Metabolite Profiling and Network Analysis Reveal Coordinated Changes in Low-N Tolerant and Low-N Sensitive Maize Genotypes under Nitrogen Deficiency and Restoration Conditions
Abstract
1. Introduction
2. Results
2.1. Growth and N Status of Maize Genotypes under N Deficiency
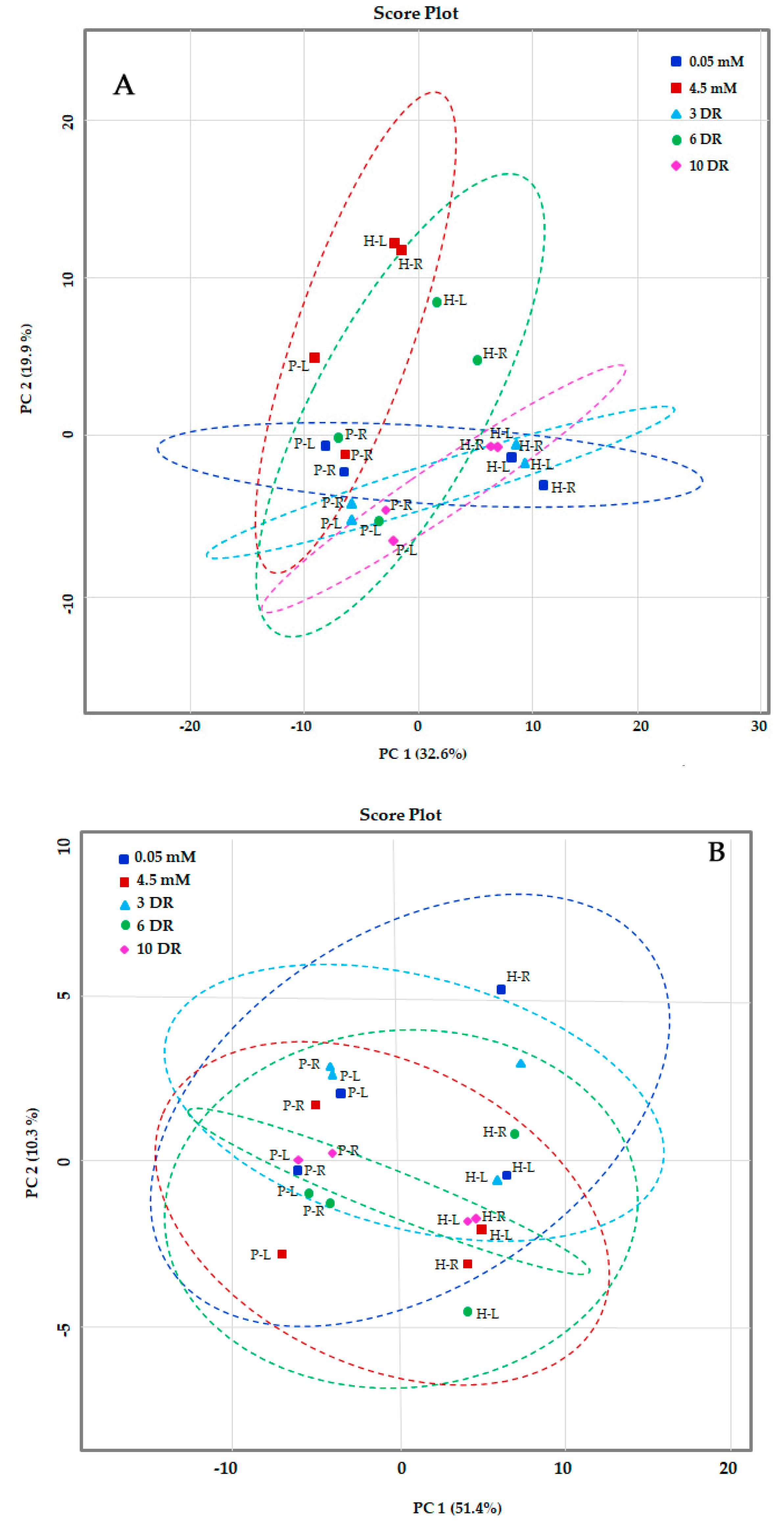
2.2. Principal Component Analysis (PCA) of Metabolites of Maize Genotypes
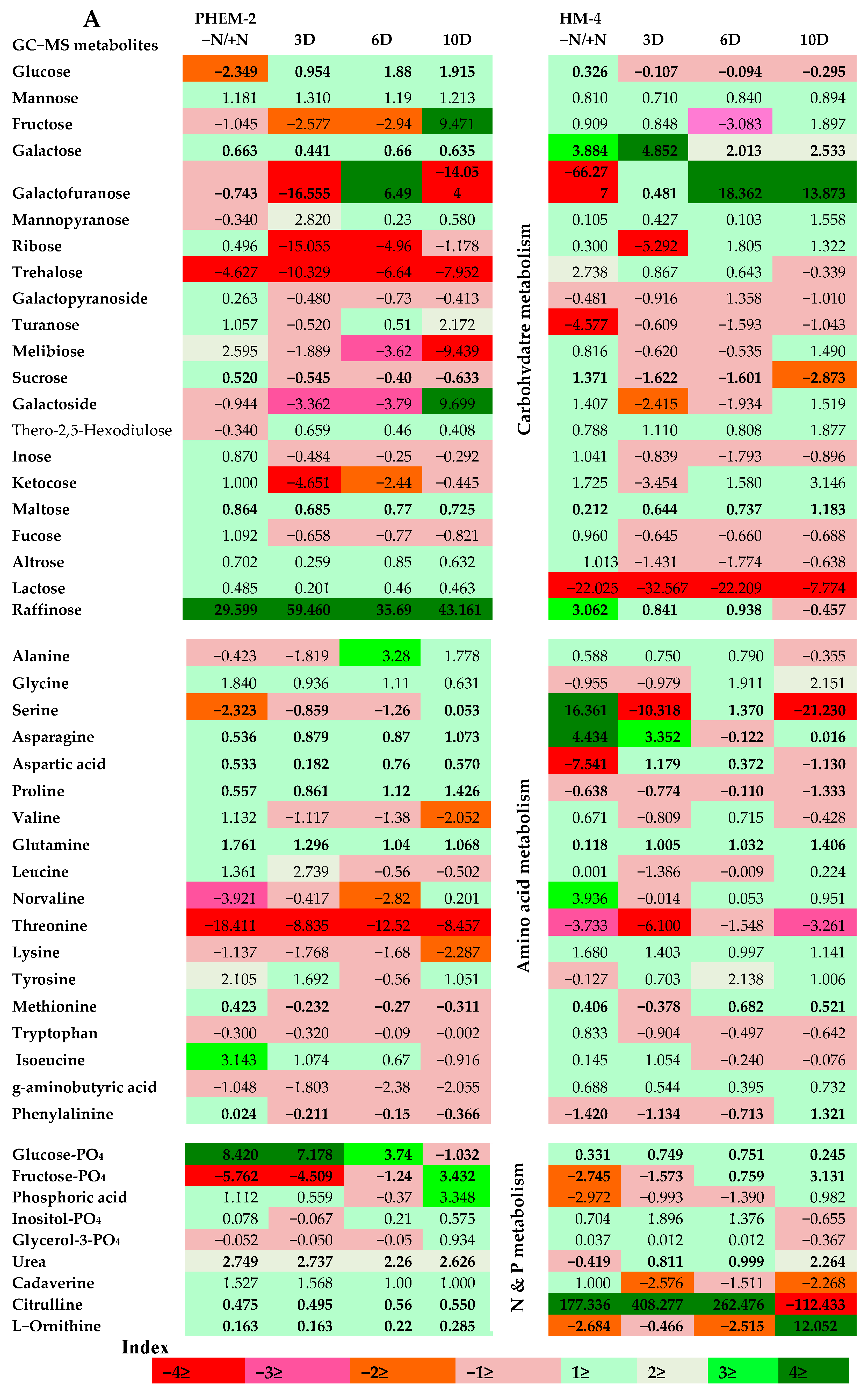
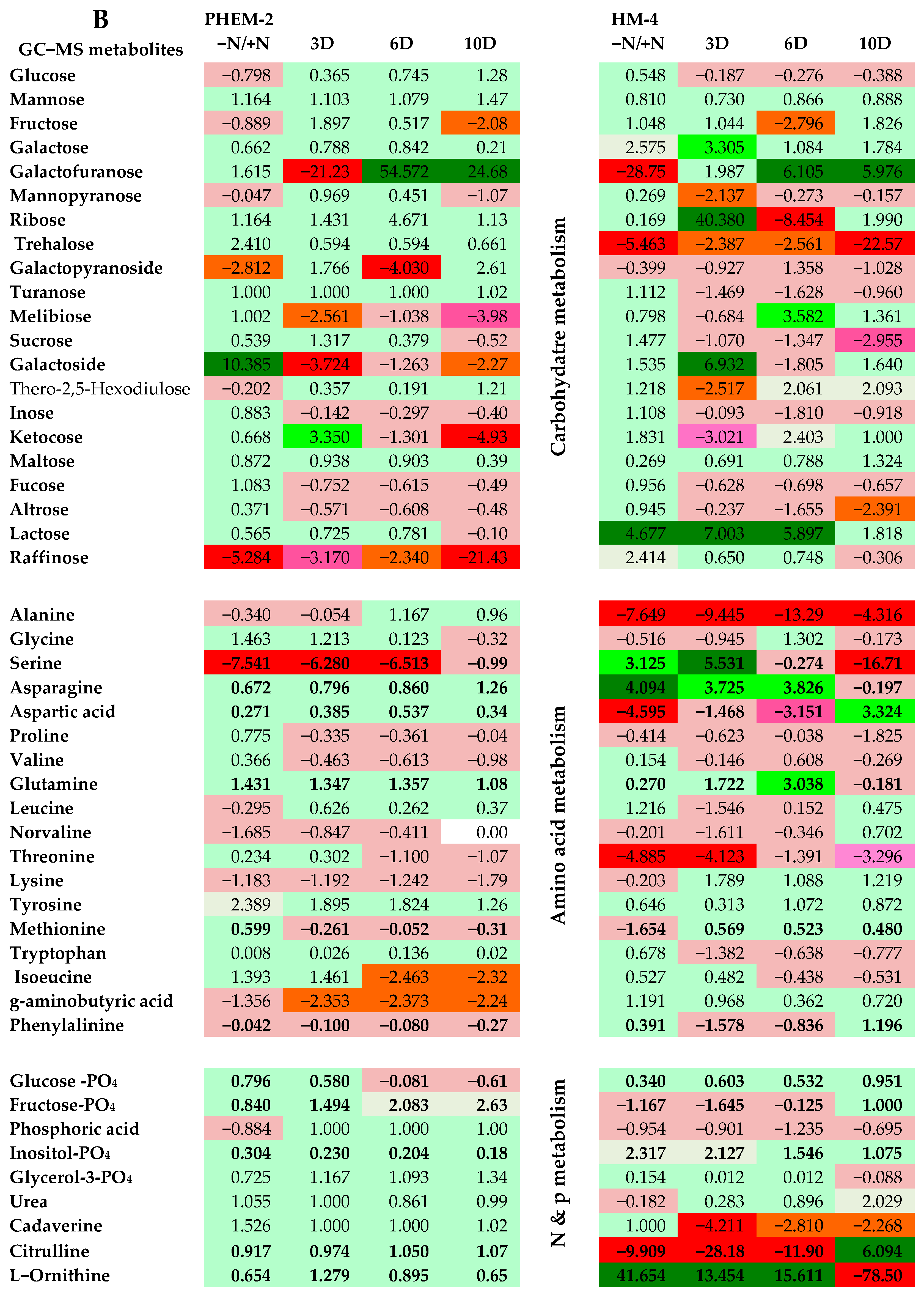
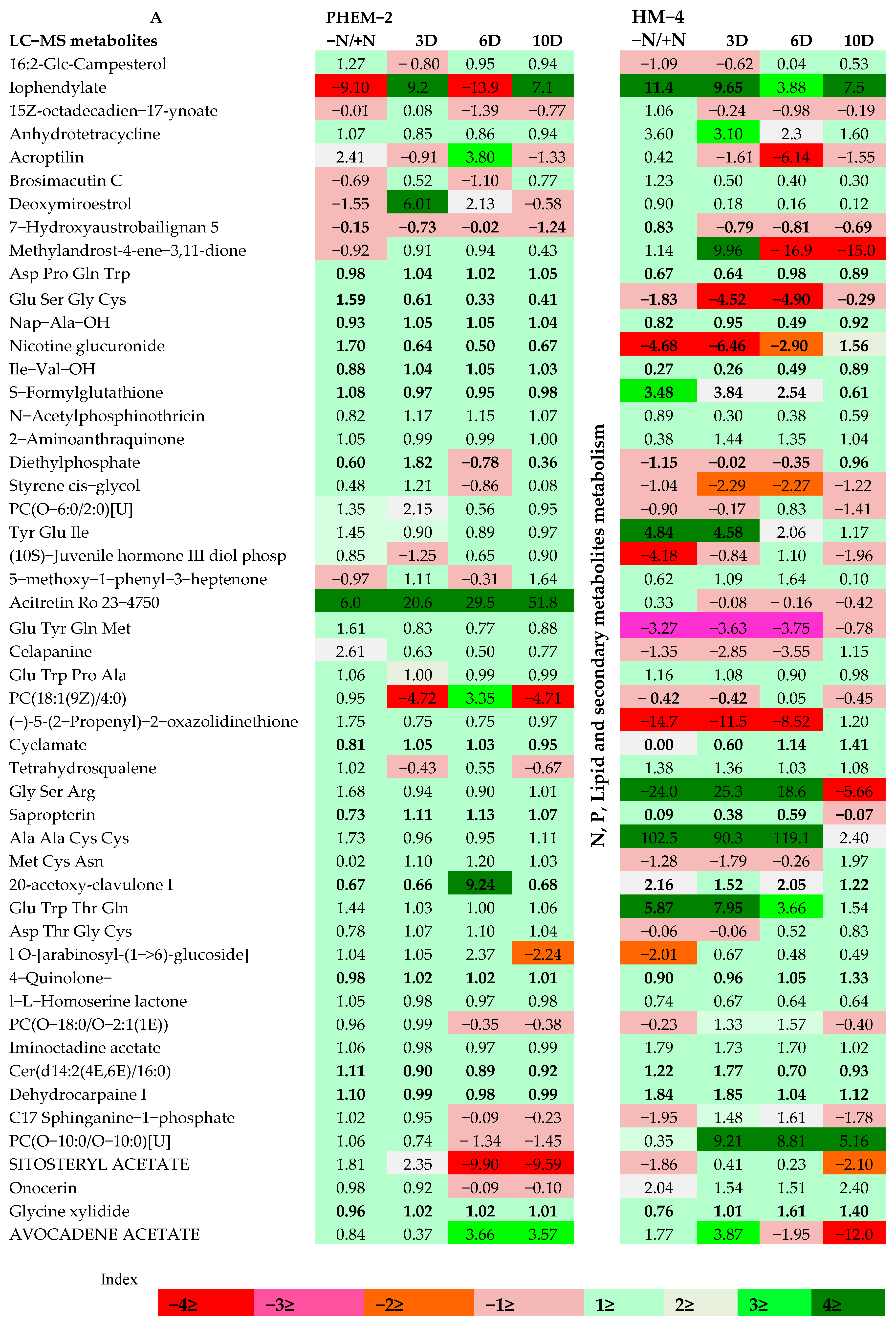
2.3. Metabolite Profiling of the Leaf and Root of Maize Genotypes under N Treatments
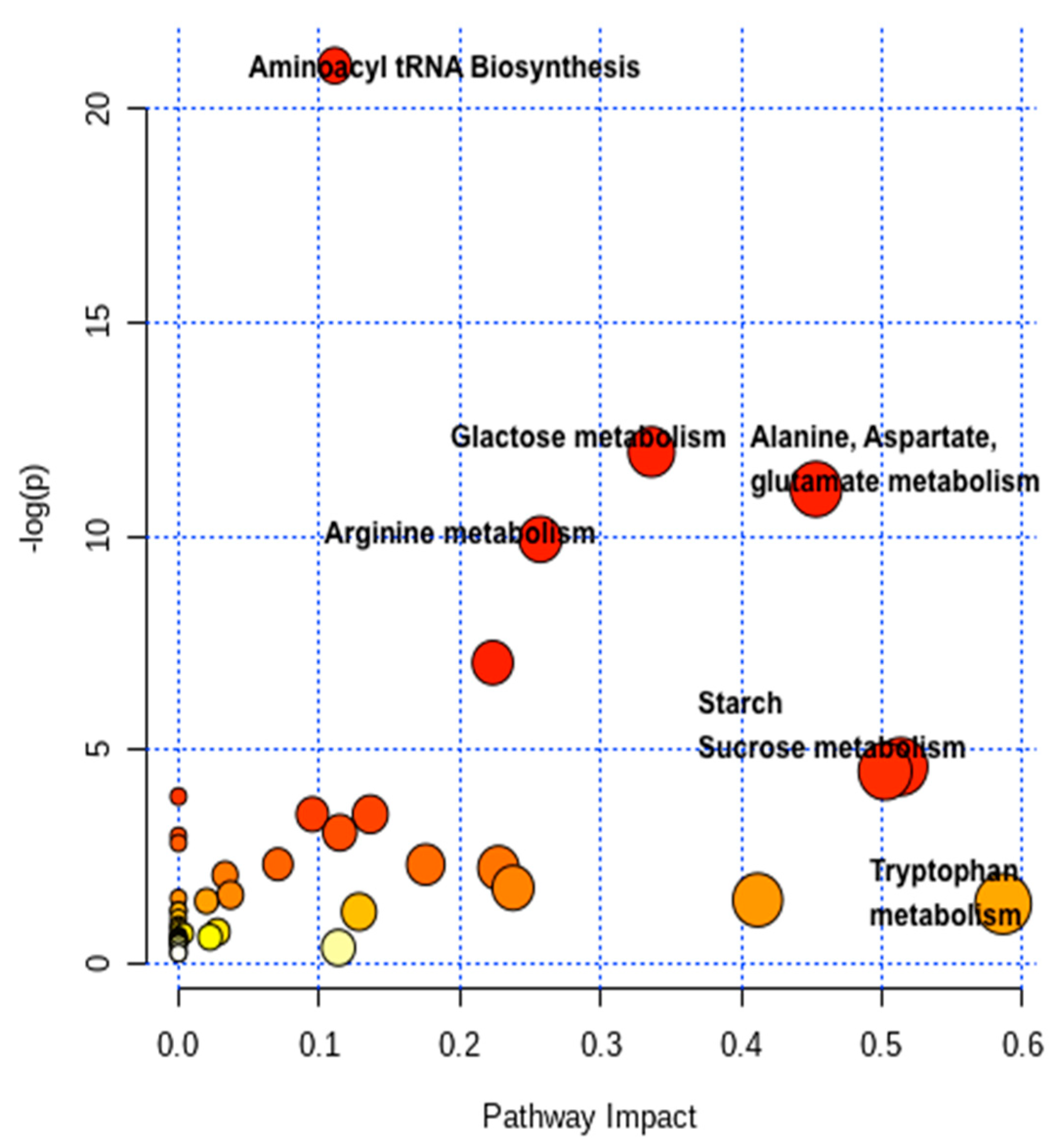
2.4. Pathway Network and MESA Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Cultivation, N-Deficiency and N-Restoration Treatments
4.2. Sample Preparation
4.3. Metabolite Extraction
4.4. Normalization and Metabolite Identification
4.5. PCA and Statistical Analysis for Metabolite Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NUE | Nitrogen Use Efficiency |
| GC–MS | Gas Chromatography-Mass Spectrometry |
| LNT | Low-N tolerant (PEHM-2) |
| LNS | Low-N sensitive (HM-4) |
| HMW | High Molecular Weight Compounds |
| LMW | Low Molecular Weight Compounds |
| PCA | Principle component Analysis |
| PC1 | Principle Component 1 |
| FDR | False Discovery Rate |
References
- Prinsi, B.; Negri, A.S.; Pesaresi, P.; Cocucci, M.; Espen, L. Evaluation of protein pattern changes in roots and leaves of Zea mays plants in response to nitrate availability by two-dimensional gel electrophoresis analysis. BMC Plant Biol. 2009, 9, 113. [Google Scholar] [CrossRef]
- Sutton, M.A.; Howard, C.M.; Erisman, J.W.; Bealey, W.J.; Billen, G.; Bleeker, A.; Bouwman, A.F.; Grennfelt, P.; Van Grinsven, H.; Grizzetti, B. The challenge to integrate nitrogen science and policies: The European nitrogen assessment approach. In The European Nitrogen Assessment; Sutton, M.A., Howard, C.M., Erisman, J.M., Billen, G., Bleeker, A., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 82–96. [Google Scholar]
- Hakeem, K.R.; Ahmad, A.; Iqbal, M.; Gücel, S.; Öztürk, M. Nitrogen-efficient rice cultivars can reduce nitrate pollution. Environ. Sci. Pollut. Res. 2011, 18, 1184–1193. [Google Scholar] [CrossRef]
- Burney, J.A.; Davis, S.J.; Lobell, D.B. Greenhouse gas mitigation by agricultural intensification. Proc. Natl. Acad. Sci. USA 2010, 107, 12052–12057. [Google Scholar] [CrossRef] [PubMed]
- Woods, J.; Williams, A.; Hughes, J.K.; Black, M.; Murphy, R. Energy and the food system. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 2991–3006. [Google Scholar] [CrossRef] [PubMed]
- Shcherbak, I.; Millar, N.; Robertson, G.P. Global meta-analysis of the nonlinear response of soil nitrous oxide (N2O) emissions to fertilizer nitrogen. Proc. Natl. Acad. Sci. USA 2014, 111, 9199–9204. [Google Scholar] [CrossRef] [PubMed]
