef] [PubMed]
- Rešetnik, I.; Šatović, Z.; Schneeweiss, G.M.; Liber, Z. Phylogenetic relationships in Brassicaceae tribe Alysseae inferred from nuclear ribosomal and chloroplast DNA sequence data. Mol. Phylogenet. Evol. 2013, 69, 772–786. [Google Scholar] [CrossRef]
- Li, Y.; Feng, Y.; Lv, G.; Liu, B.; Qi, A. The phylogeny of Alyssum (Brassicaceae) inferred from molecular data. Nord. J. Bot. 2015, 33, 715–721. [Google Scholar] [CrossRef]
- Bani, A.; Pavlova, D.; Echevarria, G.; Mullaj, A.; Reeves, R.D.; Morel, J.L.; Sulçe, S. Nickel hyperaccumulation by species of Alyssum and Thlaspi (Brassicaceae) from ultramafic soils of the Balkans. Bot. Serb. 2010, 34, 3–14. [Google Scholar]
- Quintela-Sabarís, C.; Marchand, L.; Smith, J.A.C.; Kidd, P. Using AFLP genome scanning to explore serpentine adaptation and nickel hyperaccumulation in Alyssum serpyllifolium. Plant Soil 2017, 416, 391–408. [Google Scholar] [CrossRef]
- Kim, C.; Shin, H.; Choi, H. Genetic diversity and population structure of diploid and polyploid species of Isoëtes in East Asia based on Amplified Fragment Length Polymorphism Markers. Int. J. Plant Sci. 2009, 170, 496–504. [Google Scholar] [CrossRef]
- Zalapa, J.E.; Brunet, J.; Guries, R.P. The extent of hybridization and its impact on the genetic diversity and population structure of an invasive tree, Ulmus pumila (Ulmaceae). Evol. Appl. 2010, 3, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Hamlin, J.A.P.; Arnold, M.L. Determining population structure and hybridization for two iris species. Ecol. Evol. 2014, 4, 743–755. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Bettarini, I.; Colzi, I.; Coppi, A.; Falsini, S.; Echevarria, G.; Pazzagli, L.; Selvi, F.; Gonnelli, C. Unravelling soil and plant metal relationships in Albanian nickel hyperaccumulators in the genus Odontarrhena (syn. Alyssum sect. Odontarrhena, Brassicaceae). Plant Soil 2019, 440, 135–149. [Google Scholar] [CrossRef]
- Choler, P.; Erschbamer, B.; Tribsch, A.; Gielly, L.; Taberlet, P. Genetic introgression as a potential to widen a species’ niche: Insights from alpine Carex curvula. Proc. Natl. Acad. Sci. USA 2003, 101, 171–176. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Kim, S.-C.; Randell, R.A.; Whitney, K.D.; Gross, B.L.; Lexer, C.; Clay, K. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 2007, 129, 149–165. [Google Scholar] [CrossRef]
- Pfennig, K.S.; Kelly, A.L.; Pierce, A.A. Hybridization as a facilitator of species range expansion. Proc. R. Soc. B Boil. Sci. 2016, 283, 20161329. [Google Scholar] [CrossRef]
- Guo, Q. Plant hybridization: The role of human disturbance and biological invasion. Divers. Distrib. 2014, 20, 1345–1354. [Google Scholar] [CrossRef]
- Todesco, M.; Pascual, M.A.; Owens, G.L.; Ostevik, K.L.; Moyers, B.T.; Hübner, S.; Heredia, S.M.; Hahn, M.A.; Caseys, C.; Bock, D.G.; et al. Hybridization and extinction. Evol. Appl. 2016, 9, 892–908. [Google Scholar] [CrossRef] [PubMed]
- Lamont, B.B.; He, T.; Enright, N.J.; Krauss, S.L.; Miller, B.P. Anthropogenic disturbance promotes hybridization between Banksia species by altering their biology. J. Evol. Biol. 2003, 16, 551–557. [Google Scholar] [CrossRef]
