In Silico Identification of MYB and bHLH Families Reveals Candidate Transcription Factors for Secondary Metabolic Pathways in Cannabis sativa L.
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification of Cannabis MYB and bHLH Genes
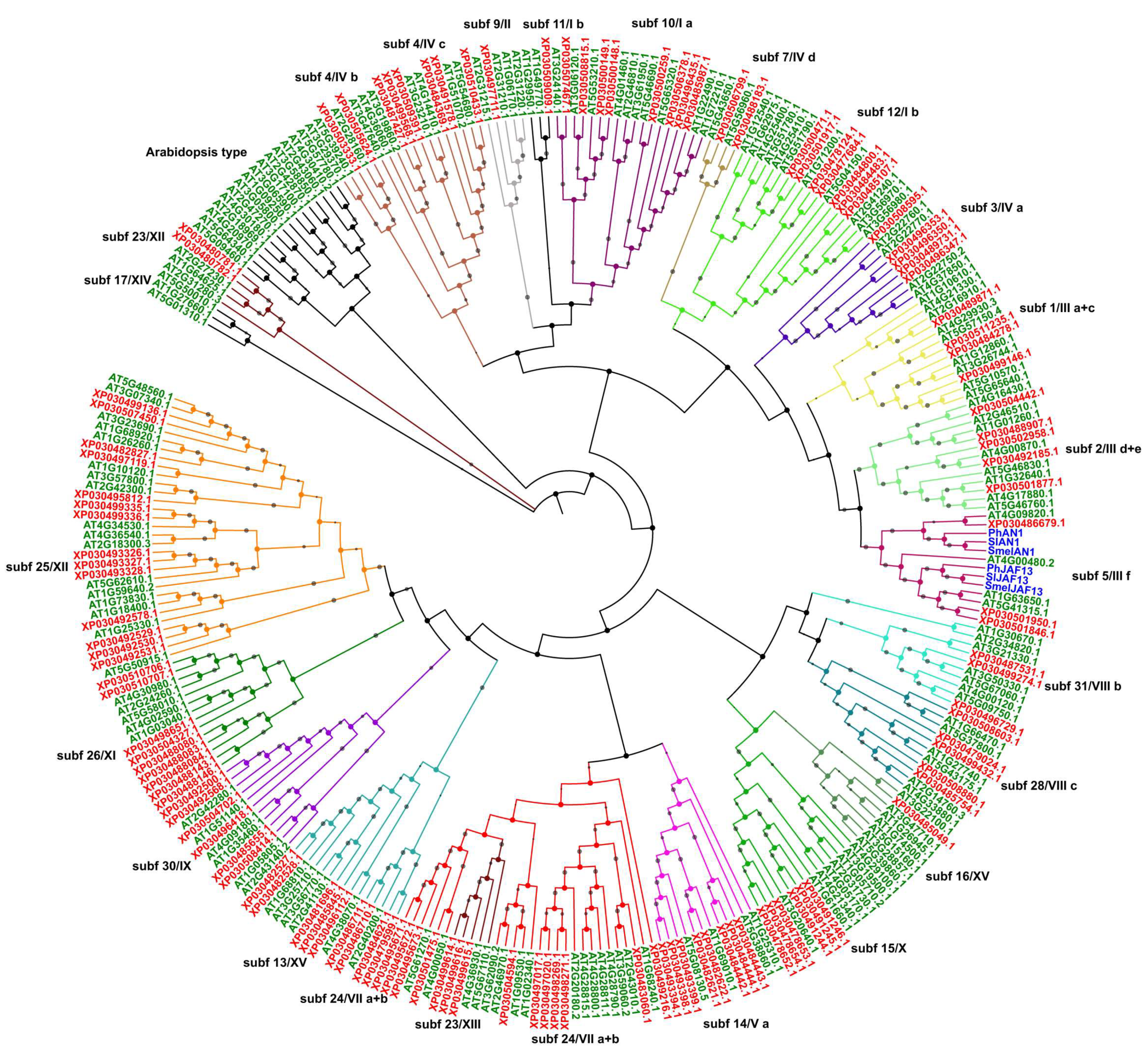
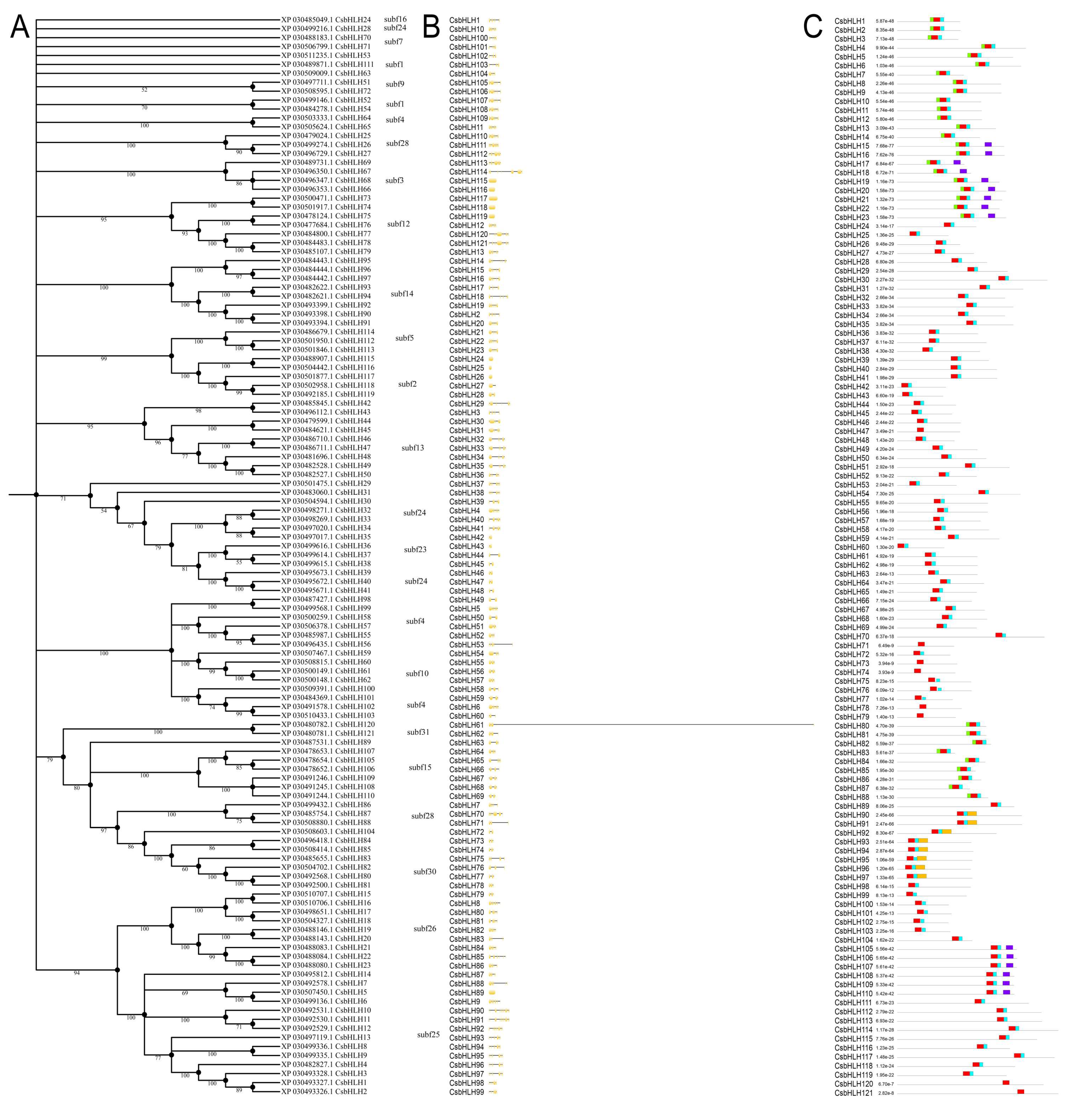
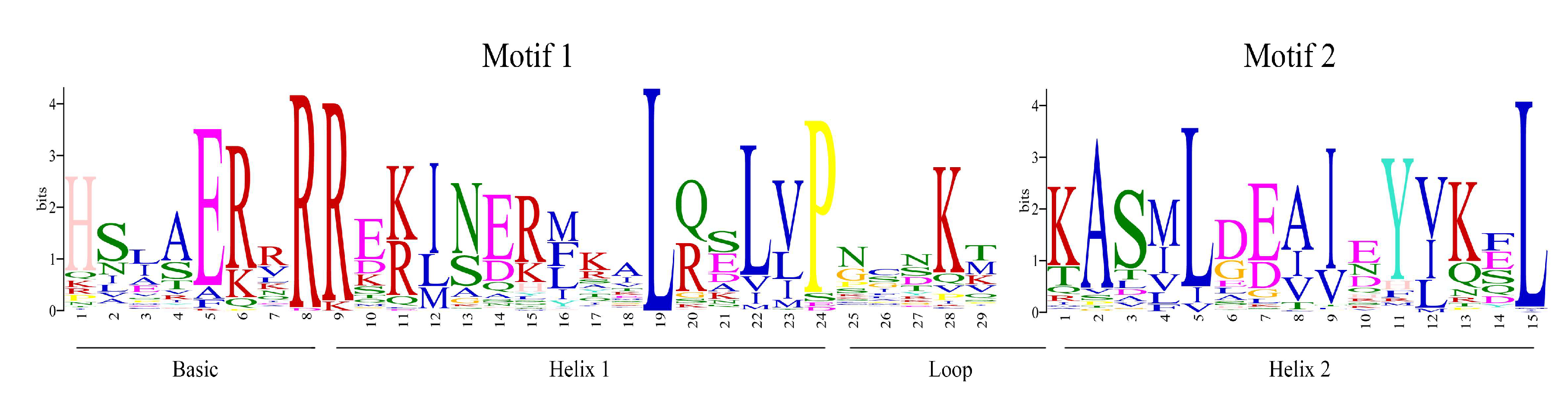
2.2. Phylogenetic Analysis, Gene Structures, and Motif Composition of bHLH Genes in Cannabis
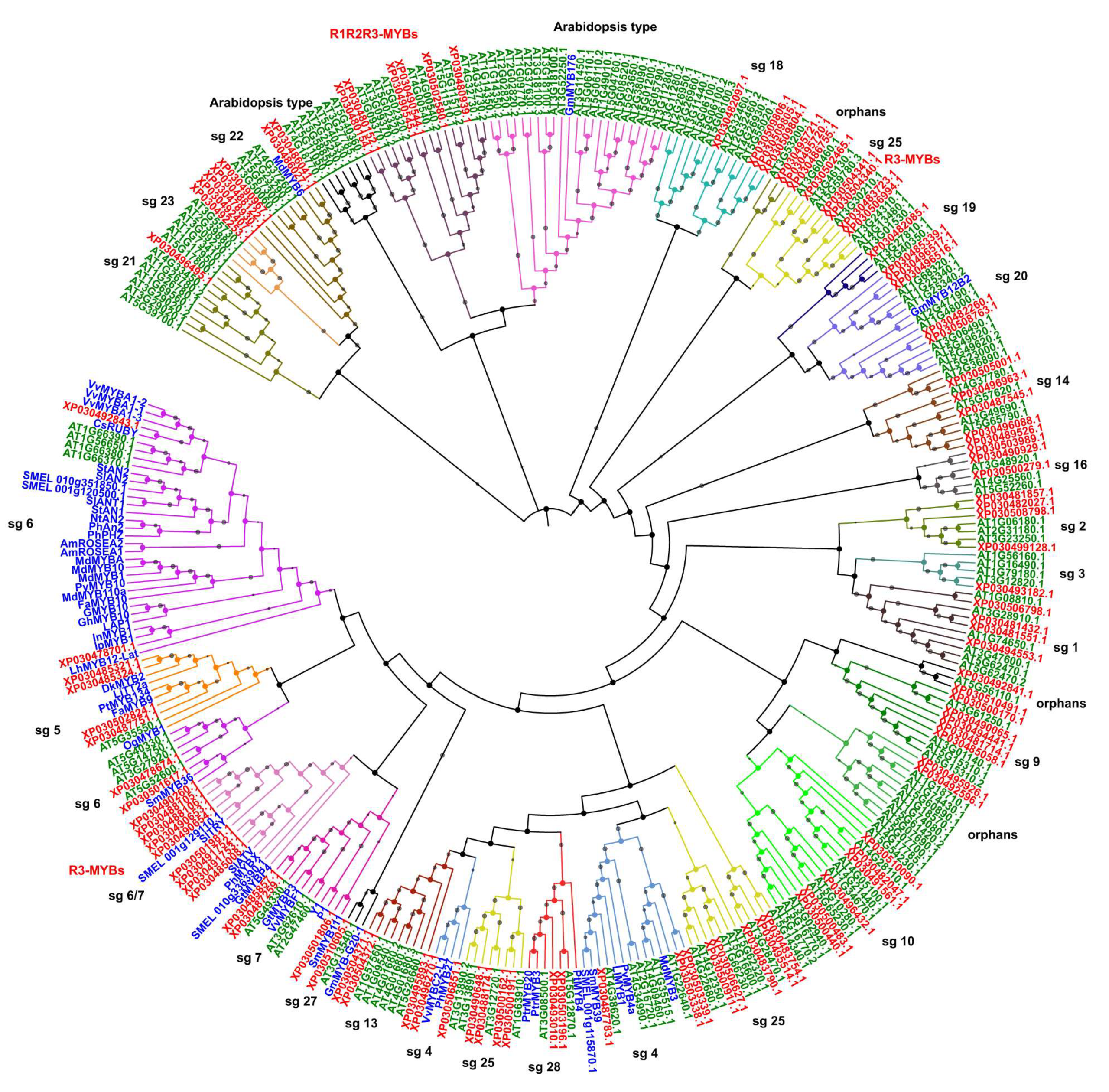
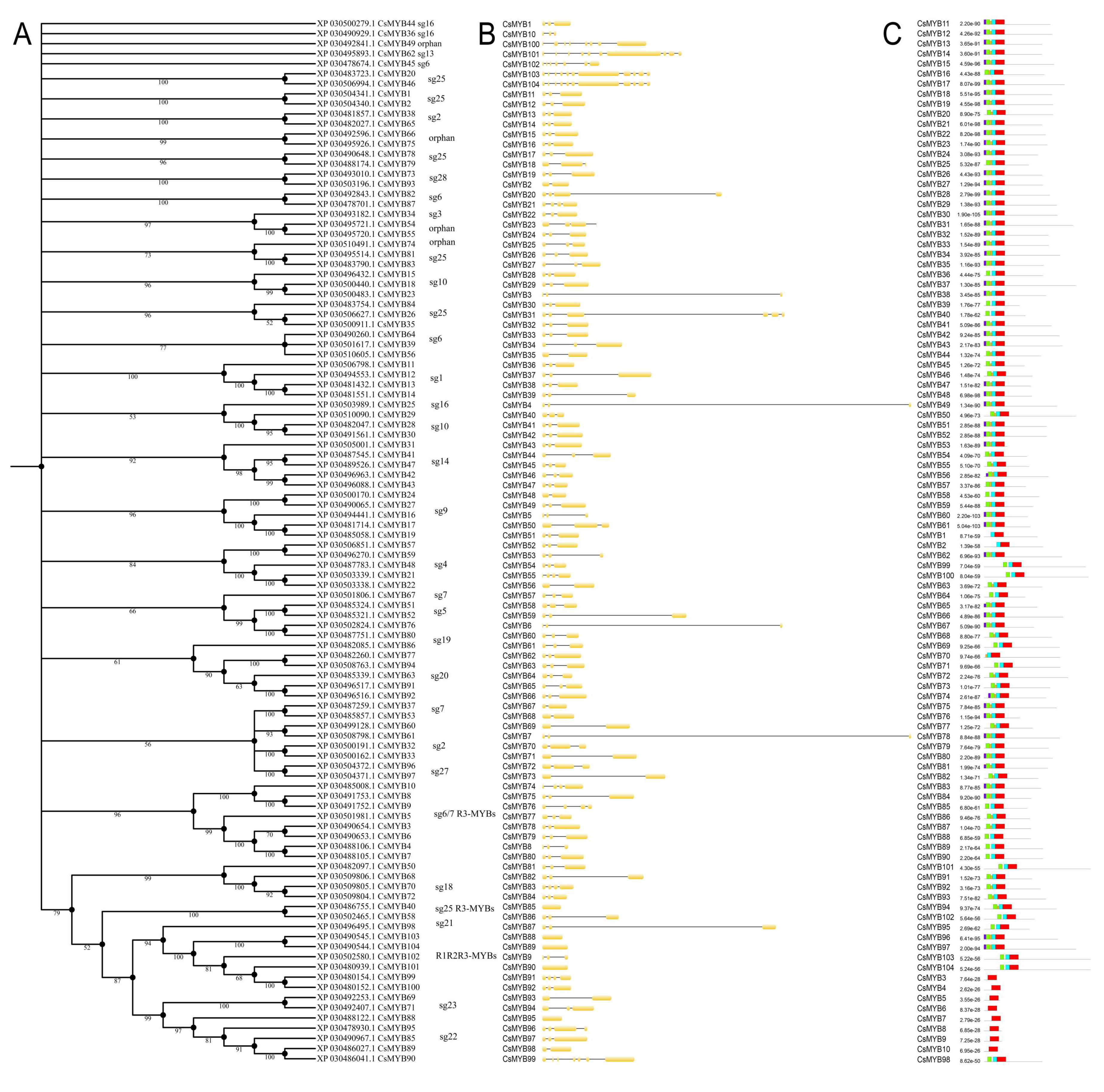
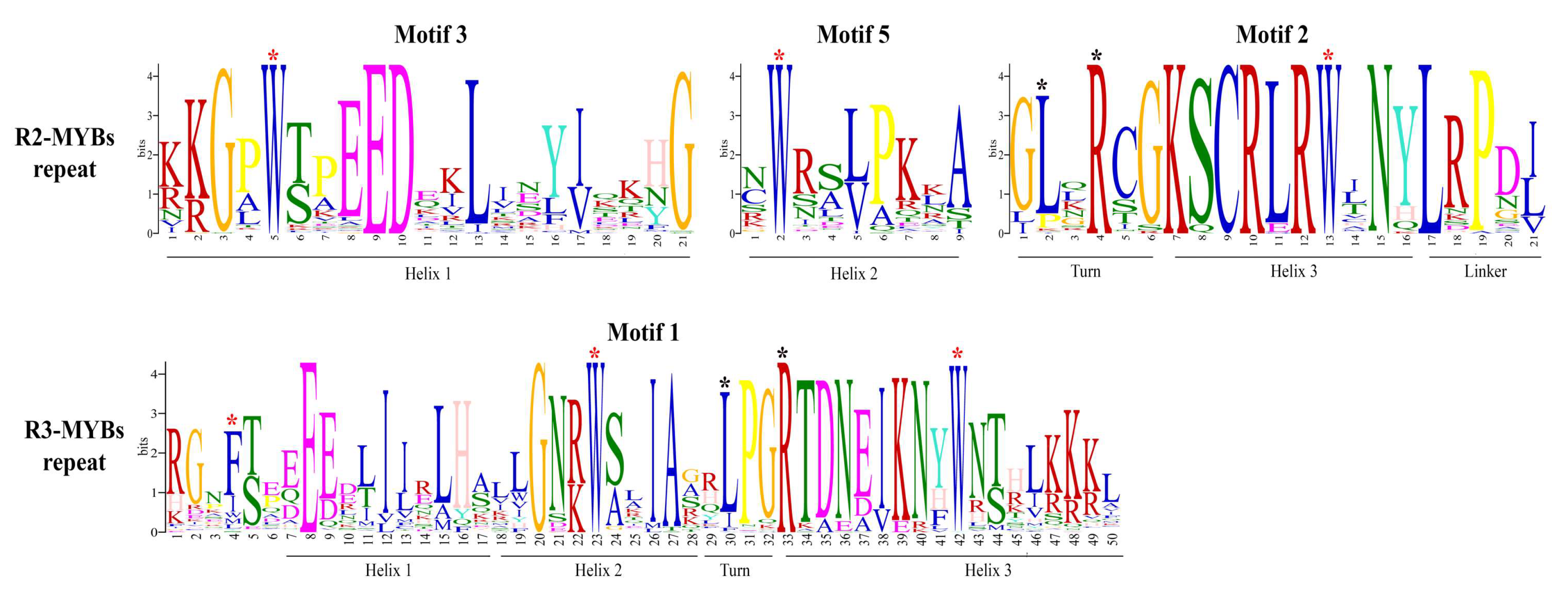
2.3. Phylogenetic Analysis, Gene Structure, and Motif Composition of MYB Genes in Cannabis
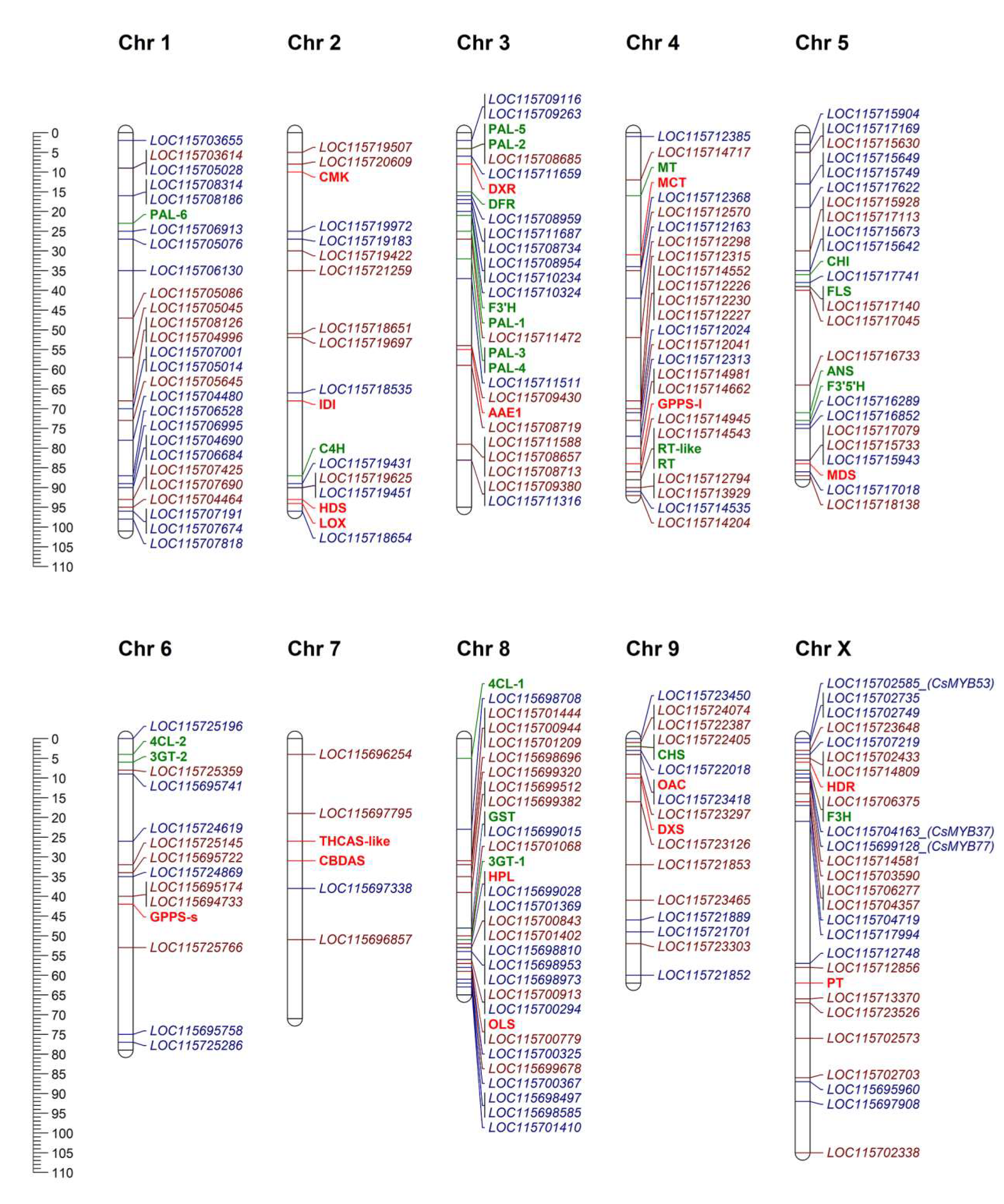
2.4. Chromosomal Distribution of Genomic Loci for CsMYBs, CsbHLHs, and Biosynthetic Enzymes for Flavonoids and Cannabinoids
3. Materials and Methods
3.1. Database Searches and Identification of the MYB and bHLH Genes in Cannabis sativa
3.2. Gene Structure Analysis
3.3. Conserved Motif Identification and Prediction of Subcellular Localization
3.4. Comparative Phylogenetic Analysis
3.5. Identification of Putative Cannabis Flavonoid and Cannabinoid Biosynthetic Genes, Chromosomal Mapping of TFs and Structural Genes on cs10 Assembly
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.C. General Aspects of Plant Transcription Factor Families; Elsevier Inc.: Amsterdam, The Netherlands, 2016; ISBN 9780128011270. [Google Scholar]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; MacHemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Castillon, A.; Shen, H.; Huq, E. Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Bócsa, I.; Karus, M. The Cultivation of Hemp: Botany, Varieties, Cultivation and Harvesting; Hemptech: Auckland, New Zealand, 1998. [Google Scholar]
