Genome Survey Sequencing of In Vivo Mother Plant and In Vitro Plantlets of Mikania cordata
Abstract
1. Introduction
2. Results
2.1. Micropropagation of M. cordata
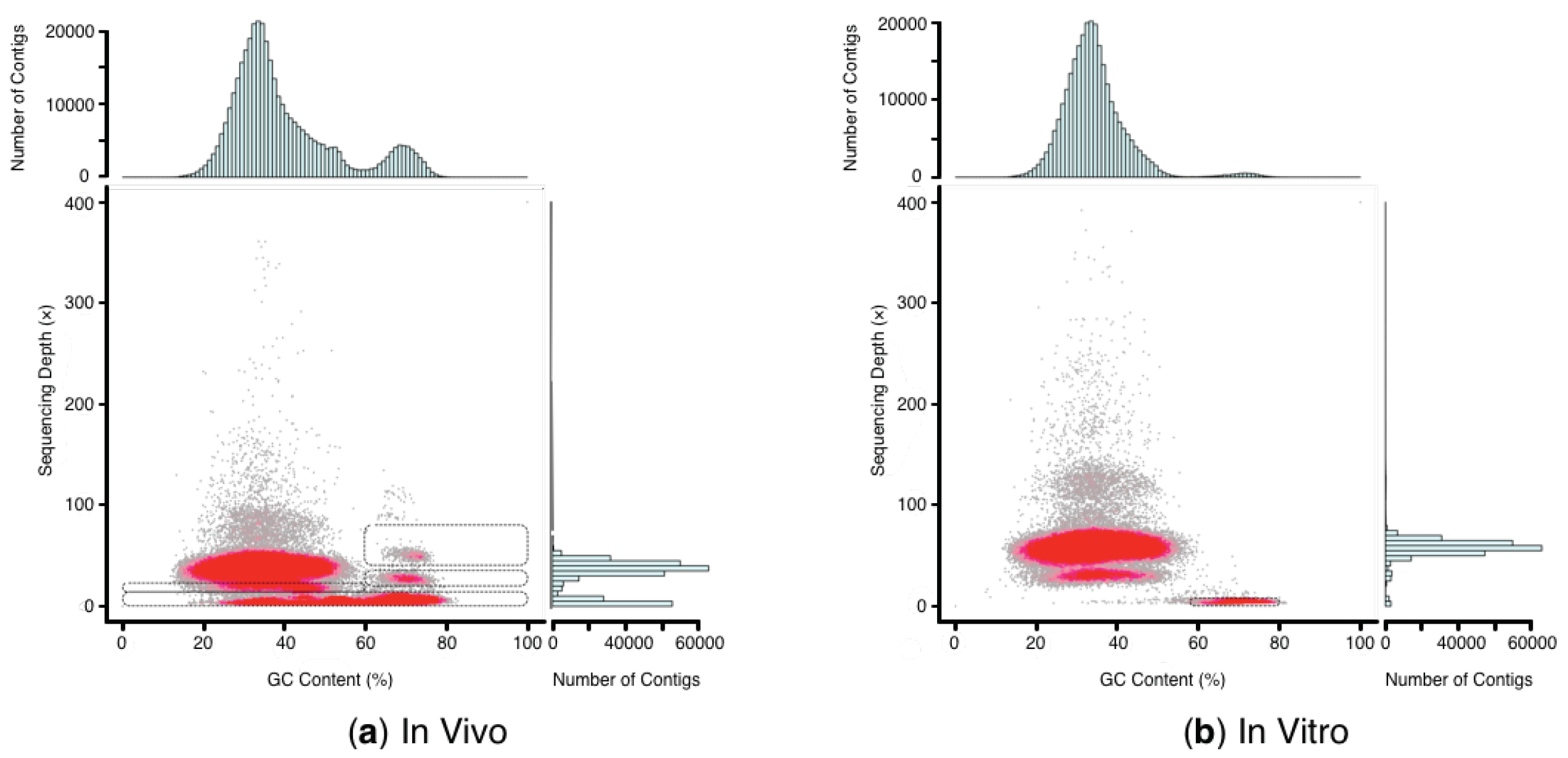
2.2. Sequencing, Contigs Assembly, and Preliminary GC Depth Profiling
2.3. Identification and Filtration of Genomic Contaminants
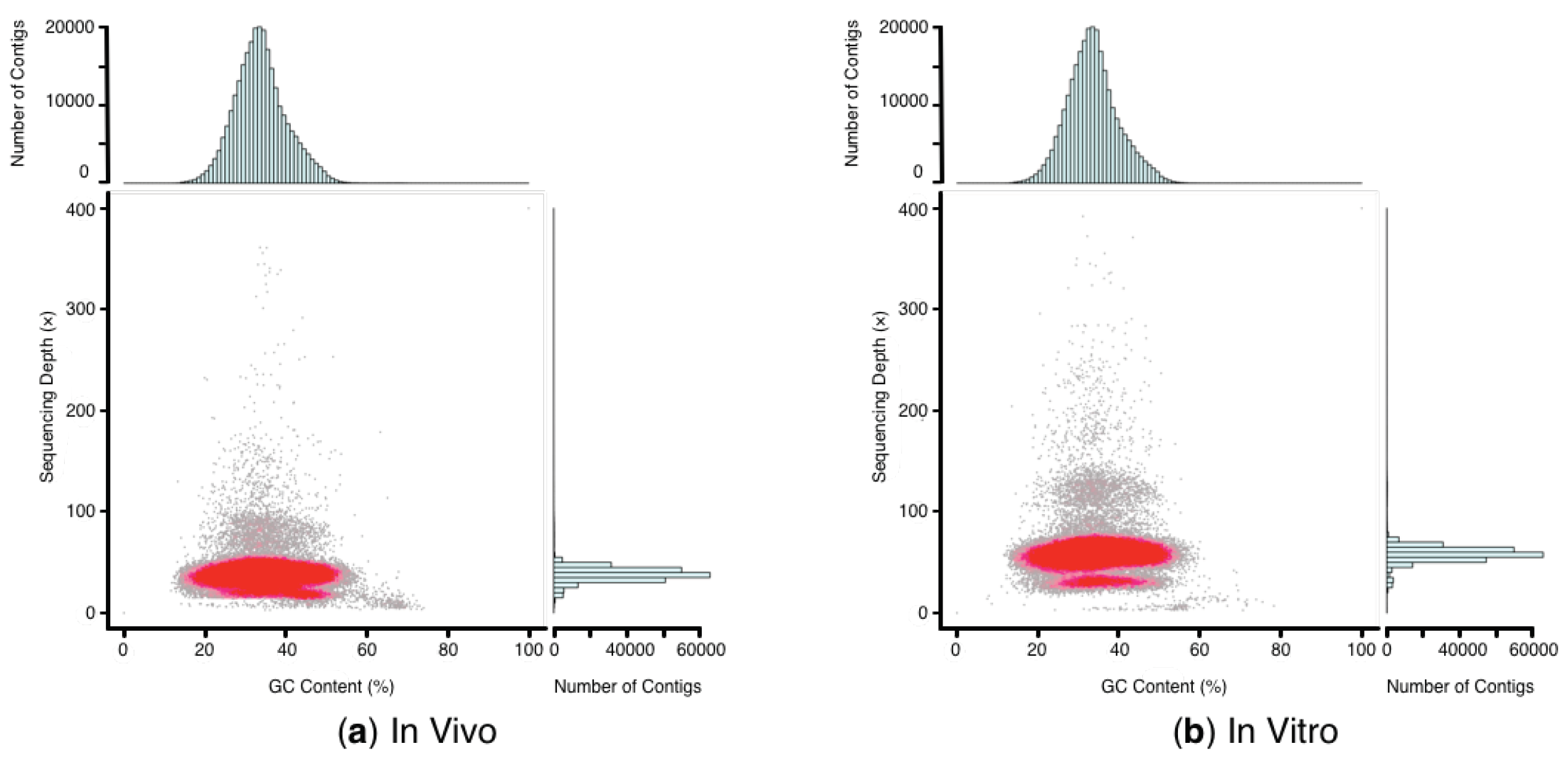
2.4. Genome Assembly and GC Depth Profiling after In Silico Decontamination
2.5. Estimation of Genome Size, Heterozygosity, and Repetitive Rate Based on 17-mer Analysis
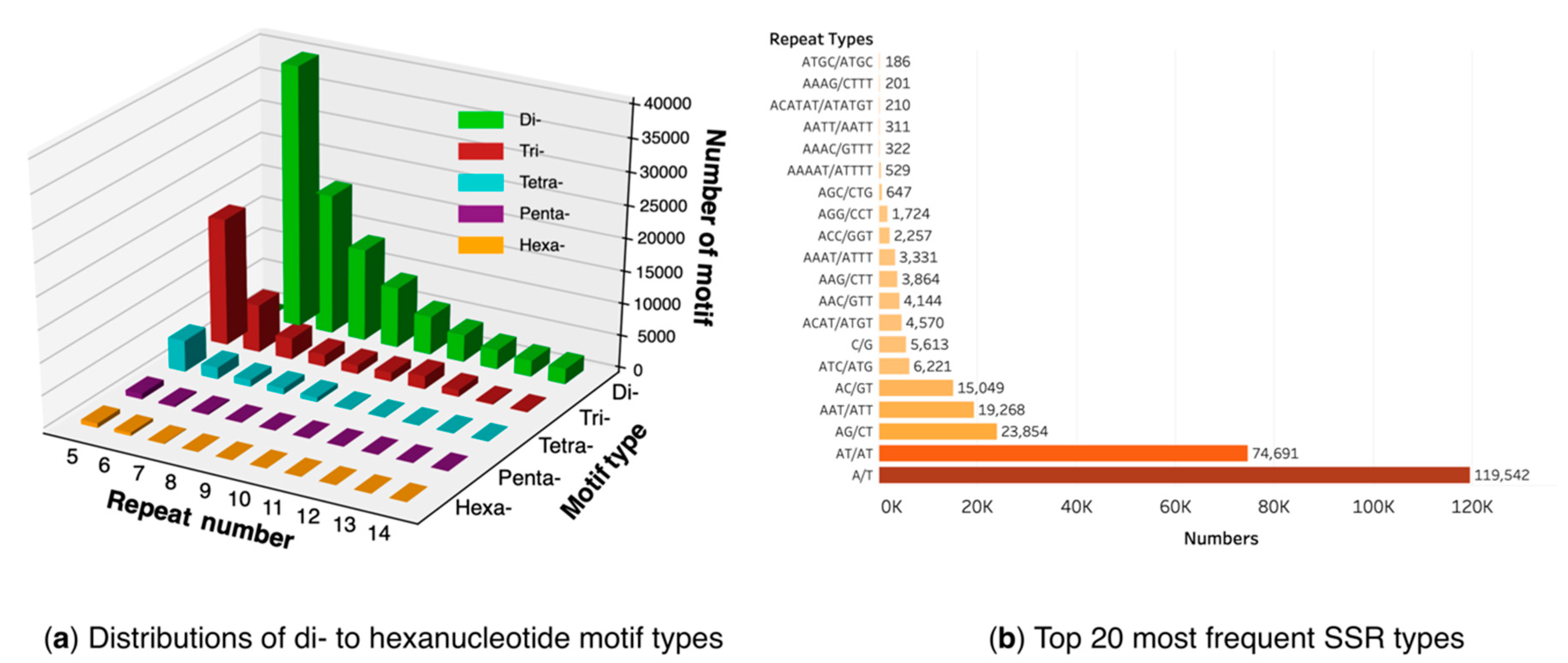
2.6. SSR Identification
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Micropropagation of M. cordata
4.3. DNA Extraction, Library Construction, and Genome Sequencing
4.4. Contigs Assembly and Preliminary GC Depth Profiling
4.5. Identification and Filtration of Genomic Contaminants
4.6. Genome Assembly and GC Depth Profiling after In Silico Decontamination
4.7. Estimation of Genome Size, Heterozygosity, and Repetitive Rate Based on 17-mer Analysis
4.8. SSR Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BA | 6-Benzylaminopurine |
| BLAST | Basic Local Alignment Search Tool |
| CAM | Crassulacean Acid Metabolism |
| CTX | Cefotaxime |
| MS | Murashige and Skoog |
| PGR | Plant Growth Regulator |
| SSR | Simple sequence repeat |
| TIM | Timentin |
References
- Zhu, S.; Chen, Y.L.; Chen, Y.S.; Lin, Y.R.; Liu, S.W.; Ge, X.J.; Gao, T.G.; Zhu, S.X.; Liu, Y.; Humphries, C.; et al. Asteraceae (Compositae). In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2011; Volume 20–21, pp. 879–891. [Google Scholar]
- Rahmatullah, M.; Mukti, I.J.; Haque, A.; Mollik, M.A.H.; Parvin, K.; Jahan, R.; Chowdhury, M.H.; Rahman, T. An ethnobotanical survey and pharmacological evaluation of medicinal plants used by the Garo tribal community living in Netrakona district, Bangladesh. Adv. Nat. Appl. Sci. 2009, 3, 402–418. [Google Scholar]
- Mercado, B. Notes on some growth characteristics of Mikania cordata (Burm. f.) B.L. Robinson. Biotropia-Southeast Asian J. Trop. Biol. 1994, 7, 30–40. [Google Scholar]
- Holm, L.G.; Plucknett, D.L.; Pancho, J.V.; Herberger, J.P. The World’s Worst Weeds. Distribution and Biology; University Press of Hawaii: Honolulu, HI, USA, 1977; pp. 320–327. [Google Scholar]
