Atypical Splicing Accompanied by Skipping Conserved Micro-Exons Produces Unique WRINKLED1, An AP2 Domain Transcription Factor in Rice Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Analysis
2.2. Expression Analysis
2.3. Vector Construction
2.4. Transient Assay
3. Results
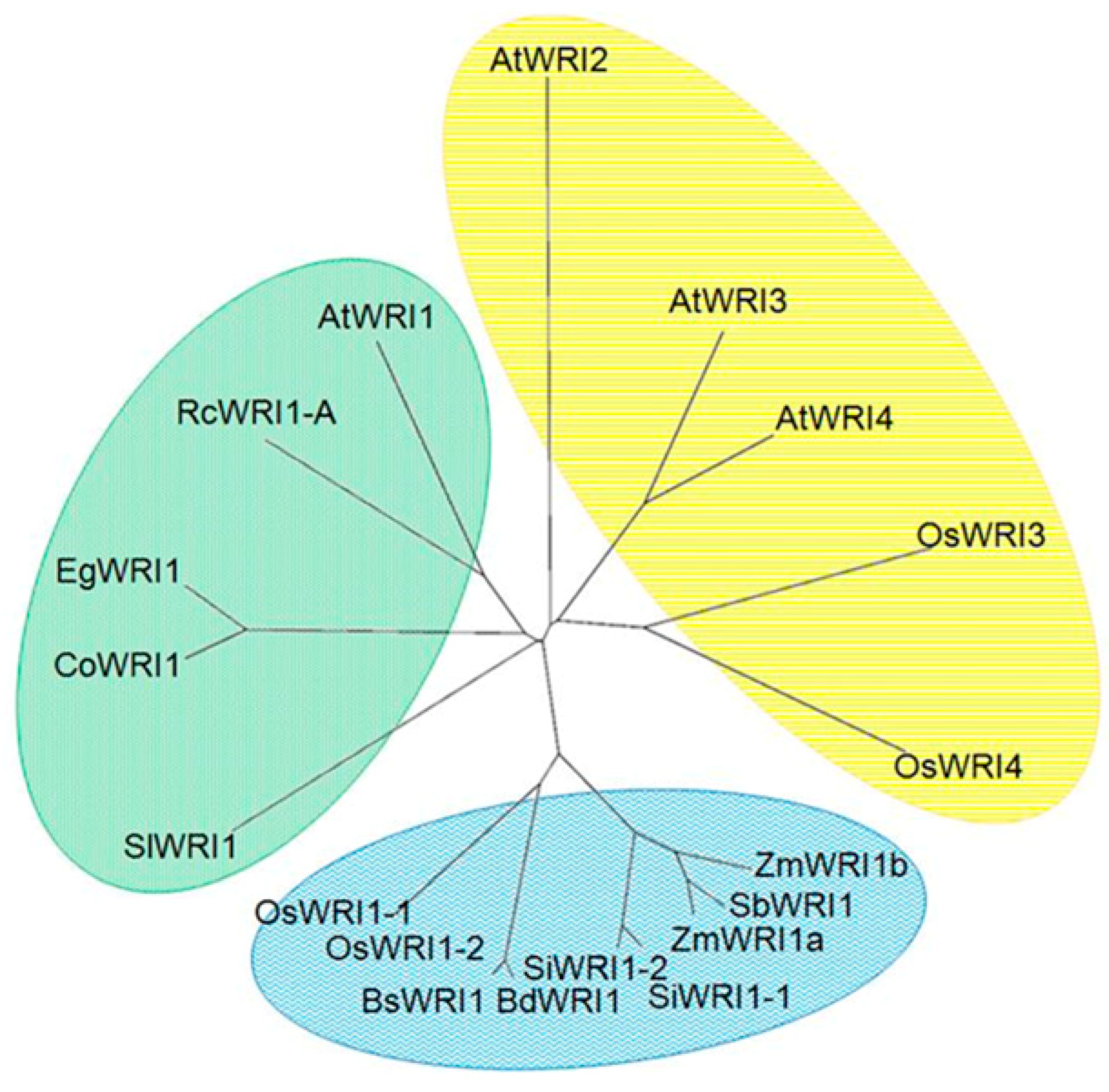
3.1. Molecular Characterization of WRI1 like Transcription Factors in Rice
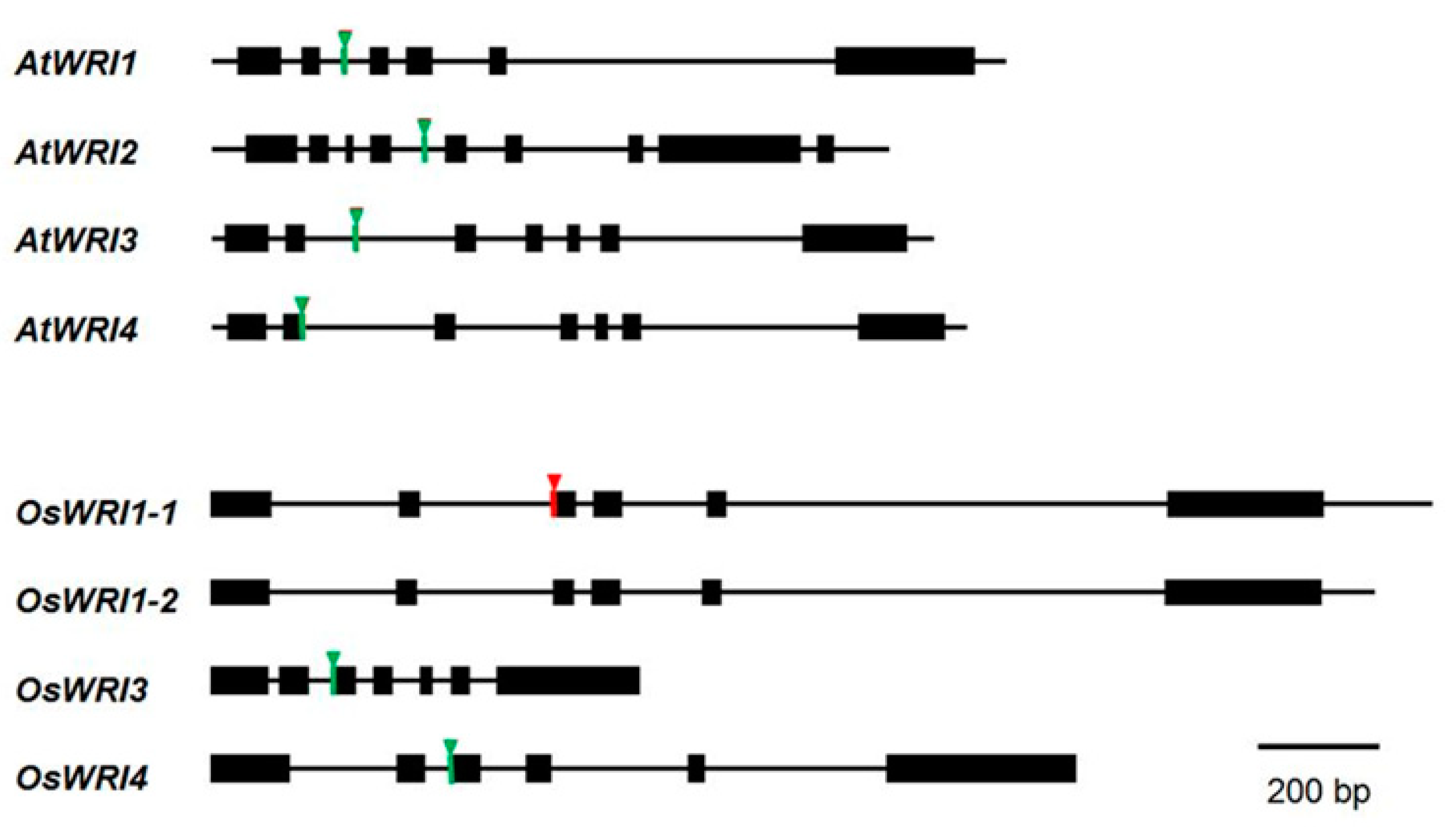
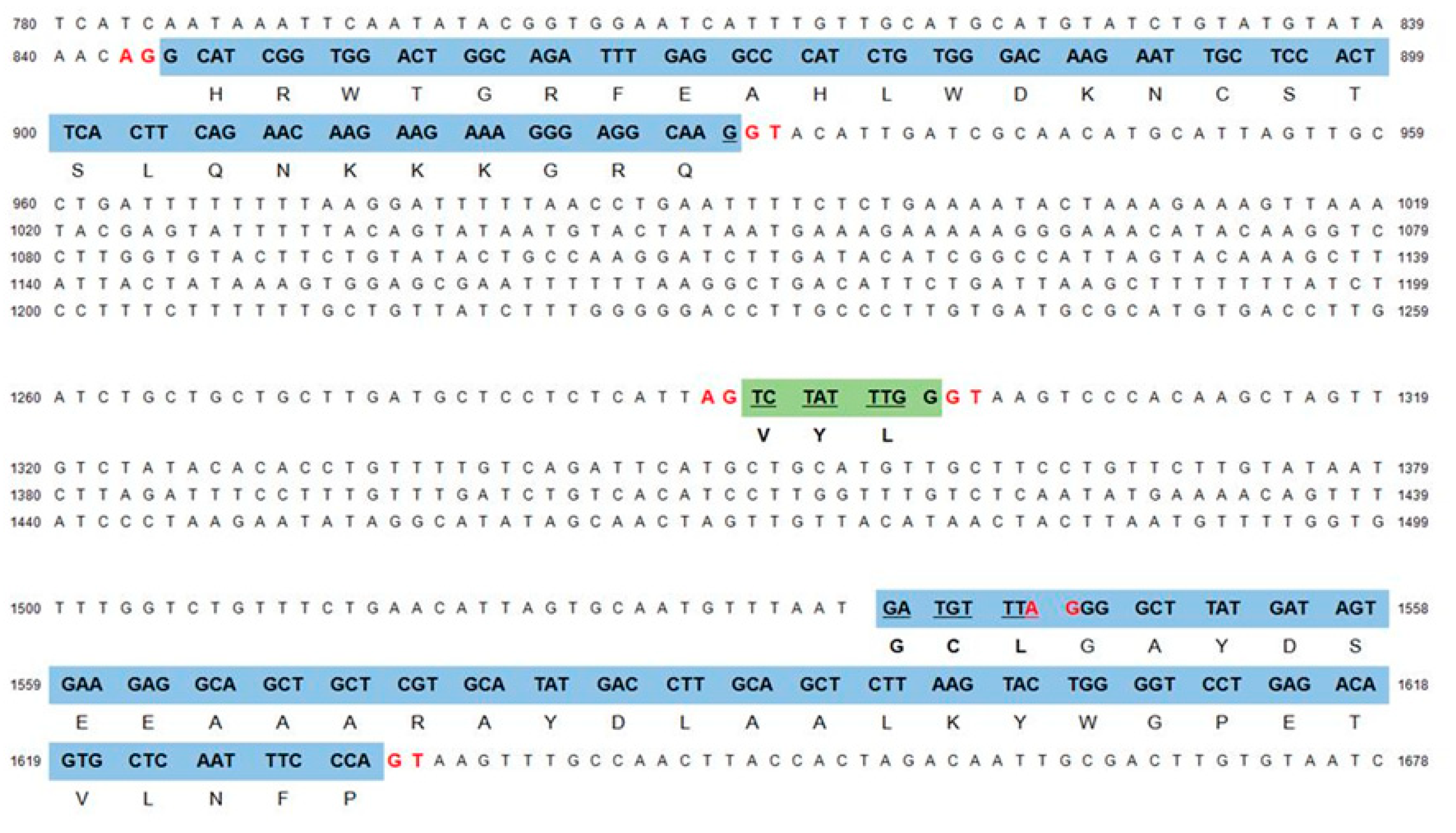
3.2. Genome Structure
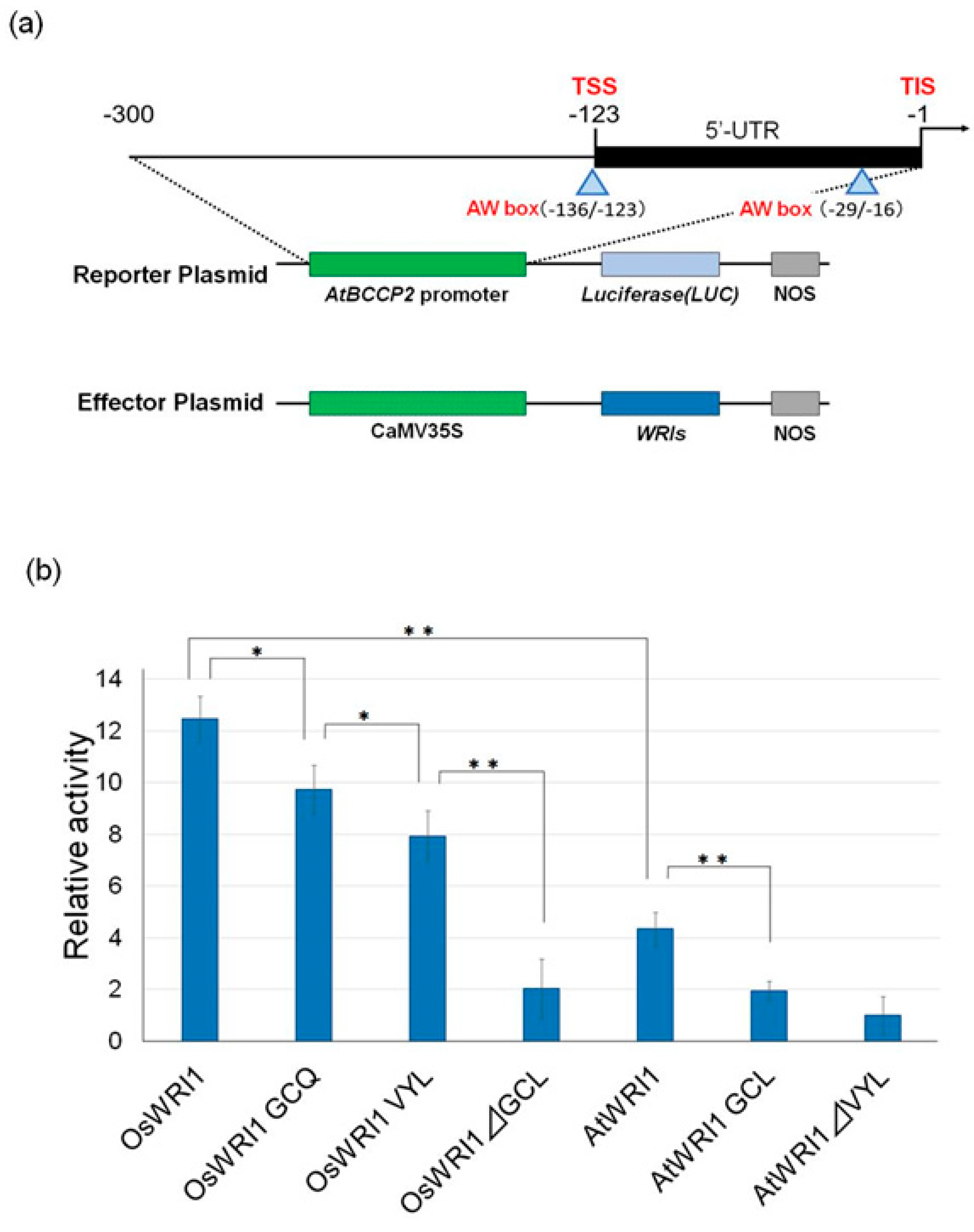
3.3. The Activities of OsWRI1-1 and Its Mutant.
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arab. Book 2013, 11, e0133. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Rose, J.K. The Formation and Function of Plant Cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Cernac, A.; Benning, C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [PubMed]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.Q.; Gerentes, D.; Perez, P.; Smyth, D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar] [PubMed]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar]
- Wilson, K.; Long, D.; Swinburne, J.; Coupland, G. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 1996, 8, 659–671. [Google Scholar] [PubMed]
- Finkelstein, R.R.; Wang, M.L.; Lynch, T.J.; Rao, S.; Goodman, H.M. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 1998, 10, 1043–1054. [Google Scholar] [CrossRef]