- Aliferis, K.A.; Bernard-Perron, D. Cannabinomics: Application of Metabolomics in Cannabis (Cannabis sativa L.) Research and Development. Front. Plant Sci. 2020, 11, 554. [Google Scholar] [CrossRef]
- Citti, C.; Linciano, P.; Russo, F.; Luongo, L.; Iannotta, M.; Maione, S.; Laganà, A.; Capriotti, A.L.; Forni, F.; Vandelli, M.A.; et al. A novel phytocannabinoid isolated from Cannabis sativa L. with an in vivo cannabimimetic activity higher than Δ9-tetrahydrocannabinol: Δ9-Tetrahydrocannabiphorol. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Chandra, S.; Lata, H.; ElSohly, M.A. (Eds.) Cannabis Sativa L.—Botany and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–474. [Google Scholar] [CrossRef]
- Andre, C.M.; Hausman, J.F.; Guerriero, G. Cannabis sativa: The plant of the thousand and one molecules. Front. Plant Sci. 2016, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rodziewicz, P.; Loroch, S.; Marczak, Ł.; Sickmann, A.; Kayser, O. Cannabinoid synthases and osmoprotective metabolites accumulate in the exudates of Cannabis sativa L. glandular trichomes. Plant Sci. 2019, 284, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Flores-Sanchez, I.J.; Verpoorte, R. Secondary metabolism in cannabis. Phytochem. Rev. 2008, 7, 615–639. [Google Scholar] [CrossRef]
- Barrett, M.L.; Scutt, A.M.; Evans, F.J. Cannflavin A and B, prenylated flavones from Cannabis sativa L. Experientia 1986, 42, 452–453. [Google Scholar] [CrossRef]
- Rea, K.A.; Casaretto, J.A.; Al-Abdul-Wahid, M.S.; Sukumaran, A.; Geddes-McAlister, J.; Rothstein, S.J.; Akhtar, T.A. Biosynthesis of cannflavins A and B from Cannabis sativa L. Phytochemistry 2019, 164, 162–171. [Google Scholar] [CrossRef]