- Kong, G.; Wu, Q.G.; Hu, Q.M.; Ye, W.H. Further supplementary data on Mikania micrantha HBK (Asteraceae). J. Trop. Subtrop. Bot. 2000, 8, 128–130. [Google Scholar]
- Parker, C. The Mikania problem. Pans Pest Artic. News Summ. 1972, 18, 312–315. [Google Scholar] [CrossRef]
- Siwakoti, M. Mikania weed: A challenge for conservationists. Our Nat. 2007, 5, 70–74. [Google Scholar] [CrossRef]
- Lowe, S.; Browne, M.; Boudjelas, S.; De Poorter, M. 100 of the World’s Worst Invasive Alien Species: A Selection from the Global Invasive Species Database; Invasive Species Specialist Group: Auckland, New Zealand, 2000. [Google Scholar]
- Deng, X.; Ye, W.H.; Feng, H.L.; Yang, Q.H.; Cao, H.L.; Xu, K.Y.; Zhang, Y. Gas exchange characteristics of the invasive species Mikania micrantha and its indigenous congener M. cordata (Asteraceae) in South China. Bot. Bull. Acad. Sin. 2004, 45, 213–220. [Google Scholar]
- Vila, M.; Weiner, J. Are invasive plant species better competitors than native plant species?—Evidence from pair-wise experiments. Oikos 2004, 105, 229–238. [Google Scholar] [CrossRef]
- Su, Y.; Huang, L.; Wang, Z.; Wang, T. Comparative chloroplast genomics between the invasive weed Mikania micrantha and its indigenous congener Mikania cordata: Structure variation, identification of highly divergent regions, divergence time estimation, and phylogenetic analysis. Mol. Phylogenet. Evol. 2018, 126, 181–195. [Google Scholar] [CrossRef]
- Guo, W.; Liu, Y.; Ng, W.L.; Liao, P.C.; Huang, B.H.; Li, W.; Li, C.; Shi, X.; Huang, Y. Comparative transcriptome analysis of the invasive weed Mikania micrantha with its native congeners provides insights into genetic basis underlying successful invasion. BMC Genom. 2018, 19, 1–17. [Google Scholar] [CrossRef]
- Liu, B.; Yan, J.; Li, W.; Yin, L.; Li, P.; Yu, H.; Xing, L.; Cai, M.; Wang, H.; Zhao, M.; et al. Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Wei, N.; Cronn, R.; Liston, A.; Ashman, T.L. Functional trait divergence and trait plasticity confer polyploid advantage in heterogeneous environments. New Phytol. 2019, 221, 2286–2297. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.B.; Lee, C.E. Evolutionary origins of genomic adaptations in an invasive copepod. Nat. Ecol. Evol. 2020, 4, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, D.K. Endophytes: Biology and Biotechnology; Springer: New York, NY, USA, 2017. [Google Scholar]