- Kim, S.; Soltis, P.S.; Wall, K.; Soltis, D.E. Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 2006, 23, 107–120. [Google Scholar] [CrossRef]
- Maeo, K.; Tokuda, T.; Ayame, A.; Mitsui, N.; Kawai, T.; Tsukagoshi, H.; Ishiguro, S.; Nakamura, K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009, 60, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Ma, W.; Yang, H.; Ma, G.; Mantyla, J.J.; Benning, C. The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J. Exp. Bot. 2017, 68, 4627–4634. [Google Scholar] [CrossRef] [PubMed]
- To, A.; Joubes, J.; Barthole, G.; Lecureuil, A.; Scagnelli, A.; Jasinski, S.; Lepiniec, L.; Baud, S. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 2012, 24, 5007–5023. [Google Scholar] [CrossRef] [PubMed]
- Park, C.S.; Go, Y.S.; Suh, M.C. Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 2016, 88, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lee, J.H.; Weber, H.; Tohge, T.; Witt, S.; Roje, S.; Fernie, A.R.; Hellmann, H. Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 2013, 25, 2253–2264. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Liu, H.; Shanklin, J. Phosphorylation of WRINKLED1 by KIN10 Results in Its Proteasomal Degradation, Providing a Link between Energy Homeostasis and Lipid Biosynthesis. Plant Cell 2017, 29, 871–889. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Mantyla, J.J.; Yang, Y.; Ohlrogge, J.B.; Benning, C. 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 2016, 88, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Kong, Q.; Grix, M.; Mantyla, J.J.; Yang, Y.; Benning, C.; Ohlrogge, J.B. Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 2015, 83, 864–874. [Google Scholar] [CrossRef]
- Kong, Q.; Ma, W. WRINKLED1 transcription factor: How much do we know about its regulatory mechanism? Plant Sci. 2018, 272, 153–156. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Arondel, V.; Kilaru, A.; Bates, P.D.; Thrower, N.A.; Benning, C.; Ohlrogge, J.B. Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE 2013, 8, e68887. [Google Scholar] [CrossRef]
- Ji, X.J.; Mao, X.; Hao, Q.T.; Liu, B.L.; Xue, J.A.; Li, R.Z. Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves. Int. J. Mol. Sci. 2018, 19, 146. [Google Scholar] [CrossRef]
- Sun, R.; Ye, R.; Gao, L.; Zhang, L.; Wang, R.; Mao, T.; Zheng, Y.; Li, D.; Lin, Y. Characterization and Ectopic Expression of CoWRI1, an AP2/EREBP Domain-Containing Transcription Factor from Coconut (Cocos nucifera L.) Endosperm, Changes the Seeds Oil Content in Transgenic Arabidopsis thaliana and Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Sun, Y.; Qu, J.; Syah, R.; Lim, C.H.; Alfiko, Y.; Rahman, N.E.B.; Suwanto, A.; Yue, G.; Wong, L.; et al. Transcriptome and functional analysis reveals hybrid vigor for oil biosynthesis in oil palm. Sci. Rep. 2017, 7, 439. [Google Scholar] [CrossRef] [PubMed]