- Docimo, T.; Consonni, R.; Coraggio, I.; Mattana, M. Early phenylpropanoid biosynthetic steps in Cannabis sativa: Link between genes and metabolites. Int. J. Mol. Sci. 2013, 14, 13626–13644. [Google Scholar] [CrossRef]
- Sakamoto, K.; Akiyama, Y.; Fukui, K.; Kamada, H.; Satoh, S. Characterization; Genome sizes and morphology of sex chromosomes in hemp (Cannabis sativa L.). Cytologia 1998, 63, 459–464. [Google Scholar] [CrossRef]
- Romero, P.; Peris, A.; Vergara, K.; Matus, J.T. Comprehending and improving cannabis specialized metabolism in the systems biology era. Plant Sci. 2020, 298, 110571. [Google Scholar] [CrossRef]
- Allen, K.D.; McKernan, K.; Pauli, C.; Roe, J.; Torres, A.; Gaudino, R. Genomic characterization of the complete terpene synthase gene family from Cannabis sativa. PLoS ONE 2019, 14, e0222363. [Google Scholar] [CrossRef]
- Van Bakel, H.; Stout, J.M.; Cote, A.G.; Tallon, C.M.; Sharpe, A.G.; Hughes, T.R.; Page, J.E. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011, 12. [Google Scholar] [CrossRef]
- Gao, S.; Wang, B.; Xie, S.; Xu, X.; Zhang, J.; Pei, L.; Yu, Y.; Yang, W.; Zhang, Y. A high-quality reference genome of wild Cannabis sativa. Hortic. Res. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.; Orsburn, B. The First Publicly Available Annotated Genome for Cannabis plants. BioRxiv 2019. [Google Scholar] [CrossRef]