- Stankowski, S. Practical Conservation Biology; CSIRO Publishing: Collingwood, Australia, 2005. [Google Scholar]
- Soltis, D.E.; Soltis, P.; Tate, J.A. Advances in the study of polyploidy since plant speciation. New Phytol. 2003, 161, 173–191. [Google Scholar] [CrossRef]
- Chapman, M.; Abbott, R.J. Introgression of fitness genes across a ploidy barrier. New Phytol. 2009, 186, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, M.H.; Ehrich, D.; Schmickl, R.E.; Koch, M.A.; Brysting, A.K. Interspecific and interploidal gene flow in Central European Arabidopsis (Brassicaceae). BMC Evol. Biol. 2011, 11, 346. [Google Scholar] [CrossRef]
- Osborne, O.G.; Chapman, M.; Nevado, B.; Filatov, D.A. Maintenance of species boundaries despite ongoing gene flow in ragworts. Genome Biol. Evol. 2016, 8, 1038–1047. [Google Scholar] [CrossRef]
- Chapman, M.A.; Hiscock, S.J.; Filatov, D. Genomic divergence during speciation driven by adaptation to altitude. Mol. Biol. Evol. 2013, 30, 2553–2567. [Google Scholar] [CrossRef]
- Matute, D.R. The role of founder effects on the evolution of reproductive isolation. J. Evol. Biol. 2013, 26, 2299–2311. [Google Scholar] [CrossRef]
- Gustafson, D.J.; Romano, G.; Latham, R.E.; Morton, J.K. Amplified Fragment Length Polymorphism analysis of genetic relationships among the serpentine barrens endemic Cerastium velutinum Rafinesque var. villosissimum Pennell (Caryophyllaceae) and closely related Cerastium Species. J. Torrey Bot. Soc. 2003, 130, 218. [Google Scholar] [CrossRef]
- Rajakaruna, N.; Baldwin, B.G.; Chan, R.; DesRochers, A.M.; Bohm, B.A.; Whitton, J. Edaphic races and phylogenetic taxa in the Lasthenia californica complex (Asteraceae: Heliantheae): An hypothesis of parallel evolution. Mol. Ecol. 2003, 12, 1675–1679. [Google Scholar] [CrossRef]
- Berglund, A.-B.N.; Dahlgren, S.; Westerbergh, A. Evidence for parallel evolution and site-specific selection of serpentine tolerance in Cerastium alpinum during the colonization of Scandinavia. New Phytol. 2003, 161, 199–209. [Google Scholar] [CrossRef]
- Grdiša, M.; Radosavljević, I.; Liber, Z.; Stefkov, G.; Ralli, P.; Chatzopoulou, P.S.; Carović-Stanko, K.; Šatović, Z. Divergent selection and genetic structure of Sideritis scardica populations from southern Balkan Peninsula as revealed by AFLP fingerprinting. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Feng, X.-J.; Jiang, G.-F.; Fan, Z. Identification of outliers in a genomic scan for selection along environmental gradients in the bamboo locust, Ceracris kiangsu. Sci. Rep. 2015, 5, 13758. [Google Scholar] [CrossRef]
- Rannala, B. The sampling theory of neutral alleles in an island population of fluctuating size. Theor. Popul. Biol. 1996, 50, 91–104. [Google Scholar] [CrossRef]
- Russello, M.A.; Kirk, S.L.; Frazer, K.K.; Askey, P.J. Detection of outlier loci and their utility for fisheries management. Evol. Appl. 2011, 5, 39–52. [Google Scholar] [CrossRef]
