Development of 454 New Kompetitive Allele-Specific PCR (KASP) Markers for Temperate japonica Rice Varieties
Abstract
1. Introduction
2. Results
2.1. Prediction of Effects of SNPs on Gene Function
2.2. Development of KASP Markers
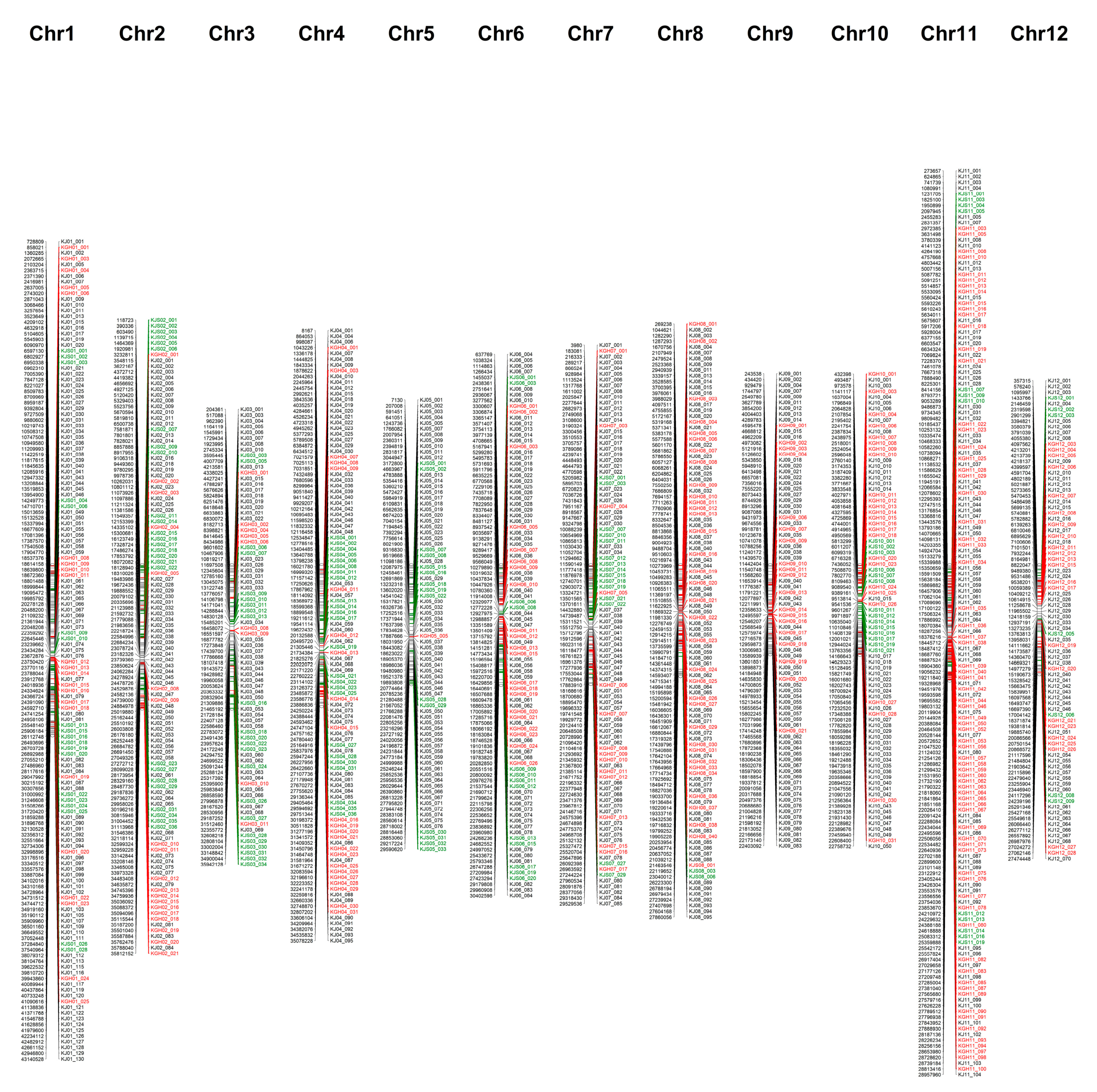
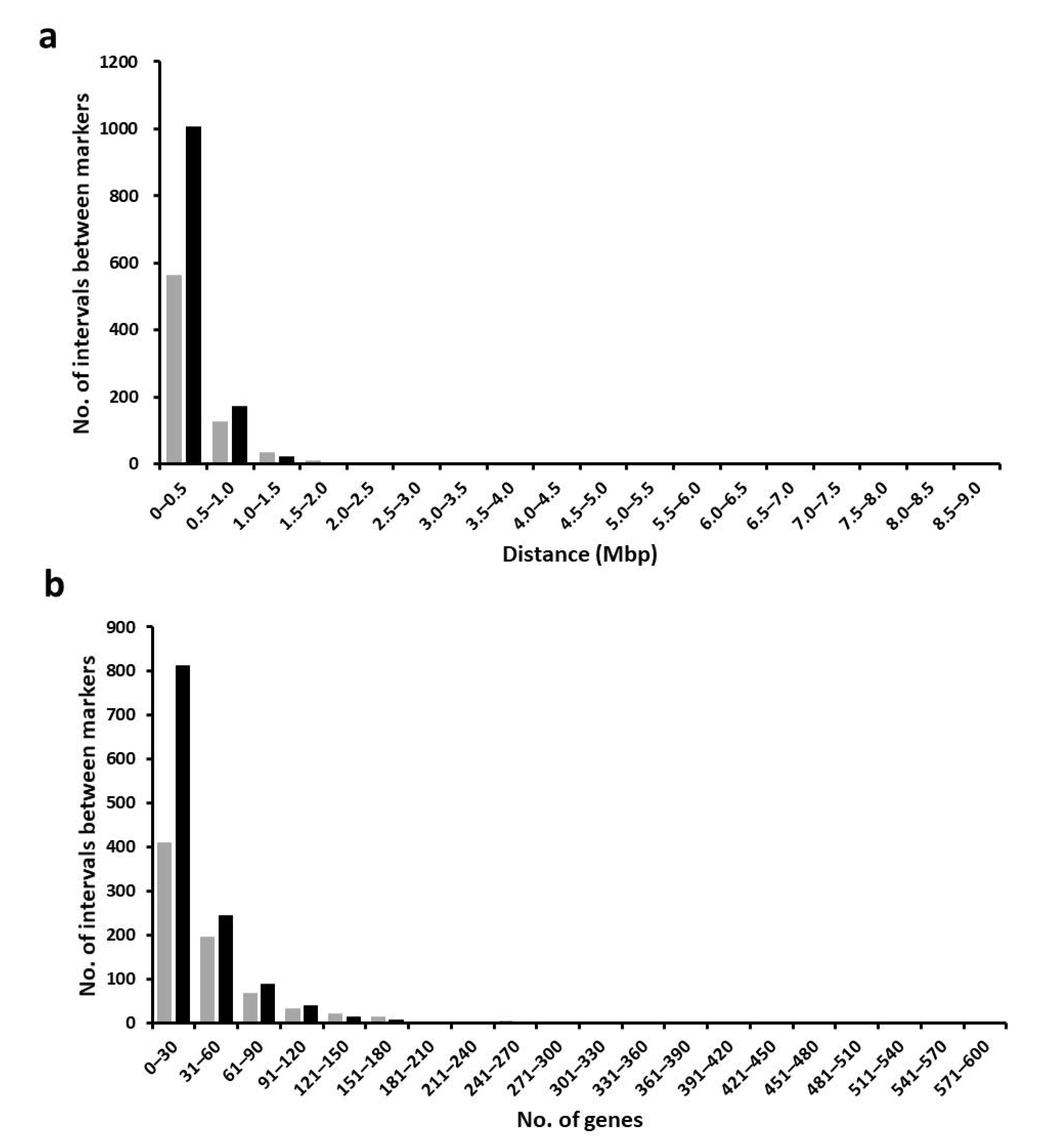
2.3. Integration of 1225 KASP Markers
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Genomic DNA Extraction
4.2. SNP Impact Prediction and Selection of SNPs for KASP Marker Design
4.3. KASP Marker Assay
4.4. Construction of a Physical Map and Phylogenetic Tree
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Garris, A.J.; Tai, T.H.; Coburn, J.; Kresovich, S.; McCouch, S. Genetic structure and diversity in Oryza sativa L. Genetics 2005, 169, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Colowit, P.M.; Mackill, D.J. Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci. 2002, 42, 601–607. [Google Scholar] [CrossRef]
- Gao, L.; Innan, H. Nonindependent domestication of the two rice subspecies, Oryza sativa ssp. indica and ssp. japonica, demonstrated by multilocus microsatellites. Genetics 2008, 179, 965–976. [Google Scholar] [CrossRef]
- Zhang, Q.; Saghai Maroof, M.A.; LIl, T.Y.; Shen, B.Z. Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor. Appl. Genet. 1992, 83, 495–499. [Google Scholar] [CrossRef]
- Hori, K.; Yamamoto, T.; Yano, M. Genetic dissection of agronomically important traits in closely related temperate japonica rice cultivars. Breed. Sci. 2017, 67, 427–434. [Google Scholar] [CrossRef]
- Mo, Y.; Jeong, J.-M.; Kim, B.-K.; Kwon, S.-W.; Jeung, J.-U. Utilization of elite Korean japonica rice varieties for association mapping of heading time, culm length, and amylose and protein content. Korean J. Crop Sci. 2020, 65, 1–21. [Google Scholar] [CrossRef]
- McCouch, S.R.; Zhao, K.; Wright, M.; Tung, C.-W.; Ebana, K.; Thomson, M.; Reynolds, A.; Wang, D.; DeClerck, G.; Ali, M.L.; et al. Development of genome-wide SNP assays for rice. Breed. Sci. 2010, 60, 524–535. [Google Scholar] [CrossRef]
- Jeong, I.-S.; Yoon, U.-H.; Lee, G.-S.; Ji, H.-S.; Lee, H.-J.; Han, C.-D.; Hahn, J.-H.; An, G.; Kim, T.-H. SNP-based analysis of genetic diversity in anther-derived rice by whole genome sequencing. Rice 2013, 6, 6. [Google Scholar] [CrossRef]