- Laverty, K.U.; Stout, J.M.; Sullivan, M.J.; Shah, H.; Gill, N.; Holbrook, L.; Deikus, G.; Sebra, R.; Hughes, T.R.; Page, J.E.; et al. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Res. 2019, 29, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, I.; Pellino, M.; Rigault, P.; van Velzen, R.; Ebersbach, J.; Ashnest, J.R.; Mau, M.; Schranz, M.E.; Alcorn, J.; Laprairie, R.B.; et al. The Genomics of Cannabis and Its Close Relatives. Annu. Rev. Plant Biol. 2020, 71, 713–739. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Dowling, C.A.; Shi, J.; Ryan, L.; Hunt, D.; O’Reilly, E.; Perry, A.S.; Kinnane, O.; McCabe, P.F.; Melzer, R. The Cream of the Crop: Biology, Breeding and Applications of Cannabis sativa. Authorea 2020, 1–69. [Google Scholar] [CrossRef]
- Grassa, C.; Wenger, J.; Dabney, C.; Poplawski, S.; Motley, S.T.; Michael, T.; Schwartz, C.J.; Weiblen, G. A complete Cannabis chromosome assembly and adaptive admixture for elevated cannabidiol (CBD) content. bioRxiv 2018, 458083. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Sun, H.; Fan, H.J.; Ling, H.Q. Genome-wide identification and characterization of the gene family in tomato. BMC Genom. 2015, 16. [Google Scholar] [CrossRef]
- Tian, S.; Li, L.; Wei, M.; Yang, F. Genome-wide analysis of basic helix–loop–helix superfamily members related to anthocyanin biosynthesis in eggplant (Solanum melongena L.). PeerJ 2019, 7, e7768. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, Y.; Kim, S.U.; Chen, Z.; Nie, G.; Cheng, S.; Ye, J.; Xu, F. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Moglia, A.; Florio, F.E.; Iacopino, S.; Guerrieri, A.; Milani, A.M.; Comino, C.; Barchi, L.; Marengo, A.; Cagliero, C.; Rubiolo, P.; et al. Identification of a new R3 MYB type repressor and functional characterization of the members of the MBW transcriptional complex involved in anthocyanin biosynthesis in eggplant (S. melongena L.). PLoS ONE 2020, 15, e0232986, Erratum in: PLoS ONE 2020, 15, e0235081. [Google Scholar] [CrossRef]