- Wang, T.; Chen, G.; Zan, Q.; Wang, C.; Su, Y.-j. AFLP genome scan to detect genetic structure and candidate loci under selection for local adaptation of the invasive weed Mikania micrantha. PLoS ONE 2012, 7, e41310. [Google Scholar] [CrossRef]
- Coppi, A.; Lastrucci, L.; Cappelletti, D.; Cerri, M.; Ferranti, F.; Ferri, V.; Foggi, B.; Gigante, D.; Venanzoni, R.; Viciani, D.; et al. AFLP Approach Reveals Variability in Phragmites australis: Implications for Its Die-Back and Evidence for Genotoxic Effects. Front. Plant Sci. 2018, 9, 386. [Google Scholar] [CrossRef]
- Kruckeberg, A.R. The Influences of Lithology on Plant Life. In Geology and Plant Life: The Effects of Landforms and Rock Type on Plants; University Wash Press: Seattle, WA, USA; London, UK, 2002; pp. 160–181. [Google Scholar]
- Brady, K.U.; Kruckeberg, A.R.; Bradshaw, H.D., Jr. Evolutionary ecology of plant adaptation to serpentine soils. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 243–266. [Google Scholar] [CrossRef]
- Kazakou, E.; Dimitrakopoulos, P.G.; Baker, A.J.M.; Reeves, R.D.; Troumbis, A.Y. Hypotheses, mechanisms and trade-offs of tolerance and adaptation to serpentine soils: From species to ecosystem level. Biol. Rev. 2008, 83, 495–508. [Google Scholar] [CrossRef]
- Arnold, B.J.; Lahner, B.; Dacosta, J.M.; Weisman, C.M.; Hollister, J.D.; Salt, D.E.; Bomblies, K.; Yant, L. Borrowed alleles and convergence in serpentine adaptation. Proc. Natl. Acad. Sci. USA 2016, 113, 8320–8325. [Google Scholar] [CrossRef]
- Selby, J.P.; Willis, J.H. Major QTL controls adaptation to serpentine soils in Mimulus guttatus. Mol. Ecol. 2018, 27, 5073–5087. [Google Scholar] [CrossRef]
- Ohsawa, T.; Ide, Y. Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Glob. Ecol. Biogeogr. 2008, 17, 152–163. [Google Scholar] [CrossRef]
- Brooks, R.R. Serpentine and Its Vegetation: A Multidisciplinary Approach; Dioscorides Press: Portland, OR, USA, 1987. [Google Scholar]
- Selvi, F. Diversity, geographic variation and conservation of the serpentine flora of Tuscany (Italy). Biodivers. Conserv. 2007, 16, 1423–1439. [Google Scholar] [CrossRef]
- Cacho, N.I.; Strauss, S.Y. Occupation of bare habitats, an evolutionary precursor to soil specialization in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 15132–15137. [Google Scholar] [CrossRef]
- Whiting, S.N.; Reeves, R.D.; Richards, D.; Johnson, M.S.; Cooke, J.A.; Malaisse, F.; Paton, A.; Smith, J.A.C.; Angle, J.S.; Chaney, R.L.; et al. Research priorities for conservation of metallophyte biodiversity and their potential for restoration and site remediation. Restor. Ecol. 2004, 12, 106–116. [Google Scholar] [CrossRef]
- Rusterholz, H.-P.; Aydin, D.; Baur, B. Population structure and genetic diversity of relict populations of Alyssum montanum on limestone cliffs in the Northern Swiss Jura mountains. Alp. Bot. 2012, 122, 109–117. [Google Scholar] [CrossRef][Green Version]
- Dilek, Y.; Furnes, H. Structure and geochemistry of Tethyan ophiolites and their petrogenesis in subduction rollback systems. Lithos 2009, 113, 1–20. [Google Scholar] [CrossRef]