- Jeong, H.; Pan, J.G.; Park, S.H. Contamination as a major factor in poor Illumina assembly of microbial isolate genomes. bioRxiv 2016. [Google Scholar] [CrossRef]
- Dittami, S.M.; Corre, E. Detection of bacterial contaminants and hybrid sequences in the genome of the kelp Saccharina japonica using Taxoblast. PeerJ 2017, 5, e4073. [Google Scholar] [CrossRef] [PubMed]
- Reiter, T.; Brown, C.T. Microbial contamination in the genome of the domesticated olive. bioRxiv. 2018. [Google Scholar] [CrossRef]
- Simion, P.; Belkhir, K.; François, C.; Veyssier, J.; Rink, J.C.; Manuel, M.; Philippe, H.; Telford, M.J. A software tool ‘CroCo’ detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 2018, 16, 1–9. [Google Scholar] [CrossRef]
- Qiu, H.; Cai, G.; Luo, J.; Bhattacharya, D.; Zhang, N. Extensive horizontal gene transfers between plant pathogenic fungi. BMC Biol. 2016, 14, 1–11. [Google Scholar] [CrossRef]
- Cheng, S.; Xian, W.; Fu, Y.; Marin, B.; Keller, J.; Wu, T.; Sun, W.; Li, X.; Xu, Y.; Zhang, Y.; et al. Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 2019, 179, 1057–1067. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Y.; Huang, P.; Ma, Y.; Qing, Z.; Tang, Q.; Cao, H.; Cheng, P.; Zheng, Y.; Yuan, Z.; et al. The genome of medicinal plant Macleaya cordata provides new insights into benzylisoquinoline alkaloids metabolism. Mol. Plant 2017, 10, 975–989. [Google Scholar] [CrossRef]
- Jayatilake, P.L.; Munasinghe, H. Antimicrobial activity of cultivable endophytic and rhizosphere fungi associated with “mile-a-minute,” Mikania cordata (Asteraceae). Biomed Res. Int. 2020, 2020, 1–7. [Google Scholar] [CrossRef]
- Elavazhagan, T.; Jayakumar, S.; Balakrishnan, V.; Chitravadivu, C. Isolation of endophytic bacteria from the invasive alien weed, Mikania micrantha and their molecular characterization. Am. Eurasian J. Sci. Res. 2009, 4, 154–158. [Google Scholar]
- Pereira, A.M.S.; França, S.; Câmara, F.L.A. Vegetative propagation of Mikania glomerata: Micro-propagation and cuttings. Acta Hortic. 1999, 502, 347–352. [Google Scholar] [CrossRef]
- Da Silva Mendes, A.F.; Cidade, L.C.; de Oliveira, M.L.P.; Otoni, W.C.; Soares-Filho, W.D.S.; Costa, M.G.C. Evaluation of novel beta-lactam antibiotics in comparison to cefotaxime on plant regeneration of Citrus sinensis L. Osb. Plant CellTissue Organ Cult. 2009, 97, 331–336. [Google Scholar] [CrossRef]