- Montefiori, M.; Brendolise, C.; Dare, A.P.; Kui, L.W.; Davies, K.M.; Hellens, R.P.; Allan, A.C. In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 2015, 66, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P.; et al. Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef] [PubMed]
- Spelt, C.; Quattrocchio, F.; Mol, J.N.M.; Koes, R. Anthocyanin1 of Petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 2000, 12, 1619–1631. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; de Marcos, A.; Illescas-Miranda, J.; Mena, M.; Fenoll, C. The Tomato Genome Encodes SPCH, MUTE, and FAMA Candidates That Can Replace the Endogenous Functions of Their Arabidopsis Orthologs. Front. Plant Sci. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A. Selection pressures on stomatal evolution. New Phytol. 2002, 153, 371–386. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, R.; Xu, J.; Yan, J.; Guo, L.; Song, J.; Feng, R.; Yu, M. Genome-wide identification and classification of MYB superfamily genes in peach. PLoS ONE 2018, 13, e0199192. [Google Scholar] [CrossRef]
- Guo, X.J.; Wang, J.R. Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat. BMC Plant Biol. 2017, 17, 1–12. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Saitoh, M.; Hoshino, A.; Nitasaka, E.; Iida, S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 2006, 47, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Li, Y.; Ai, Q.; Yu, D. MYB82 functions in regulation of trichome development in Arabidopsis. J. Exp. Bot. 2014, 65, 3215–3223. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lu, S. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genom. 2014, 15. [Google Scholar] [CrossRef]