- Li, Y.-M.; Chaney, R.L.; Brewer, E.P.; Angle, A.J.S.; Nelkin, J. Phytoextraction of nickel and cobalt by hyperaccumulator Alyssum Species grown on nickel-contaminated soils. Environ. Sci. Technol. 2003, 37, 1463–1468. [Google Scholar] [CrossRef]
- Tappero, R.; Peltier, E.; Gräfe, M.; Heidel, K.; Ginder-Vogel, M.; Livi, K.J.T.; Rivers, M.L.; Marcus, M.A.; Chaney, R.L.; Sparks, D.L. Hyperaccumulator Alyssum murale relies on a different metal storage mechanism for cobalt than for nickel. New Phytol. 2007, 175, 641–654. [Google Scholar] [CrossRef]
- Bani, A.; Echevarria, G.; Sulçe, S.; Morel, J.L.; Mullai, A. In-situ phytoextraction of Ni by a native population of Alyssum murale on an ultramafic site (Albania). Plant Soil 2007, 293, 79–89. [Google Scholar] [CrossRef]
- Bani, A.; Echevarria, G.; Sulçe, S.; Morel, J.L. Improving the agronomy of Alyssum murale for extensive phytomining: A five-year field study. Int. J. Phytoremediation 2014, 17, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Bani, A.; Echevarria, G.; Zhang, X.; Benizri, E.; Laubie, B.; Morel, J.L.; Simonnot, M.-O. The effect of plant density in nickel-phytomining field experiments with Alyssum murale in Albania. Aust. J. Bot. 2015, 63, 72–77. [Google Scholar] [CrossRef]
- Calderon-Aguilera, L.E.; Rivera-Monroy, V.H.; Porter-Bolland, L.; Martínez-Yrízar, A.; Ladah, L.B.; Martínez-Ramos, M.; Alcocer, J.; Santiago-Pérez, A.L.; Hernandez-Arana, H.A.; Reyes-Gómez, V.M.; et al. An assessment of natural and human disturbance effects on Mexican Chaneya ecosystems: Current trends and research gaps. Biodivers. Conserv. 2012, 21, 589–617. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Cheng, T.; Xu, C.; Lei, L.; Li, C.; Zhang, Y.; Zhou, S. Barcoding the kingdom Plantae: New PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016, 16, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Rosati, L.; Coppi, A.; Farris, E.; Fascetti, S.; Becca, G.; Peregrym, M.; Tan, K.; Selvi, F. The genus Gymnospermium (Berberidaceae) in Italy: Identity and relationships of the populations at the western limit of the genus range. Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2018, 153, 796–808. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Simmons, M.P.; Ochoterena, H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 2000, 49, 369–381. [Google Scholar] [CrossRef]
- Borchsenius, F. FastGap 1.2. 2009. Available online: http://www.aubot.dk/FastGap_home.htm (accessed on 23 September 2014).
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Page, R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Tamura, K.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Leipold, M.; Tausch, S.; Hirtreiter, M.; Poschlod, P.; Reisch, C. Sampling for conservation genetics: How many loci and individuals are needed to determine the genetic diversity of plant populations using AFLP? Conserv. Genet. Resour. 2018, 12, 99–108. [Google Scholar] [CrossRef]
- Meyer, C.-L.; Vitalis, R.; Saumitou-Laprade, P.; Castric, V. Genomic pattern of adaptive divergence in Arabidopsis halleri, a model species for tolerance to heavy metal. Mol. Ecol. 2009, 18, 2050–2062. [Google Scholar] [CrossRef]