- Thomson, M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Jeong, I.-S.; Kim, T.-H.; Lee, S.-B.; Suh, S.-C.; Ji, H. Genome-wide detection of DNA polymorphisms between two Korean japonica rice varieties. Plant Breed. Biotechnol. 2015, 3, 208–215. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nagasaki, H.; Yonemaru, J.-I.; Ebana, K.; Nakajima, M.T.; Shibaya, T.; Yano, M. Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genom. 2010, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, H.; Ebana, K.; Shibaya, T.; Yonemaru, J.-I.; Yano, M. Core single-nucleotide polymorphisms—A tool for genetic analysis of the Japanese rice population. Breed. Sci. 2010, 60, 648–655. [Google Scholar] [CrossRef][Green Version]
- Thomson, M.J.; Singh, N.; Dwiyanti, M.S.; Wang, D.R.; Wright, M.H.; Perez, F.A.; DeClerck, G.; Chin, J.H.; Malitic-Layaoen, G.A.; Juanillas, V.M.; et al. Large-scale development of a rice 6 K SNP array for genetics and breeding applications. Rice 2017, 10, 40. [Google Scholar] [CrossRef]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and persepectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using kompetitive allele specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Steele, K.A.; Quinton-Tulloch, M.J.; Amgai, R.B.; Dhakal, R.; Khatiwada, S.P.; Vyas, D.; Heine, M.; Witcombe, J.R. Accelerating public sector rice breeding with high-density KASP markers derived from whole genome sequencing of indica rice. Mol. Breed. 2018, 38, 38. [Google Scholar] [CrossRef]
- Cheon, K.-S.; Baek, J.; Cho, Y.-I.; Jeong, Y.-M.; Lee, Y.-Y.; Oh, J.; Won, Y.J.; Kang, D.-Y.; Oh, H.; Kim, S.L.; et al. Single nucleotide polymorphism (SNP) discovery and kompetitive allele-specific PCR (KASP) marker development with Korean japonica rice varieties. Plant Breed. Biotechnol. 2018, 6, 391–403. [Google Scholar] [CrossRef]
- Cheon, K.-S.; Jeong, Y.-M.; Lee, Y.-Y.; Oh, J.; Kang, D.-Y.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; Baek, J.; et al. Kompetitive allele-specific PCR marker development and quantitative trait locus mapping for bakanae disease resistance in Korean japonica rice varieties. Plant Breed. Biotechnol. 2019, 7, 208–219. [Google Scholar] [CrossRef]