- Dyson, T. World food trends and prospects to 2025. Proc. Natl. Acad. Sci. USA 1999, 96, 5929–5936. [Google Scholar] [CrossRef]
- FAO. World Agriculture towards 2030/2050: The 2012 Revision; Paper No. 12–03, ESA Working; FAO: Rome, Italy, 2012. [Google Scholar]
- FAI. Fertilizer Association of India; FAI House: New Delhi, India, 2014; p. 3. [Google Scholar]
- Raun, W.R.; Johnson, G.V. Improving Nitrogen Use Efficiency for Cereal Production. Agron. J. 1999, 91, 357–363. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004, 45, 386–391. [Google Scholar] [CrossRef]
- Meyer, R.C.; Steinfath, M.; Lisec, J.; Becher, M.; Witucka-Wall, H.; Törjék, O.; Fiehn, O.; Änne, E.; Willmitzer, L.; Selbig, J.; et al. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 4759–4764. [Google Scholar] [CrossRef]
- Lisec, J.; Meyer, R.C.; Steinfath, M.; Redestig, H.; Becher, M.; Witucka-Wall, H.; Fiehn, O.; Törjék, O.; Selbig, J.; Altmann, T.; et al. Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J. 2007, 53, 960–972. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Fukushima, A.; Redestig, H.; Saito, K. Metabolomic approaches toward understanding nitrogen metabolism in plants. J. Exp. Bot. 2011, 62, 1439–1453. [Google Scholar] [CrossRef]
- Tohge, T.; Mettler, T.; Arrivault, S.; Carroll, A.J.; Stitt, M.; Fernie, A.R. From models to crop species: Caveats and solutions for translational metabolomics. Front. Plant Sci. 2011, 2, 61. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Takahashi, Y.; Izutsu, Y.; Maeno, M. Identification and expression of ventrally associated leucine-zipper (VAL) in Xenopus embryo. Int. J. Dev. Biol. 2010, 54, 203–208. [Google Scholar] [CrossRef]
- Krapp, A.; Berthomé, R.; Orsel, M.; Mercey-Boutet, S.; Yu, A.; Castaings, L.; Elftieh, S.; Major, H.; Renou, J.-P.; Daniel-Vedele, F. Arabidopsis Roots and Shoots Show Distinct Temporal Adaptation Patterns toward Nitrogen Starvation. Plant Physiol. 2011, 157, 1255–1282. [Google Scholar] [CrossRef] [PubMed]
- Castro-Santos, P.; Laborde, C.M.; Díaz-Peña, R. Genomics, proteomics and metabolomics: Their emerging roles in the discovery and validation of rheumatoid arthritis biomarkers. Clin. Exp. Rheumatol. 2015, 33, 279–286. [Google Scholar] [PubMed]
- Guy, C.; Kopka, J.; Moritz, T. Plant metabolomics coming of age. Physiol. Plant. 2008, 132, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for plant stress response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Kralova, K.; Jampílek, J.; Ostrovský, I. Metabolomics-useful tool for study of plant responses to abiotic stresses. Ecol. Chem. Eng. 2012, S19, 133–161. [Google Scholar]