- Huang, Y.F.; Vialet, S.; Guiraud, J.L.; Torregrosa, L.; Bertrand, Y.; Cheynier, V.; This, P.; Terrier, N. A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 2014, 201, 795–809. [Google Scholar] [CrossRef]
- Colanero, S.; Perata, P.; Gonzali, S. The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in Eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef]
- Marks, M.D.; Tian, L.; Wenger, J.P.; Omburo, S.N.; Soto-Fuentes, W.; He, J.; Gang, D.R.; Weiblen, G.D.; Dixon, R.A. Identification of candidate genes affecting Δ 9—Tetrahydrocannabinol biosynthesis in Cannabis sativa. J. Exp. Bot. 2009, 60, 3715–3726. [Google Scholar] [CrossRef]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef]
- Chen, P.; Takatsuka, H.; Takahashi, N.; Kurata, R.; Fukao, Y.; Kobayashi, K.; Ito, M.; Umeda, M. Arabidopsis R1R2R3-Myb proteins are essential for inhibiting cell division in response to DNA damage. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Livingston, S.J.; Quilichini, T.D.; Booth, J.K.; Wong, D.C.J.; Rensing, K.H.; Laflamme-Yonkman, J.; Castellarin, S.D.; Bohlmann, J.; Page, J.E.; Samuels, A.L. Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J. 2020, 101, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Peng, R.; Tian, Y.; Han, H.; Xu, J.; Yao, Q. Genome-wide identification and analysis of the MYB transcription factor superfamily in solanum lycopersicum. Plant Cell Physiol. 2016, 57, 1657–1677. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xue, C.; Li, J.; Qiao, X.; Li, L.; Yu, L.; Huang, Y.; Wu, J. Genome-wide identification, evolution and functional divergence of MYB transcription factors in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol. 2016, 57, 824–847. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef]