- Kang, D.-Y.; Cheon, K.-S.; Oh, J.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; Choi, I.; Baek, J.; Kim, K.-H.; et al. Rice genome resequencing reveals a major quantitative trait locus for resistance to bakanae disease caused by Fusarium fujikuroi. Int. J. Mol. Sci. 2019, 20, 2598. [Google Scholar] [CrossRef]
- Cheon, K.-S.; Won, Y.J.; Jeong, Y.-M.; Lee, Y.-Y.; Kang, D.-Y.; Oh, J.; Oh, H.; Kim, S.L.; Kim, N.; Lee, E.; et al. QTL mapping for pre-harvest sprouting resistance in japonica rice varieties utilizing genome re-sequencing. Mol. Genet. Genom. 2020, 295, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-W.; Shin, D.; Cho, J.-H.; Lee, J.-Y.; Kwon, Y.; Park, D.; Ko, J.-M.; Lee, J.-H. Accelerated development of rice stripe virusresistant, near-isogenic rice lines through marker-assisted backcrossing. PLoS ONE 2019, 14, e0225974. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Lee, K.-M.; Kim, K.-Y.; Kim, J.-J.; Shin, W.-C.; Baek, M.-K.; Kim, C.-S.; Park, S.-G.; Lee, C.-M.; Suh, J.-P.; et al. Development of near-Isogenic line of japonica rice cultivar Saenuri without lipoxygenase-3. Korean J. Breed. Sci. 2019, 51, 190–200. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Ruden, D.M.; Lu, X. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Saxena, R.K.; Penmetsa, R.V.; Upadhyaya, H.D.; Kumar, A.; Carrasquilla-Garcia, N.; Schlueter, J.A.; Farmer, A.; Whaley, A.M.; Sarma, B.K.; May, G.D.; et al. Large-scale development of cost-effective single-nucleotide polymorphism marker assays for genetic mapping in pigeonpea and comparative mapping in legumes. DNA Res. 2012, 19, 449–461. [Google Scholar] [CrossRef]
- Hiremath, P.J.; Kumar, A.; Penmetsa, R.V.; Farmer, A.; Schlueter, J.A.; Chamarthi, S.K.; Whaley, A.M.; Carrasquilla-Garcia, N.; Gaur, P.M.; Upadhyaya, H.D.; et al. Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 2012, 10, 716–732. [Google Scholar] [CrossRef]