- Zhang, J.; Shen, H.; Xu, W.; Xia, Y.; Barr, D.B.; Mu, X.; Wang, X.; Liu, L.; Huang, Q.; Tian, M. Urinary metabolomics revealed arsenic internal dose-related metabolic alterations: A proof-of-concept study in a Chinese male cohort. Environ. Sci. Technol. 2014, 48, 12265–12274. [Google Scholar] [CrossRef]
- Gibbons, H.; O’Gorman, A.; Brennan, L. Metabolomics as a tool in nutritional research. Curr. Opin. Lipidol. 2015, 26, 30–34. [Google Scholar] [CrossRef]
- Patel, S.; Ahmed, S. Emerging field of metabolomics: Big promise for cancer biomarker identification and drug discovery. J. Pharm. Biomed. Anal. 2015, 107, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Jorge, T.F.; Rodrigues, J.A.; Caldana, C.; Schmidt, R.; van Dongen, J.T.; Thomas-Oates, J.; Antonio, C. Mass spectrometry-based plant metabolomics: Metabolite responses to abiotic stress. Mass Spectrom. Rev. 2016, 35, 620–649. [Google Scholar] [CrossRef] [PubMed]
- Amiour, N.; Imbaud, S.; Clément, G.; Agier, N.; Zivy, M.; Valot, B.; Balliau, T.; Armengaud, P.; Quilleré, I.; Cañas, R.; et al. The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. J. Exp. Bot. 2012, 63, 5017–5033. [Google Scholar] [CrossRef]
- Rossella, G.; Anaclerio, F.; Passera, C. Effects of nitrogen deprivation on the ATP-sulphurylase and O-acetylserine sulphydrylase activities of mesophyll protoplasts and bundle sheath strands of maize leaves. Biol. Plant. 1986, 28, 114–119. [Google Scholar]
- Atanasova, E. Effect of nitrogen sources on the nitrogenous forms and accumulation of amino acid in head cabbage. Plant Soil Environ. 2008, 54, 66–71. [Google Scholar] [CrossRef]
- Oaks, A. Re-evaluation of nitrogen assimilation in roots. Bioscience 1992, 42, 103–111. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Valadier, M.H.; Carrayol, E.; Reisdorf-Cren, M.; Hirel, B. Diurnal changes in the expression of glutamate dehydrogenase and nitrate reductase are involved in the C/N balance of tobacco source leaves. Plant Cell Environ. 2002, 25, 1451–1462. [Google Scholar] [CrossRef]
- Foyer, C.H.; Parry, M.; Noctor, G. Markers and signals associated with nitrogen assimilation in higher plants. J. Exp. Bot. 2003, 54, 585–593. [Google Scholar] [CrossRef]
- Bouche, N.; Fromm, H. GABA in plants: Just a metabolite? Trends Plant Sci. 2004, 9, 110–115. [Google Scholar] [CrossRef]
- Banerjee, A.; Sharma, R.; Chisti, Y.; Banerjee, U.C. Botryococcus braunii: A Renewable Source of Hydrocarbons and Other Chemicals. Crit. Rev. Biotechnol. 2002, 22, 245–279. [Google Scholar] [CrossRef]
- Dayananda, C.; Sarada, R.; Shamala, T.R.; Ravishankar, G.A. Influence of nitrogen sources on growth, hydrocarbon and fatty acid production by Botryococcus braunii. Asian J. Plant Sci. 2006, 5, 799–804. [Google Scholar] [CrossRef]