- Goto, S.; Thakur, R.; Ishii, K. Determination of genetic stability in long-term micropropagated shoots of Pinus thunbergii Parl. using RAPD markers. Plant Cell Rep. 1998, 18, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Senapati, S.; Aparajita, S.; Rout, G. An assessment of genetic fidelity of in vitro grown plantlets of rose (Rosa hybrida) through molecular markers. Afr. J. Biotechnol. 2012, 11, 16532–16538. [Google Scholar]
- Schatz, M.C.; Witkowski, J.; McCombie, W.R. Current challenges in de novo plant genome sequencing and assembly. Genome Biol. 2012, 13, 1–7. [Google Scholar] [CrossRef]
- Michael, T.P.; Jupe, F.; Bemm, F.; Motley, S.T.; Sandoval, J.P.; Lanz, C.; Loudet, O.; Weigel, D.; Ecker, J.R. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef]
- Summerell, B.A.; Leslie, J.F. Fifty years of Fusarium: How could nine species have ever been enough? Fungal Divers 2011, 50, 135. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Poonguzhali, S.; Senthilkumar, M.; Seshadri, S.; Chung, H.; Yang, J.; Sundaram, S.; Sa, T. Growth promotion and induction of systemic resistance in rice cultivar Co-47 (Oryza sativa L.) by Methylobacterium spp. Bot. Bull. Acad. Sin. 2004, 45, 315–324. [Google Scholar]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.; Kloepper, J. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Skirvin, R.; Motoike, S.; Norton, M.; Ozgur, M.; Al-Juboory, K.; McMeans, O. Establishment of contaminant-free perennial plants in vitro. Vitr. Cell. Dev. Biol. Plant 1999, 35, 278–280. [Google Scholar] [CrossRef]
- Hong, L.; Ye, W.H.; Shen, H.; Cao, H.L.; Liu, W. Tissue culture and somatic embryogenesis of Mikania micrantha HBK. J. Zhejiang Univ. 2005, 5, 572–578. [Google Scholar]
- Eed, A.M.; Reddy, S.A.; Reddy, K.M.; Silva, J.; Reddy, P.V.; Beghum, H.; Venkatsubbaiah, P.Y. Effect of antibiotics and fungicides on the in vitro production of Citrus limonia Osbeck nodal segment and shoot tip explants. Asian Australas J. Plant Sci. Biotechnol. 2010, 4, 66–70. [Google Scholar]
- Brauer, V.S.; Rezende, C.P.; Pessoni, A.M.; De Paula, R.G.; Rangappa, K.S.; Nayaka, S.C.; Gupta, V.K.; Almeida, F. Antifungal agents in agriculture: Friends and foes of public health. Biomolecules 2019, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Nauerby, B.; Billing, K.; Wyndaele, R. Influence of the antibiotic timentin on plant regeneration compared to carbenicillin and cefotaxime in concentrations suitable for elimination of Agrobacterium tumefaciens. Plant Sci. 1997, 123, 169–177. [Google Scholar] [CrossRef]