- Pariasca-Tanaka, J.; Lorieux, M.; He, C.; McCouch, S.; Thomson, M.J.; Wissuwa, M. Development of a SNP genotyping panel for detecting polymorphisms in Oryza glaberrima/O. sativa interspecific crosses. Euphytica 2015, 201, 67–78. [Google Scholar] [CrossRef]
- Yang, G.; Chen, S.; Chen, L.; Sun, K.; Huang, C.; Zhou, D.; Huang, Y.; Wang, J.; Liu, Y.; Wang, H.; et al. Development of a core SNP arrays based on the KASP method for molecular breeding of rice. Rice 2019, 12, 21. [Google Scholar] [CrossRef]
- Zhao, W.; Chung, J.-W.; Ma, K.-H.; Kim, T.-S.; Kim, S.-M.; Shin, D.-I.; Kim, C.-H.; Koo, H.-M.; Park, Y.-J. Analysis of genetic diversity and population structure of rice cultivars from Korea, China and Japan using SSR markers. Gene Genom. 2009, 31, 283–292. [Google Scholar] [CrossRef]
- Song, M.-T.; Lee, J.-H.; Cho, Y.-S.; Jeon, Y.-H.; Lee, S.-B.; Ku, J.-H.; Choi, S.-H.; Hwang, H.-G. Narrow genetic background of Korean rice germplasm as revealed by DNA fingerprinting with SSR markers and their pedigree information. Korean J. Genet. 2002, 24, 397–403. [Google Scholar]
- Kwon, Y.-S.; Manigbas, N.L.; Kim, D.H.; Yi, G. Phylogenic analysis of 246 Korean rice varieties using core sets of microsatellite markers. Philipp. J. Crop Sci. 2017, 42, 27–40. [Google Scholar]
- Project, International Rice Genome Sequencing. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with windows interface. Mol. Biol. Evol. 2010, 27, 747–752. [Google Scholar] [CrossRef]


| Impact of SNP Effects a | Sum b | ||||
|---|---|---|---|---|---|
| High | Moderate | Modifier | Low | ||
| 1 | 54 (7.7%) | 1839 (9.1%) | 41,820 (6.0%) | 1805 (9.1%) | 45,518 (6.1%) |
| 2 | 43 (6.1%) | 981 (4.9%) | 19,906 (2.8%) | 982 (5.0%) | 21,912 (3.0%) |
| 3 | 18 (2.6%) | 450 (2.2%) | 19,924 (2.8%) | 460 (2.3%) | 20,852 (2.8%) |
| 4 | 45 (6.4%) | 1566 (7.8%) | 37,565 (5.4%) | 1498 (7.6%) | 40,674 (5.5%) |
| 5 | 11 (1.6%) | 438 (2.2%) | 15,094 (2.2%) | 474 (2.4%) | 16,017 (2.2%) |
| 6 | 53 (7.5%) | 1570 (7.8%) | 87,427 (12.5%) | 1465 (7.4%) | 90,515 (12.2%) |
| 7 | 32 (4.6%) | 1118 (5.5%) | 36,872 (5.3%) | 1181 (6.0%) | 39,203 (5.3%) |
| 8 | 96 (13.7%) | 2209 (10.9%) | 101,463 (14.5%) | 2252 (11.4%) | 106,020 (14.3%) |
| 9 | 41 (5.8%) | 1009 (5.0%) | 36,503 (5.2%) | 1033 (5.2%) | 38,586 (5.2%) |
