Role of ClpP in the Biogenesis and Degradation of RuBisCO and ATP Synthase in Chlamydomonas reinhardtii
Abstract
1. Introduction
2. Results
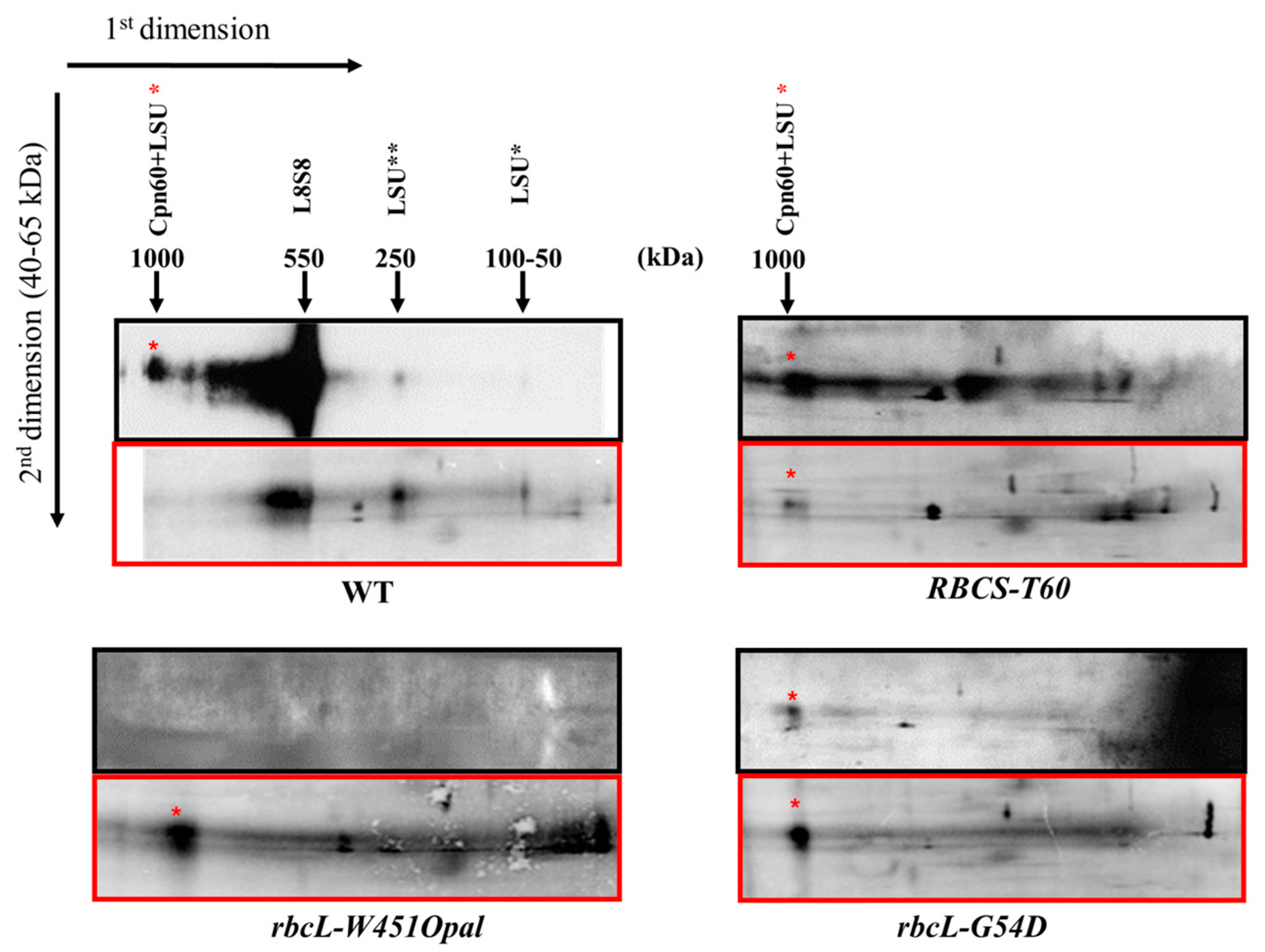