- Slocum, R.D. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 2005, 43, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Reinbothe, H.; Mothes, K. Urea, ureides, and guanidines in plants. Annu. Rev. Plant Physiol. 1962, 13, 129–151. [Google Scholar] [CrossRef]
- Witte, C.P. Urea metabolism in plants. Plant Sci. 2011, 180, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.M.; Zhang, J.H. Photosynthetic CO2 assimilation, chlorophyll fluorescence and photoinhibition as affected by nitrogen deficiency in maize plants. Plant Sci. 2000, 151, 135–143. [Google Scholar] [CrossRef]
- Yeesang, C.; Cheirsilp, B. Effect of nitrogen, salt, and iron content in the growth medium and light intensity on lipid production by microalgae isolated from freshwater sources in Thailand. Bioresour. Technol. 2011, 102, 3034–3040. [Google Scholar] [CrossRef]
- Schaller, H. The role of sterols in plant growth and development. Prog. Lipid Res. 2003, 42, 163–175. [Google Scholar] [CrossRef]
- Souter, M.; Topping, J.; Pullen, M.; Friml, J.; Palme, K.; Hackett, R.; Grierson, D.; Lindsey, K. Hydra Mutants of Arabidopsis Are Defective in Sterol Profiles and Auxin and Ethylene Signaling. Plant Cell 2002, 14, 1017–1031. [Google Scholar] [CrossRef]
- Neumann, G.; Römheld, V. Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil 1999, 211, 121–130. [Google Scholar] [CrossRef]
- Sas, L.; Rengel, Z.; Tang, C. Excess cation uptake, and extrusion of protons and organic acid anions by Lupinus albus under phosphorus deficiency. Plant Sci. 2001, 160, 1191–1198. [Google Scholar] [CrossRef]
- Haase, S.; Neumann, G.; Kania, A.; Kuzyakov, Y.; Römheld, V.; Kandeler, E. Elevation of atmospheric CO2 and N-nutritional status modify nodulation, nodule-carbon supply, and root exudation of Phaseolus vulgaris L. Soil Biol. Biochem. 2007, 39, 2208–2221. [Google Scholar] [CrossRef]
- Edwards, P.A.; Ericsson, J. Sterols and Isoprenoids: Signaling Molecules Derived from the Cholesterol Biosynthetic Pathway. Annu. Rev. Biochem. 1999, 68, 157–185. [Google Scholar] [CrossRef] [PubMed]
- Matthew, T.; Zhou, W.; Rupprecht, J.; Lim, L.; Thomas-Hall, S.R.; Doebbe, A.; Kruse, O.; Hankamer, B.; Marx, U.C.; Smith, S.M.; et al. The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J. Biol. Chem. 2009, 284, 23415–23425. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.T.; Ullrich, N.; Joo, S.; Waffenschmidt, S.; Goodenough, U. Algal lipid bodies: Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 2009, 8, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Dean, A.P.; Sigee, D.C.; Estrada, B.; Pittman, J.K. Using FTIR spectroscopy for rapid determination of lipid accumulation in response to nitrogen limitation in freshwater microalgae. Bioresour. Technol. 2010, 101, 4499–4507. [Google Scholar] [CrossRef]