- Li, W.; Ding, Z.; Ruan, M.; Yu, X.; Peng, M.; Liu, Y. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- The cs10 Genome Assembly. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_900626175.2/ (accessed on 3 August 2020).
- The Pfam Database. Available online: http://pfam.xfam.org/ (accessed on 3 August 2020).
- The ScanProsite Database. Available online: https://prosite.expasy.org/scanprosite/ (accessed on 3 August 2020).
- CDD Database. Available online: https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi (accessed on 3 August 2020).
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- GSDS. Available online: http://gsds.cbi.pku.edu.cn (accessed on 3 August 2020).
- MEME Suite. version 5.1.1. Available online: http://meme-suite.org/tools/meme (accessed on 3 August 2020).
- WebLogo Software. Available online: http://weblogo.berkeley.edu/logo.cgi (accessed on 3 August 2020).
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- WoLF PSORT Software. Available online: https://wolfpsort.hgc.jp/ (accessed on 3 August 2020).
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- The TAIR Database. Available online: https://www.arabidopsis.org (accessed on 3 August 2020).
- Matus, J.T.; Aquea, F.; Arce-Johnson, P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008, 8, 1–15. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.F.; van der Woude, K.; Mol, J.N.M.; Koes, R. Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998, 13, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- The iTOL Platform. Available online: https://itol.embl.de/ (accessed on 3 October 2020).