- Meudt, H.M.; Clarke, A.C. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci. 2007, 12, 106–117. [Google Scholar] [CrossRef]
- Dufresne, F.; Stift, M.; Vergilino, R.; Mable, B.K. Recent progress and challenges in population genetics of polyploid organisms: An overview of current state-of-the-art molecular and statistical tools. Mol. Ecol. 2014, 23, 40–69. [Google Scholar] [CrossRef]
- Čertner, M.; Kolář, F.; Frajman, B.; Winkler, M.; Schönswetter, P. Massive introgression weakens boundaries between a regionally endemic allopolyploid and a widespread congener. Perspect. Plant Ecol. Evol. Syst. 2020, 42, 125502. [Google Scholar] [CrossRef]
- Bonin, A.; Ehrich, D.; Manel, S. Statistical analysis of amplified fragment length polymorphism data: A toolbox for molecular ecologists and evolutionists. Mol. Ecol. 2007, 16, 3737–3758. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar]
- Schneider, S.; Roessli, D.; Excoffier, L. Arlequin: A Software for Population Genetics Data Analysis; Genetics and Biometry Laboratory, University of Geneva: Geneva, Switzerland, 2000. [Google Scholar]
- Stift, M.; Kolář, F.; Meirmans, P.G. Structure is more robust than other clustering methods in simulated mixed-ploidy populations. Heredity 2019, 123, 429–441. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. Structure harvester: A website and program for visualizing Structure output and implementing the Evanno Mehtod. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2005, 23, 254–267. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Beaumont, M.A.; Nichols, R.A. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. B Boil. Sci. 1996, 263, 1619–1626. [Google Scholar] [CrossRef]
- Beaumont, M.A.; Balding, D.J. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 2004, 13, 969–980. [Google Scholar] [CrossRef]
- Rannala, B.; Hartigan, J.A. Estimating gene flow in island populations. Genet. Res. 1996, 67, 147–158. [Google Scholar] [CrossRef]
- Burr, T.L. Quasi-Equilibrium theory for the distribution of rare alleles in a subdivided population: Justification and implications. Theor. Popul. Biol. 2000, 57, 297–306. [Google Scholar] [CrossRef]
- Foll, M. BayeScan v2.1 User Manual. 2012. Available online: http://cmpg.unibe.ch/software/BayeScan/ (accessed on 11 November 2016).
- Yang, A.; Dick, C.W.; Yao, X.; Huang, H. Impacts of biogeographic history and marginal population genetics on species range limits: A case study of Liriodendron chinense. Sci. Rep. 2016, 6, 25632. [Google Scholar] [CrossRef]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Guillaume Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-4. 2019. Available online: http://vegan.r-forge.r-project.org (accessed on 1 April 2017).
- Whitlock, M.C.; McCauley, D.E. Indirect measures of gene flow and migration: FST≠1/(4Nm+1). Heredity 1999, 82, 117–125. [Google Scholar] [CrossRef]
- Pagès, J. Analyse factorielle multiple appliquée aux variables qualitatives et aux données mixtes. Rev. Stat. Appl. 2002, 4, 5–37. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org (accessed on 1 April 2017).