- Leifert, C.; Ritchie, J.; Waites, W. Contaminants of plant-tissue and cell cultures. World J. Microbiol. Biotechnol. 1991, 7, 452–469. [Google Scholar] [CrossRef]
- Costa, M.; Nogueira, F.; Figueira, M.; Otoni, W.; Brommonschenkel, S.; Cecon, P. Influence of the antibiotic timentin on plant regeneration of tomato (Lycopersicon esculentum Mill.) cultivars. Plant Cell Rep. 2000, 19, 327–332. [Google Scholar] [CrossRef]
- Yin, L.; Liu, B.; Wang, H.; Zhang, Y.; Wang, S.; Jiang, F.; Ren, Y.; Liu, H.; Liu, C.; Wan, F.; et al. The rhizosphere microbiome of Mikania micrantha provides insight into adaptation and invasion. Front. Microbiol. 2020, 11, 1462. [Google Scholar] [CrossRef]
- Mora Polanco, I.Y. Bactéria Endofítica Associada a Mikania Glomerata (Asteraceae): Investigação Microbiológica, Busca de Substâncias Bioativas e Avaliação do Seu Efeito In Vitro na Modulação de Metabólitos Secundários; University of Campinas: São Paulo, Brazil, 2018. [Google Scholar]
- Jiang, L.; Zhang, Y.; Guo, Q.; Liu, Y.; Li, C. Cytological study on Mikania cordata (Asteraceae), a native plant in China. Guihaia 2018, 38, 324–331. [Google Scholar]
- Kelly, L.J.; Leitch, I.J. Exploring giant plant genomes with next-generation sequencing technology. Chromosome Res. 2011, 19, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, S.; Liu, C.; Liu, Y.; Zhao, X.; Yang, C.; Qu, G.Z. Genome survey sequencing of Betula platyphylla. Forests 2019, 10, 826. [Google Scholar] [CrossRef]
- Motalebipour, E.Z.; Kafkas, S.; Khodaeiaminjan, M.; Çoban, N.; Gözel, H. Genome survey of pistachio (Pistacia vera L.) by next generation sequencing: Development of novel SSR markers and genetic diversity in Pistacia species. BMC Genom. 2016, 17, 998. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.J.; Liu, M.X.; Lu, X.Y.; Sun, S.S.; Cheng, Y.W.; Ya, H.Y. Genome survey sequencing and identification of genomic SSR markers for Rhododendron micranthum. Biosci. Rep. 2020, 40, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vallès, J.; Canela, M.