| 10 | 65 (9.2%) | 1781 (8.8%) | 68,970 (9.9%) | 1624 (8.2%) | 72,440 (9.8%) |
| 11 | 186 (26.5%) | 5459 (27.1%) | 152,754 (21.8%) | 5158 (26.0%) | 163,557 (22.1%) |
| 12 | 59 (8.4%) | 1759 (8.7%) | 81,568 (11.7%) | 1886 (9.5%) | 85,272 (11.5%) |
| Sum c | 703 (0.1%) | 20,179 (2.7%) | 699,866 (94.5%) | 19,818 (2.7%) | 740,566 (100%) |
| Name | Type | No. | % |
|---|---|---|---|
| KGH | Assay | 357 | |
| Polymorphism | 283 | 79.3 | |
| Monomorphism | 73 | 20.4 | |
| No amplification | 1 | 0.3 | |
| KJS | Assay | 284 | |
| Polymorphism | 171 | 60.2 | |
| Monomorphism | 104 | 36.6 | |
| No amplification | 9 | 3.2 | |
| Total | Assay | 641 | |
| Polymorphism | 454 | 70.8 | |
| Monomorphism | 177 | 27.6 | |
| No amplification | 10 | 1.6 |
| Impact of SNP Effect | SNP Effect | Marker Set | |
|---|---|---|---|
| 454 KASPs | 1225 KASPs | ||
| HIGH | Frameshift | 7 | 7 |
| Splice_acceptor | 25 | 25 | |
| Splice_donor | 29 | 29 | |
| Start_lost | 12 | 12 | |
| Stop_gained | 147 | 147 | |
| Stop_lost | 63 | 64 | |
| Sum | 283 | 284 | |
| MODERATE | Missense | 7 | 34 |
| MODIFIER | 5_prime_UTR | 1 | 9 |
| 3_prime_UTR | 4 | 25 | |
| Downstream_gene | 4 | 31 | |
| Upstream_gene | 131 | 708 | |
| Intergenic_region | 16 | 97 | |
| Intron_variant | 1 | 12 | |
| Non_coding_transcript_exon | 2 | 4 | |
| Sum | 159 | 886 | |
| LOW | 5_prime_UTR_premature_start_codon_gain a | 0 | 3 |
| Synonymous | 4 | 16 | |
| Splice_region | 1 | 2 | |
| Sum | 5 | 21 | |
| Name | Assay (n) | Polymorphism (n, %) | Monomorphism (n, %) | No Amplification (n, %) | Reference | |
|---|---|---|---|---|---|---|
| 1st | KJ | 506 | 400 (79.1) | 89 (17.6) | 17 (3.4) | [18] |
| 2nd | KJ | 504 | 371 (73.6) | 126 (25.0) | 7 (1.4) | [19] |
| 3rd | KGH, KJS | 641 | 454 (70.8) | 177 (27.6) | 10 (1.6) | this study |
| Total | 1651 | 1225 (74.2) | 392 (23.7) | 35 (2.1) | ||
| DA | HA | HY | IP | JU | JN | GH | NP | OD | SG | SN | UB | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DA | - | |||||||||||
| HA | 523 (42.7) | - | ||||||||||
| HY | 483 (39.4) | 544 (44.4) | - | |||||||||
| IP | 579 (47.3) | 526 (42.9) | 600 (49.0) | - | ||||||||
| JU | 546 (44.6) | 487 (39.8) | 688 (56.2) | 440 (35.9) | - | |||||||