- Doebbe, A.; Keck, M.; La Russa, M.; Mussgnug, J.H.; Hankamer, B.; Tekce, E.; Niehaus, K.; Kruse, O. The interplay of proton, electron and metabolite supply for photosynthetic H2 production in Chlamydomonas reinhardtii. J. Biol. Chem. 2010, 285, 30247–30260. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, D.; Hu, G.; Sommerfeld, M.; Hu, Q. Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol. Bioeng. 2010, 107, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Moellering, E.R.; Benning, C. RNAi silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 2010, 9, 97–106. [Google Scholar] [CrossRef]
- Work, V.H.; Radakovits, R.; Jinkerson, R.E.; Meuser, J.E.; Elliot, L.G.; Vinyard, D.J.; Laurens, L.M.L.; Dismukes, G.C.; Posewitz, M.C. Increased lipid accumulation in the Chlamydomonas reinhardtii sta7–10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot. Cell 2010, 9, 1251–1261. [Google Scholar] [CrossRef]
- Fan, J.; Andre, C.; Xu, C. A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 2011, 585, 1985–1991. [Google Scholar] [CrossRef]
- Kropat, J.; Hong-Hermesdorf, A.; Casero, D.; Ent, P.; Castruita, M.; Pellegrini, M.; Merchant, S.S.; Malasarn, D. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011, 66, 770–780. [Google Scholar] [CrossRef]
- James, G.O.; Hocart, C.H.; Hillier, W.; Chen, H.; Kordbacheh, F.; Price, G.D.; Djordjevic, M.A. Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresour. Technol. 2011, 102, 3343–3351. [Google Scholar] [CrossRef] [PubMed]
- Siaut, M.; Cuine, S.; Cagnon, C.; Fessler, B.; Nguyen, M.; Carrier, P.; Beyly, A.; Beisson, F.; Triantaphylides, C.; Li-Beisson, Y.; et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: Characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Scheible, W.R.; Gonzalez-Fontes, A.; Lauerer, M.; Müller-Röber, B.; Caboche, M.; Stitt, M. Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 1997, 9, 783–798. [Google Scholar] [CrossRef] [PubMed]
- Cordoba, E.; Salmi, M.; León, P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 2009, 60, 2933–2943. [Google Scholar] [CrossRef]
- Kanayama, Y.; Yamada, K.; Kato, K.; Morigushi, R. Biochemical and molecular aspects of sorbitol metabolism in Rosaceae fruit tree and other plants. In Phytochemistry Research Progress; Matsumoto, T., Ed.; Nova Science Publishers Inc.: New York, NY, USA, 2008; pp. 75–86. [Google Scholar]
- Fritz, C.; Palacios-Rojas, N.P.; Feil, R.; Stitt, M. Regulation of secondary metabolism by the carbon–nitrogen status of tobacco: Nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J. 2006, 46, 533–548. [Google Scholar] [CrossRef]
- Margna, U. Control at the level of substrate supply—An alternative in the regulation of phenylpropanoid accumulation in plant cells. Phytochemistry 1977, 16, 419–426. [Google Scholar] [CrossRef]