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 2–5. [Google Scholar] [CrossRef] [PubMed]
- The BLASTP Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins (accessed on 3 August 2020).
- MapChart Software. Available online: http://www.biometris.wur.nl/UK/Software/MapChart/download (accessed on 3 August 2020).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bassolino, L.; Buti, M.; Fulvio, F.; Pennesi, A.; Mandolino, G.; Milc, J.; Francia, E.; Paris, R. In Silico Identification of MYB and bHLH Families Reveals Candidate Transcription Factors for Secondary Metabolic Pathways in Cannabis sativa L. Plants 2020, 9, 1540. https://doi.org/10.3390/plants9111540
Bassolino L, Buti M, Fulvio F, Pennesi A, Mandolino G, Milc J, Francia E, Paris R. In Silico Identification of MYB and bHLH Families Reveals Candidate Transcription Factors for Secondary Metabolic Pathways in Cannabis sativa L. Plants. 2020; 9(11):1540. https://doi.org/10.3390/plants9111540
Chicago/Turabian StyleBassolino, Laura, Matteo Buti, Flavia Fulvio, Alessandro Pennesi, Giuseppe Mandolino, Justyna Milc, Enrico Francia, and Roberta Paris. 2020. "In Silico Identification of MYB and bHLH Families Reveals Candidate Transcription Factors for Secondary Metabolic Pathways in Cannabis sativa L." Plants 9, no. 11: 1540. https://doi.org/10.3390/plants9111540
APA StyleBassolino, L., Buti, M., Fulvio, F., Pennesi, A., Mandolino, G., Milc, J., Francia, E., & Paris, R. (2020). In Silico Identification of MYB and bHLH Families Reveals Candidate Transcription Factors for Secondary Metabolic Pathways in Cannabis sativa L. Plants, 9(11), 1540. https://doi.org/10.3390/plants9111540