Á.; Garcia, S.; Hidalgo, O.; Pellicer, J.; Sánchez-Jiménez, I.; Siljak-Yakovlev, S.; Vitales, D.; Garnatje, T. Genome size variation and evolution in the family Asteraceae. Caryologia Int. J. Cytol. Cytosystematics Cytogenet. 2013, 66, 221–235. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.A.; Molinari, N.A.; Petrov, D.A. The large genome constraint hypothesis: Evolution, ecology and phenotype. Ann. Bot. 2005, 95, 177–190. [Google Scholar] [CrossRef]
- Kubešová, M.; Moravcova, L.; Suda, J.; Jarošík, V.; Pyšek, P. Naturalized plants have smaller genomes than their non-invading relatives: A flow cytometric analysis of the Czech alien flora. Preslia 2010, 82, 81–96. [Google Scholar]
- Males, J.; Griffiths, H. Stomatal biology of CAM plants. Plant Physiol. 2017, 174, 550–560. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Leitch, I.J.; Patel, S.; Pendharkar, A.; Knight, C.A. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 2008, 179, 975–986. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, G.; Zhang, J.; Liu, H.; Chen, W.; Wang, X.; Li, Y.; Dong, Y.; Yang, S. Hybrid de novo genome assembly of the Chinese herbal fleabane Erigeron breviscapus. Gigascience 2017, 6, gix028. [Google Scholar] [CrossRef]
- Lin, T.; Xu, X.; Ruan, J.; Liu, S.; Wu, S.; Shao, X.; Wang, X.; Gan, L.; Qin, B.; Yang, Y.; et al. Genome analysis of Taraxacum kok-saghyz Rodin provides new insights into rubber biosynthesis. Natl. Sci. Rev. 2018, 5, 78–87. [Google Scholar] [CrossRef]
- Delmont, T.O.; Eren, A.M. Identifying contamination with advanced visualization and analysis practices: Metagenomic approaches for eukaryotic genome assemblies. PeerJ 2016, 4, e1839. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jiang, D.; Li, M.; Mustapha, U.F.; Tian, C.; Chen, H.; Huang, Y.; Deng, S.; Wu, T.; Zhu, C.; et al. Genome survey of male and female spotted scat (Scatophagus argus). Animals 2019, 9, 1117. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Huang, Y.; Fang, X.; Lu, L.; Zhou, R.; Ge, X.; Shi, S. Development and characterization of EST-SSR markers in the invasive weed Mikania micrantha (Asteraceae). Am. J. Bot. 2011, 98, e1–e3. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhao, J.; Liu, M.; Liu, P.; Dai, L.; Zhao, Z. Genome-wide characterization of simple sequence repeat (SSR) loci in Chinese jujube and jujube SSR primer transferability. PLoS ONE 2015, 10, e0127812. [Google Scholar] [CrossRef]