- Pouvreau, B.; Baud, S.; Vernoud, V.; Morin, V.P.C.; Gendrot, G.; Pichon, J.P.; Rouster, J.; Paul, W.; Rogowsky, P.M. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K. J Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, H.; Kaimi, R.; Colasanti, J.; Kozaki, A. Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol. Biol. 2011, 77, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Jacquemin, J.; Chaparro, C.; Laudie, M.; Berger, A.; Gavory, F.; Goicoechea, J.L.; Wing, R.A.; Cooke, R. Long-range and targeted ectopic recombination between the two homeologous chromosomes 11 and 12 in Oryza species. Mol. Biol. Evol. 2011, 28, 3139–3150. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Mitsui, N.; Tsukagoshi, H.; Nishii, T.; Morikami, A.; Nakamura, K. ACTIVATOR of Spomin: LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol. 2005, 46, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Du, C.; Song, H.; Aziz, U.; Wang, L.; Zhao, C.; Zhang, M. Genome-wide analysis reveals the evolution and structural features of WRINKLED1 in plants. Mol. Genet. Genom. 2019, 294, 329–341. [Google Scholar] [CrossRef]
- Fukuda, N.; Ikawa, Y.; Aoyagi, T.; Kozaki, A. Expression of the genes coding for plastidic acetyl-CoA carboxylase subunits is regulated by a location-sensitive transcription factor binding site. Plant Mol. Biol. 2013, 82, 473–483. [Google Scholar] [CrossRef]
- Krizek, B.A. AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Res. 2003, 31, 1859–1868. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mano, F.; Aoyanagi, T.; Kozaki, A. Atypical Splicing Accompanied by Skipping Conserved Micro-Exons Produces Unique WRINKLED1, An AP2 Domain Transcription Factor in Rice Plants. Plants 2019, 8, 207. https://doi.org/10.3390/plants8070207
Mano F, Aoyanagi T, Kozaki A. Atypical Splicing Accompanied by Skipping Conserved Micro-Exons Produces Unique WRINKLED1, An AP2 Domain Transcription Factor in Rice Plants. Plants. 2019; 8(7):207. https://doi.org/10.3390/plants8070207
Chicago/Turabian StyleMano, Fumiya, Takuya Aoyanagi, and Akiko Kozaki. 2019. "Atypical Splicing Accompanied by Skipping Conserved Micro-Exons Produces Unique WRINKLED1, An AP2 Domain Transcription Factor in Rice Plants" Plants 8, no. 7: 207. https://doi.org/10.3390/plants8070207
APA StyleMano, F., Aoyanagi, T., & Kozaki, A. (2019). Atypical Splicing Accompanied by Skipping Conserved Micro-Exons Produces Unique WRINKLED1, An AP2 Domain Transcription Factor in Rice Plants. Plants, 8(7), 207. https://doi.org/10.3390/plants8070207