| JN | 561 (45.8) | 527 (43.0) | 345 (28.2) | 466 (38.0) | 613 (50.0) | - | ||||||
| GH | 554 (45.2) | 537 (43.8) | 521 (42.5) | 388 (31.7) | 444 (36.2) | 529 (43.2) | - | |||||
| NP | 382 (31.2) | 587 (47.9) | 543 (44.3) | 662 (54.0) | 509 (41.6) | 694 (56.7) | 653 (53.3) | - | ||||
| OD | 557 (45.5) | 505 (41.2) | 661 (54.0) | 400 (32.7) | 410 (33.5) | 575 (46.9) | 530 (43.3) | 659 (53.8) | - | |||
| SG | 454 (37.1) | 577 (47.1) | 310 (25.3) | 609 (49.7) | 600 (49.0) | 449 (36.7) | 544 (44.4) | 528 (43.1) | 598 (48.8) | - | ||
| SN | 437 (35.7) | 578 (47.2) | 428 (34.9) | 590 (48.2) | 625 (51.0) | 555 (45.3) | 582 (47.5) | 441 (36.0) | 574 (46.9) | 510 (41.6) | - | |
| UB a | 316 (38.3) | 321 (38.9) | 265 (32.1) | 376 (45.5) | 370 (44.8) | 289 (35.0) | 340 (41.2) | 378 (45.8) | 324 (39.2) | 273 (33.1) | 322 (39.0) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheon, K.-S.; Jeong, Y.-M.; Oh, H.; Oh, J.; Kang, D.-Y.; Kim, N.; Lee, E.; Baek, J.; Kim, S.L.; Choi, I.; et al. Development of 454 New Kompetitive Allele-Specific PCR (KASP) Markers for Temperate japonica Rice Varieties. Plants 2020, 9, 1531. https://doi.org/10.3390/plants9111531
Cheon K-S, Jeong Y-M, Oh H, Oh J, Kang D-Y, Kim N, Lee E, Baek J, Kim SL, Choi I, et al. Development of 454 New Kompetitive Allele-Specific PCR (KASP) Markers for Temperate japonica Rice Varieties. Plants. 2020; 9(11):1531. https://doi.org/10.3390/plants9111531
Chicago/Turabian StyleCheon, Kyeong-Seong, Young-Min Jeong, Hyoja Oh, Jun Oh, Do-Yu Kang, Nyunhee Kim, Eungyeong Lee, Jeongho Baek, Song Lim Kim, Inchan Choi, and et al. 2020. "Development of 454 New Kompetitive Allele-Specific PCR (KASP) Markers for Temperate japonica Rice Varieties" Plants 9, no. 11: 1531. https://doi.org/10.3390/plants9111531
APA StyleCheon, K.-S., Jeong, Y.-M., Oh, H., Oh, J., Kang, D.-Y., Kim, N., Lee, E., Baek, J., Kim, S. L., Choi, I., Yoon, I. S., Kim, K.-H., Won, Y. J., Cho, Y.-i., Han, J.-H., & Ji, H. (2020). Development of 454 New Kompetitive Allele-Specific PCR (KASP) Markers for Temperate japonica Rice Varieties. Plants, 9(11), 1531. https://doi.org/10.3390/plants9111531