| Source of Variation | df | Sum of Squares | Variance Components | Percentage of Variation | p-Values |
|---|---|---|---|---|---|
| Among species | 5 | 491.962 | 0.23949 | 1.06 | n.s. |
| Among all populations within species | 26 | 2234.717 | 5.96533 | 26.50 | <0.0001 |
| Within all populations | 342 | 5576.420 | 16.30532 | 72.44 | <0.0001 |
| Total | 373 | 8302.599 | 22.51014 | ||
| Among diploids | 1 | 99.78 | 2.26958 | 11.19 | <0.0001 |
| Within diploids | 70 | 1260.250 | 18.00357 | 88.81 | <0.0001 |
| Total diploids | 71 | 1359.958 | 20.27315 | ||
| Among tetraploids | 25 | 2213.633 | 6.18362 | 26.99 | <0.0001 |
| Within tetraploids | 276 | 4617.503 | 16.73008 | 73.01 | <0.0001 |
| Total tetraploids | 301 | 6831.136 | 22.91371 |
| Taxa/Pop. | KPop | Mind | No. Poly | %poly | He | Hs | Out | FST |
|---|---|---|---|---|---|---|---|---|
| O. albiflora | 0.113 | — | ||||||
| 26 | 2 | 0 | 49 | 35.766 | 0.113 | 2.08 | ||
| O. chalcidica | 0.247 | 0.267 | ||||||
| 1 | 4 | 3 | 113 | 82.48 | 0.323 | — | 6.25 | — |
| 2 | 3 | 4 | 114 | 83.21 | 0.338 | — | 6.92 | — |
| 3 | 3 | 3 | 81 | 59.12 | 0.217 | — | 4.83 | — |
| 5 | 2 | 2 | 74 | 54.02 | 0.196 | — | 3.25 | — |
| 7 | 4 | 6 | 101 | 73.72 | 0.261 | — | 4.83 | — |
| 8 | 2 | 1 | 47 | 34.31 | 0.126 | — | 0.83 | — |
| 9 | 2 | 1 | 61 | 44.53 | 0.147 | — | 0.82 | — |
| 11 | 4 | 10 | 101 | 73.72 | 0.275 | — | 5.25 | — |
| 12 | 4 | 7 | 120 | 87.59 | 0.335 | — | 5.82 | — |
| 15 | 2 | 1 | 68 | 49.64 | 0.189 | — | 1.80 | — |
| 17 | 1 | 0 | 81 | 59.12 | 0.201 | — | 1.67 | — |
| 18 | 2 | 0 | 86 | 62.77 | 0.215 | — | 1.83 | — |
| 21 | 5 | 4 | 92 | 67.15 | 0.251 | — | 2.92 | — |
| 24 | 3 | 2 | 103 | 75.18 | 0.296 | — | 5.73 | — |
| 25 | 2 | 1 | 72 | 52.56 | 0.191 | — | 3.83 | — |
| 31 | 3 | 1 | 113 | 82.48 | 0.317 | — | 3.08 | — |
| 32 | 2 | 0 | 114 | 83.21 | 0.322 | — | 4.92 | — |
| O. decipiens | 0.231 | 0.224 | ||||||
| 4 | 4 | 3 | 81 | 59.12 | 0.185 | — | 2.50 | — |
| 6 | 4 | 6 | 96 | 70.07 | 0.252 | — | 5.75 | — |
| 23 | 4 | 2 | 78 | 56.93 | 0.199 | — | 2.67 | — |
| 28 | 4 | 7 | 120 | 87.59 | 0.348 | — | 5.83 | — |
| 30 | 3 | 2 | 69 | 50.37 | 0.171 | — | 2.83 | — |
| O. moravensis | 0.239 | 0.279 | ||||||
| 19 | 2 | 0 | 103 | 75.18 | 0.273 | — | 3.17 | — |
| 20 | 4 | 5 | 110 | 80.29 | 0.314 | — | 3.67 | — |
| 27 | 2 | 0 | 45 | 32.84 | 0.130 | — | 2.08 | — |
| O. rigida | 0.185 | 0.230 | ||||||
| 10 | 2 | 0 | 69 | 50.36 | 0.171 | — | 2.08 | — |
| 13 | 1 | 0 | 80 | 58.39 | 0.232 | — | 3.58 | — |
| 14 | 2 | 0 | 69 | 50.36 | 0.153 | — | 1.08 | — |
| O. smolikana | 0.293 | 0.302 | ||||||
| 16 | 2 | 0 | 95 | 69.34 | 0.267 | — | 2.70 | — |
| 22 | 2 | 4 | 113 | 82.48 | 0.352 | — | 6.45 | — |
| 29 | 1 | 0 | 94 | 68.61 | 0.259 | — | 3.50 | — |
| Taxon | Ploidy Level | Distribution | Elevation (m a.s.l.) | Habitat |
|---|---|---|---|---|
| O. albiflora (F.K.Mey.) Španiel, Al-Shehbaz, D.A.German & Marhold | 4x | Endemic E Albania | 900–1300 | Limestone cliffs |