- Nazir, M.; Pandey, R.; Siddiqi, T.O.; Ibrahim, M.M.; Qureshi, M.I.; Vengavasi, K.; Abraham, G.; Ahmad, A. Nitrogen-deficiency stress induces protein expression differentially in low-N tolerant and low-N sensitive maize genotypes. Front. Plant Sci. 2016, 7, 298. [Google Scholar] [CrossRef]
- Stamova, B.S.; Roessner, U.; Suren, S.; Laudencia-Chingcuanco, D.; Bacic, A.; Beckles, D.M. Metabolic profiling of transgenic wheat overexpressing the high-molecular-weight Dx5 glutenin subunit. Metabolomics 2009, 5, 239–252. [Google Scholar] [CrossRef]
- Zoerb, C.; Langenkamper, G.; Betsche, T.; Niehaus, K.; Barsch, A. Metabolite profiling of wheat grains (Triticum aestivum L.) from organic and conventional agriculture. J. Agric. Food Chem. 2006, 54, 8301–8306. [Google Scholar] [CrossRef] [PubMed]
- Ganie, A.H.; Ahmad, A.; Pandey, R.; Aref, I.M.; Yousuf, P.Y.; Ahmad, S.; Iqbal, M. Metabolite Profiling of Low-P Tolerant and Low-P Sensitive Maize Genotypes under Phosphorus Starvation and Restoration Conditions. PLoS ONE 2015, 10, e0129520. [Google Scholar] [CrossRef] [PubMed]
- Hajduch, M.; Ganapathy, A.; Stein, J.W.; Thelen, J.J. A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 2005, 137, 1397–1419. [Google Scholar] [CrossRef]
- Roessner, U.; Wagner, C.; Kopka, J.; Trethewey, R.N.; Willmitzer, L. Simultaneous analysis of metabolites in potato tuber by gas chromatography–mass spectrometry. Plant J. 2000, 23, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; Dehaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, non-targeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, 652–660. [Google Scholar] [CrossRef]
- Neuweger, H.; Albaum, S.P.; Dondrup, M.; Persicke, M.; Watt, T.; Niehaus, K.; Stoye, J.; Goesmann, A. MeltDB: A software platform for the analysis and integration of metabolomics experiment data. Bioinformatics 2008, 24, 2726–2732. [Google Scholar] [CrossRef]
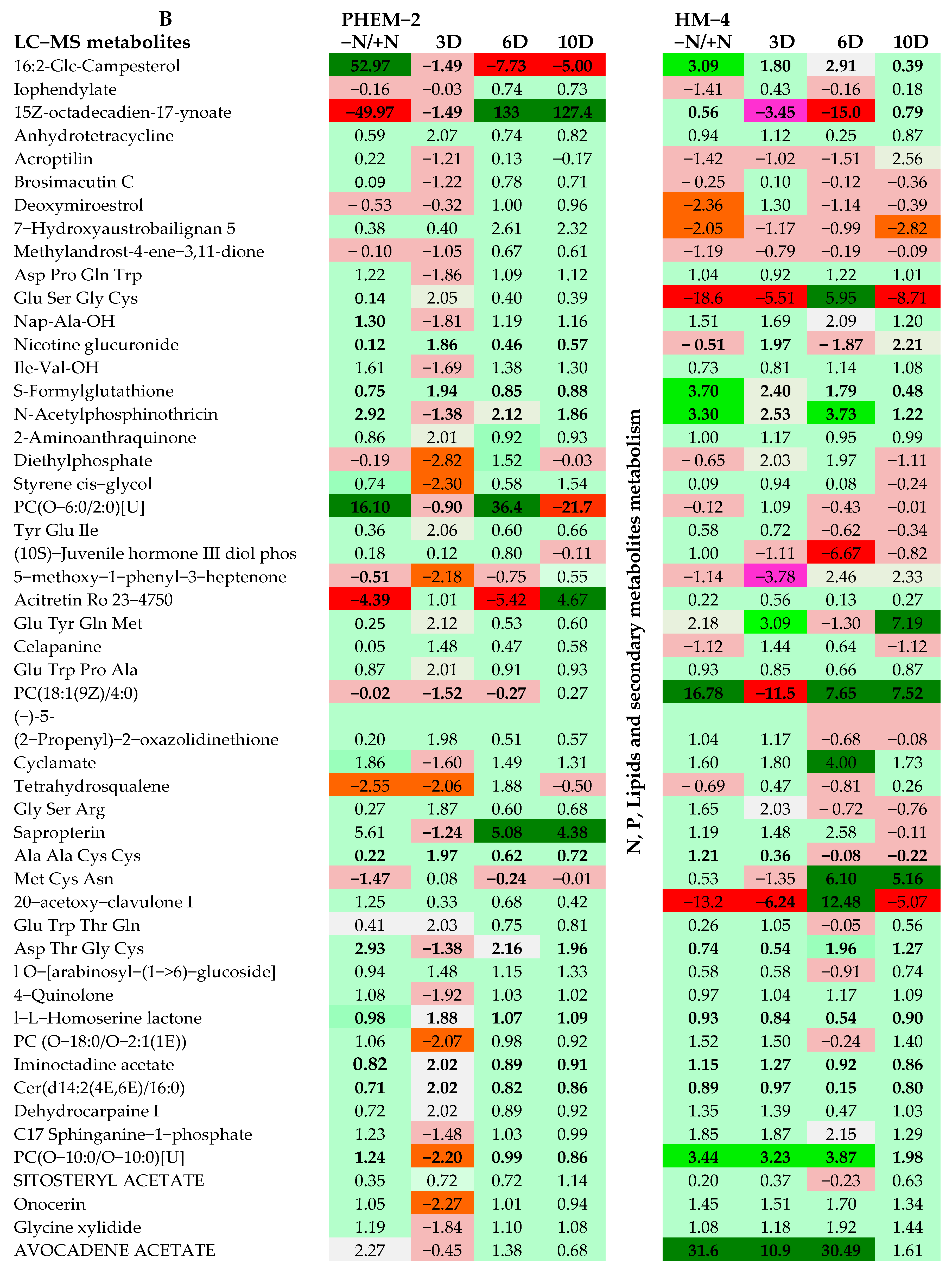
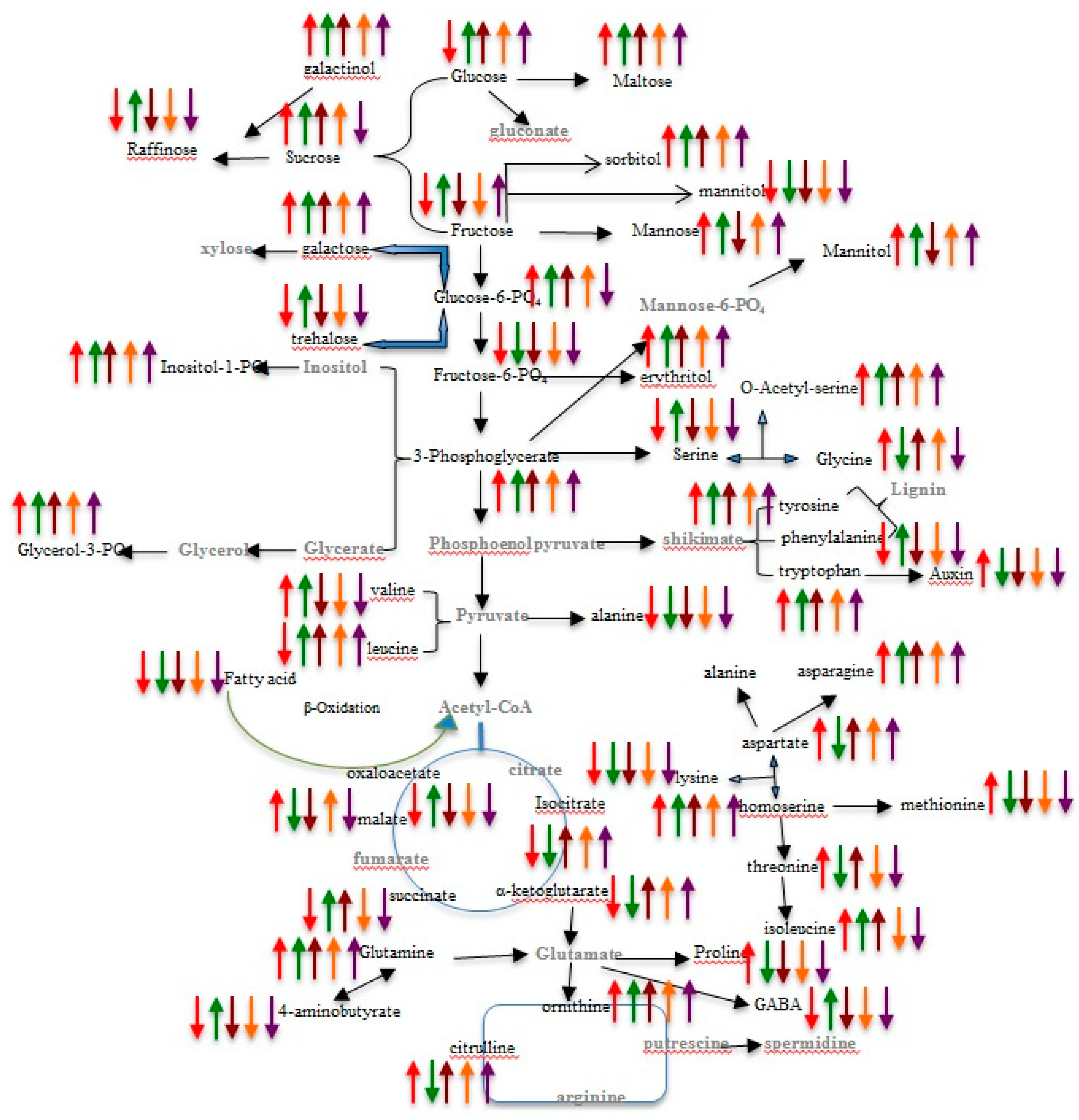
| Growth and Physiological Parameters | HM-4 Genotype | PEHM-2 Genotype | ||
|---|---|---|---|---|
| Sufficient-N (4.5 mM) | Low-N (0.05 mM) | Sufficient-N (4.5 mM) | Low-N (0.05 mM) | |
| Shoot length (cm/plant) | 16.33a | 10.07b | 15.73a | 15.11a |
| Root length (cm/plant) | 23.52b | 27.27a | 27.47a | 25.73a |
| Plant Biomass (g/plant) | 1.23a | 0.88b | 1.67a | 1.55b |
| Leaf area (cm2/plant) | 64.7a | 47.9b | 96.1a | 77.1b |
| Photosynthetic rate (µmol CO2 m−2 s−1) | 27.3a | 20.7b | 29.7a | 27.19b |
| Total Chlorophyll (mg g−1 FW) | 1.79a | 1.12b | 1.86a | 1.88a |
| NR activity (µmol g−1 FWh−1) | 5.35a | 3.74b | 5.19a | 4.75b |
| Concentration of nitrogen (%) | 3.17a | 2.07b | 3.18a | 2.87b |
| Plant N uptake (mg/plant) | 38.9a | 18.2b | 53.10a | 44.4b |
| Metabolic Pathways | Total | Expected | Hits | Raw p | -LOG (p) | Holm Adjust | FDR p | Impact |