- Tuler, A.; Carrijo, T.; Nóia, L.; Ferreira, A.; Peixoto, A.; da Silva Ferreira, M. SSR markers: A tool for species identification in Psidium (Myrtaceae). Mol. Biol. Rep. 2015, 42, 1501–1513. [Google Scholar] [CrossRef]
- Hoshino, A.A.; Bravo, J.P.; Nobile, P.M.; Morelli, K.A. Microsatellites as tools for genetic diversity analysis. In Genetic Diversity in Microorganisms; Caliskan, M., Ed.; IntechOpen: Rijeka, Croatia, 2012; pp. 149–170. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. 2010. [Google Scholar]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 2047–2217X. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]


| Media and PGR (mg·L−1) | Antibiotic (g·L−1) | Incubation (Weeks) | No. of Explants/Plantlets | Aseptic Explants/ Plantlets | Survival (>4 weeks) | Sprouting/ Rooting Plantlets | |
|---|---|---|---|---|---|---|---|
| Control | MS + BA 1.0 | None | 4 | 11 1 | 4 | 0 | 0 |
| Sprouting | MS + BA 1.0 | CTX 1.0 | 6 | 20 2 | 16 | 13 | 11 |
| Sprouting | MS + BA 1.0 | TIM 0.6 | 6 | 20 3 | 15 | 10 | 8 |
| Rooting | 1/2 MS | CTX 0.5 | 8 | 11 | 11 | 11 | 6 |
| Rooting | 1/2 MS | TIM 0.3 | 8 | 8 4 | 7 | 7 | 7 |
| Propagation 1st | 1/2 MS | TIM 0.3 | 8 | 64 5 | 60 | 60 | 60 |
| Propagation 2nd | 1/2 MS | TIM 0.3 | 8 | ∼350 6 | ∼330 | ∼330 | ∼330 |
| Libraries | Read Length (bases) | Raw Reads (Gb) | Raw Q30 (%) | Clean Reads (Gb) | Clean Q30 (%) | GC Content (%) | |
|---|---|---|---|---|---|---|---|
| In vivo | ~350 | 300 | 135.4 | 89.94 | 120.1 | 89.26 | 37.95 |
| In vitro | ~350 | 300 | 176.6 | 92.97 | 154.3 | 91.81 | 36.74 |
| Region | No. of Contigs | Species | Percentage of Contigs in Stray Region | |
|---|---|---|---|---|
| In vivo | 0−14× depth 0−100% GC content | 1,041,992 | Helianthus maximiliani | 13.23% |
| Fusarium fujikuroi1 | 10.38% | |||
| Ageratina adenophora | 5.60% | |||
| Methylobacterium radiotolerans2 | 4.15% | |||
| Methylobacterium extorquens2 | 3.86% | |||
| 14−24× depth 0−60% GC content | 152,558 | Helianthus maximiliani | 37.22% | |
| Ageratina adenophora | 13.92% | |||
| Helianthus annuus | 3.85% | |||
| Solanum lycopersicum | 3.01% | |||
| Guizotia abyssinica | 2.17% | |||
| 20−35× depth 60−100% GC content | 16,435 | Methylobacterium extorquens2 | 15.17% | |
| Methylobacterium radiotolerans2 | 14.07% | |||