| O. chalcidica (Janka) Španiel, Al-Shehbaz, D.A.German & Marhold | 4x | Balkans; throughout Albania | 50–1300 | Fields, roadsides; on various soil types, more frequently on serpentine |
| O. decipiens (Nyár.) L.Cecchi & Selvi | 4x | Balkans; C & N Albania | 220–1900 | Fields, roadsides; only on serpentine |
| O. moravensis (F.K.Mey.) L.Cecchi & Selvi | 2x | Endemic E Albania | 800–1500 | Serpentine rocks and screes |
| O. rigida (Nyár.) L.Cecchi & Selvi | 2x | Endemic C Albania | 200–1250 | Serpentine rocks and screes |
| O. smolikana (Nyár.) Španiel, Al-Shehbaz, D.A.German & Marhold subsp. glabra (Nyár.) L.Cecchi & Selvi | 4x | Endemic C and N Albania | 1100–2000 | Serpentine rocks and screes |
| Element Concentration in Soils | |||||
|---|---|---|---|---|---|
| Pop. Site | Ni (µg g−1) | Co (µg g−1) | Cr (µg g−1) | Ca (mg g−1) | Mg (mg g−1) |
| 3 (O. chalcidica) | 4602 ± 252 | 274 ± 52 | 2315 ± 139 | 1.4 ± 0.3 | 94.4 ± 33.1 |
| 5 (O. chalcidica) | 3695 ± 299 | 240 ± 64 | 1951 ± 125 | 1.1 ± 0.4 | 72.2 ± 11.5 |
| 9 (O. chalcidica) | 728 ± 13 | 63 ± 1 | 546 ± 14 | 0.15 ± 0.01 | 16.0 ± 0.6 |
| 20 (O. moravensis) | 2451 ± 256 | 118 ± 4 | 881 ± 30 | 7.0 ± 0.7 | 95.9 ± 4.3 |
| 24 (O. chalcidica) | 3860 ± 107 | 146 ± 65 | 550 ± 29 | 0.17 ± 0.03 | 145.3 ± 2.6 |
| 26 (O. albiflora) | 144 ± 6 | 19 ± 1 | 155 ± 6 | 135.8 ± 26.9 | 4.9 ± 0.9 |
| 31 (O. chalcidica) | 1098 ± 18 | 89 ± 17 | 815 ± 134 | 16.3 ± 2.0 | 109.4 ± 1.8 |
| 32 (O. chalcidica) | 4289 ± 555 | 66 ± 3 | 678 ± 98 | 0.7 ± 0.1 | 147.9 ± 3.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coppi, A.; Baker, A.J.M.; Bettarini, I.; Colzi, I.; Echevarria, G.; Pazzagli, L.; Gonnelli, C.; Selvi, F. Population Genetics of Odontarrhena (Brassicaceae) from Albania: The Effects of Anthropic Habitat Disturbance, Soil, and Altitude on a Ni-Hyperaccumulator Plant Group from a Major Serpentine Hotspot. Plants 2020, 9, 1686. https://doi.org/10.3390/plants9121686
Coppi A, Baker AJM, Bettarini I, Colzi I, Echevarria G, Pazzagli L, Gonnelli C, Selvi F. Population Genetics of Odontarrhena (Brassicaceae) from Albania: The Effects of Anthropic Habitat Disturbance, Soil, and Altitude on a Ni-Hyperaccumulator Plant Group from a Major Serpentine Hotspot. Plants. 2020; 9(12):1686. https://doi.org/10.3390/plants9121686
Chicago/Turabian StyleCoppi, Andrea, Alan J. M. Baker, Isabella Bettarini, Ilaria Colzi, Guillaume Echevarria, Luigia Pazzagli, Cristina Gonnelli, and Federico Selvi. 2020. "Population Genetics of Odontarrhena (Brassicaceae) from Albania: The Effects of Anthropic Habitat Disturbance, Soil, and Altitude on a Ni-Hyperaccumulator Plant Group from a Major Serpentine Hotspot" Plants 9, no. 12: 1686. https://doi.org/10.3390/plants9121686
APA StyleCoppi, A., Baker, A. J. M., Bettarini, I., Colzi, I., Echevarria, G., Pazzagli, L., Gonnelli, C., & Selvi, F. (2020). Population Genetics of Odontarrhena (Brassicaceae) from Albania: The Effects of Anthropic Habitat Disturbance, Soil, and Altitude on a Ni-Hyperaccumulator Plant Group from a Major Serpentine Hotspot. Plants, 9(12), 1686. https://doi.org/10.3390/plants9121686