|---|---|---|---|---|---|---|---|---|
| Aminoacyl-tRNA biosynthesis | 46 | 1.90 | 14 | 7.51 × 10−10 | 21.01 | 7.13 × 10−8 | 7.13 × 10−8 | 0.11111 |
| Galactose metabolism | 27 | 1.12 | 8 | 6.29 × 10−6 | 11.976 | 0.00059172 | 0.00029901 | 0.33624 |
| Alanine, aspartate and glutamate metabolism | 22 | 0.91 | 7 | 1.51 × 10−5 | 11.104 | 0.0014001 | 0.00047675 | 0.45324 |
| Arginine biosynthesis | 18 | 0.74 | 6 | 4.86 × 10−5 | 9.932 | 0.0044707 | 0.0011541 | 0.25729 |
| Glyoxylate and dicarboxylate metabolism | 29 | 1.20 | 6 | 0.00087103 | 7.0458 | 0.079263 | 0.01655 | 0.22338 |
| Glycine, serine and threonine metabolism | 33 | 1.36 | 5 | 0.0098897 | 4.6163 | 0.89007 | 0.14956 | 0.51346 |
| Starch and sucrose metabolism | 22 | 0.99 | 4 | 0.01102 | 4.508 | 0.98082 | 0.14956 | 0.50234 |
| Cyanoamino acid metabolism | 26 | 1.07 | 4 | 0.01991 | 3.9166 | 1 | 0.23643 | 0 |
| Pentose and glucuronate inter conversions | 17 | 0.70 | 3 | 0.030183 | 3.5005 | 1 | 0.28673 | 0.09524 |
| Butanoate metabolism | 17 | 0.70 | 3 | 0.030183 | 3.5005 | 1 | 0.28673 | 0.13636 |
| Citrate cycle (TCA cycle) | 20 | 0.82 | 3 | 0.046359 | 3.0713 | 1 | 0.39716 | 0.11468 |
| Lysine biosynthesis | 9 | 0.37 | 2 | 0.050168 | 2.9924 | 1 | 0.39716 | 0 |
| Valine, leucine and isoleucine biosynthesis | 22 | 0.91 | 3 | 0.05909 | 2.8287 | 1 | 0.43181 | 0 |
| Glutathione metabolism | 27 | 1.12 | 3 | 0.097154 | 2.3315 | 1 | 0.61964 | 0.07071 |
| Nicotinate and nicotinamide metabolism | 13 | 0.54 | 2 | 0.097838 | 2.3244 | 1 | 0.61964 | 0.17576 |
| Arginine and proline metabolism | 28 | 1.16 | 3 | 0.10573 | 2.2468 | 1 | 0.6278 | 0.22747 |
| Sulfur metabolism | 15 | 0.62 | 2 | 0.12502 | 2.0793 | 1 | 0.69862 | 0.03315 |
| Tyrosine metabolism | 18 | 0.74 | 2 | 0.16858 | 1.7803 | 1 | 0.88973 | 0.23784 |
| Fructose and mannose metabolism | 20 | 0.82 | 2 | 0.1989 | 1.615 | 1 | 0.99242 | 0.03695 |
| Carbon fixation in photosynthetic organisms | 21 | 0.87 | 2 | 0.2143 | 1.5404 | 1 | 0.99242 | 0 |
| Isoquinoline alkaloid biosynthesis | 6 | 0.25 | 1 | 0.22413 | 1.4955 | 1 | 0.99242 | 0.41176 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 22 | 0.91 | 2 | 0.22982 | 1.4704 | 1 | 0.99242 | 0.02002 |
| Tryptophan metabolism | 23 | 0.95 | 2 | 0.24542 | 1.4048 | 1 | 1 | 0.5862 |
| Monobactam biosynthesis | 8 | 0.33 | 1 | 0.28724 | 1.2474 | 1 | 1 | 0 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 8 | 0.33 | 1 | 0.28724 | 1.2474 | 1 | 1 | 0 |
| Cysteine and methionine metabolism | 46 | 1.90 | 3 | 0.29544 | 1.2193 | 1 | 1 | 0.12832 |
| Amino sugar and nucleotide sugar metabolism | 50 | 2.07 | 3 | 0.3419 | 1.0732 | 1 | 1 | 0 |
| Nitrogen metabolism | 12 | 0.49 | 1 | 0.3987 | 0.91954 | 1 | 1 | 0 |
| Selenocompound metabolism | 13 | 0.54 | 1 | 0.42377 | 0.85855 | 1 | 1 | 0 |