| Helianthus maximiliani | 6.38% | |||
| Methylobacterium nodulans2 | 3.13% | |||
| Sphingomonas sp. 2 | 1.93% | |||
| 40−80× depth 60−100% GC content | 8216 | Helianthus maximiliani | 25.51% | |
| Clavibacter michiganensis2 | 5.34% | |||
| Methylobacterium extorquens2 | 4.10% | |||
| Agathis dammara | 3.17% | |||
| Helianthus annuus | 3.10% | |||
| In vitro | 0−10× depth 57−80% GC content | 7832 | Methylobacterium radiotolerans2 | 36.16% |
| Methylobacterium extorquens2 | 16.32% | |||
| Methylobacterium sp. 2 | 8.96% | |||
| Brevundimonas subvibrioides2 | 5.34% | |||
| Caulobacter sp. 2 | 4.34% |
| Clean Reads (Gb) | GC Content (%) | Assembly (>100 bp) | Total Length (bp) | Total Number | Max Length (bp) | N50 (bp) | N Bases (%) | |
|---|---|---|---|---|---|---|---|---|
| In vivo | 109.6 | 36.94 | Contigs | 934,276,284 | 3,555,786 | 327,948 | 352 | 0 |
| Scaffolds | 1,109,673,608 | 1,443,212 | 4,285,959 | 3087 | 4.33 | |||
| In vitro | 154.2 | 36.72 | Contigs | 882,531,142 | 3,511,473 | 24,842 | 312 | 0 |
| Scaffolds | 1,048,004,173 | 1,526,309 | 60,767 | 2570 | 5.02 |
| Depth | No. of 17-mer | Genome Size (Mb) | Revised Genome Size (Mb) | Heterozygosity (%) | Repetitive Rate (%) | |
|---|---|---|---|---|---|---|
| In vivo | 53 | 97,798,286,149 | 1845.25 | 1816.81 | 0.64 | 74.72 |
| In vitro | 76 | 137,835,582,212 | 1812.83 | 1801.99 | 0.50 | 78.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Y.; Huang, X.; Li, C.; Ruan, X.; Wang, Z.; Su, Y.; Wang, T. Genome Survey Sequencing of In Vivo Mother Plant and In Vitro Plantlets of Mikania cordata. Plants 2020, 9, 1665. https://doi.org/10.3390/plants9121665
Hong Y, Huang X, Li C, Ruan X, Wang Z, Su Y, Wang T. Genome Survey Sequencing of In Vivo Mother Plant and In Vitro Plantlets of Mikania cordata. Plants. 2020; 9(12):1665. https://doi.org/10.3390/plants9121665
Chicago/Turabian StyleHong, Yongfeng, Xia Huang, Chunmei Li, Xiaoxian Ruan, Zhen Wang, Yingjuan Su, and Ting Wang. 2020. "Genome Survey Sequencing of In Vivo Mother Plant and In Vitro Plantlets of Mikania cordata" Plants 9, no. 12: 1665. https://doi.org/10.3390/plants9121665
APA StyleHong, Y., Huang, X., Li, C., Ruan, X., Wang, Z., Su, Y., & Wang, T. (2020). Genome Survey Sequencing of In Vivo Mother Plant and In Vitro Plantlets of Mikania cordata. Plants, 9(12), 1665. https://doi.org/10.3390/plants9121665