| Valine, leucine and isoleucine degradation | 37 | 1.53 | 2 | 0.45794 | 0.78102 | 1 | 1 | 0 |
| Pyrimidine metabolism | 38 | 1.57 | 2 | 0.47199 | 0.7508 | 1 | 1 | 0.02773 |
| Purine metabolism | 63 | 2.60 | 3 | 0.48919 | 0.71499 | 1 | 1 | 0.00383 |
| Sphingolipid metabolism | 17 | 0.70 | 1 | 0.51419 | 0.66517 | 1 | 1 | 0 |
| Lysine degradation | 18 | 0.74 | 1 | 0.53452 | 0.62639 | 1 | 1 | 0 |
| Beta-Alanine metabolism | 18 | 0.74 | 1 | 0.53452 | 0.62639 | 1 | 1 | 0 |
| Ascorbate and aldarate metabolism | 18 | 0.74 | 1 | 0.53452 | 0.62639 | 1 | 1 | 0.02239 |
| Pentose phosphate pathway | 19 | 0.78 | 1 | 0.55401 | 0.59057 | 1 | 1 | 0 |
| Propanoate metabolism | 20 | 0.83 | 1 | 0.5727 | 0.55739 | 1 | 1 | 0 |
| Pyruvate metabolism | 22 | 0.91 | 1 | 0.6078 | 0.49791 | 1 | 1 | 0 |
| Thiamine metabolism | 22 | 0.91 | 1 | 0.6078 | 0.49791 | 1 | 1 | 0 |
| Biosynthesis of unsaturated fatty acids | 22 | 0.91 | 1 | 0.6078 | 0.49791 | 1 | 1 | 0 |
| Pantothenate and CoA biosynthesis | 23 | 0.95 | 1 | 0.62427 | 0.47117 | 1 | 1 | 0 |
| Alpha-Linolenic acid metabolism | 27 | 1.12 | 1 | 0.68363 | 0.38034 | 1 | 1 | 0.11368 |
| Inositol phosphate metabolism | 28 | 1.16 | 1 | 0.69696 | 0.36102 | 1 | 1 | 0 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 35 | 1.45 | 1 | 0.77603 | 0.25356 | 1 | 1 | 0 |
| Phenylpropanoid biosynthesis | 35 | 1.45 | 1 | 0.77603 | 0.25356 | 1 | 1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganie, A.H.; Pandey, R.; Kumar, M.N.; Chinnusamy, V.; Iqbal, M.; Ahmad, A. Metabolite Profiling and Network Analysis Reveal Coordinated Changes in Low-N Tolerant and Low-N Sensitive Maize Genotypes under Nitrogen Deficiency and Restoration Conditions. Plants 2020, 9, 1459. https://doi.org/10.3390/plants9111459
Ganie AH, Pandey R, Kumar MN, Chinnusamy V, Iqbal M, Ahmad A. Metabolite Profiling and Network Analysis Reveal Coordinated Changes in Low-N Tolerant and Low-N Sensitive Maize Genotypes under Nitrogen Deficiency and Restoration Conditions. Plants. 2020; 9(11):1459. https://doi.org/10.3390/plants9111459
Chicago/Turabian StyleGanie, Arshid Hussain, Renu Pandey, M. Nagaraj Kumar, Viswanathan Chinnusamy, Muhammad Iqbal, and Altaf Ahmad. 2020. "Metabolite Profiling and Network Analysis Reveal Coordinated Changes in Low-N Tolerant and Low-N Sensitive Maize Genotypes under Nitrogen Deficiency and Restoration Conditions" Plants 9, no. 11: 1459. https://doi.org/10.3390/plants9111459
APA StyleGanie, A. H., Pandey, R., Kumar, M. N., Chinnusamy, V., Iqbal, M., & Ahmad, A. (2020). Metabolite Profiling and Network Analysis Reveal Coordinated Changes in Low-N Tolerant and Low-N Sensitive Maize Genotypes under Nitrogen Deficiency and Restoration Conditions. Plants, 9(11), 1459. https://doi.org/10.3390/plants9111459