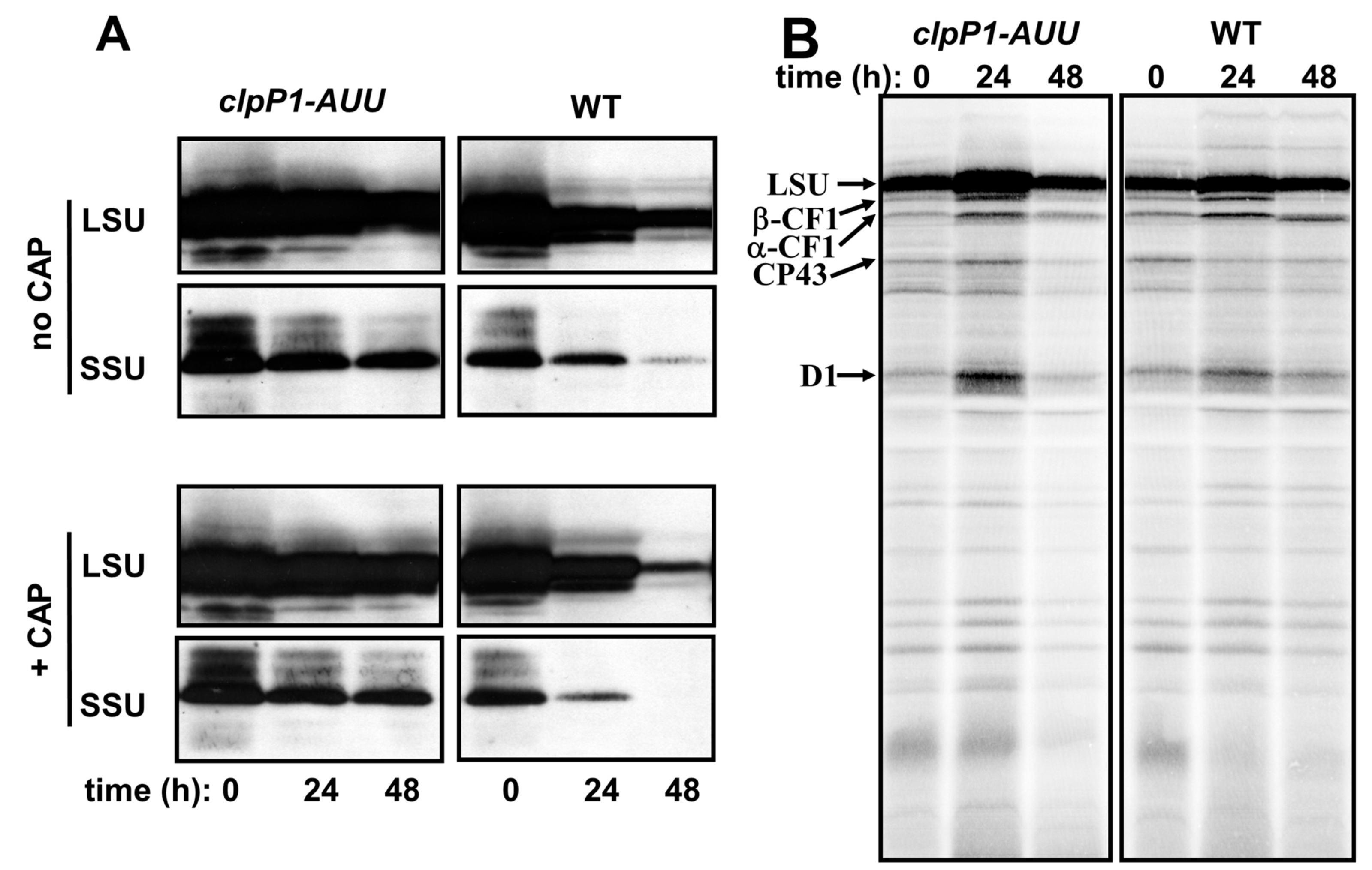
2.1. The LSU Assembly Pathway and its Perturbations in RuBisCO Mutants
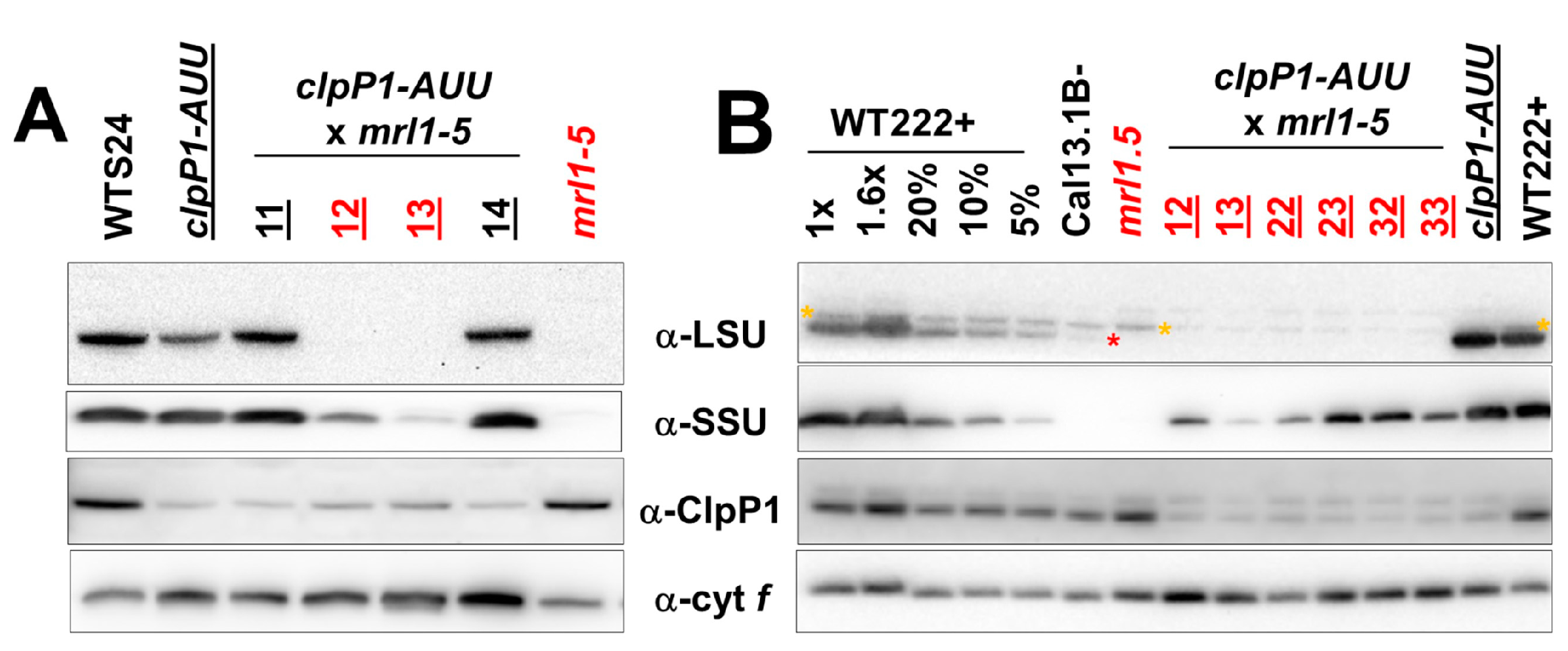
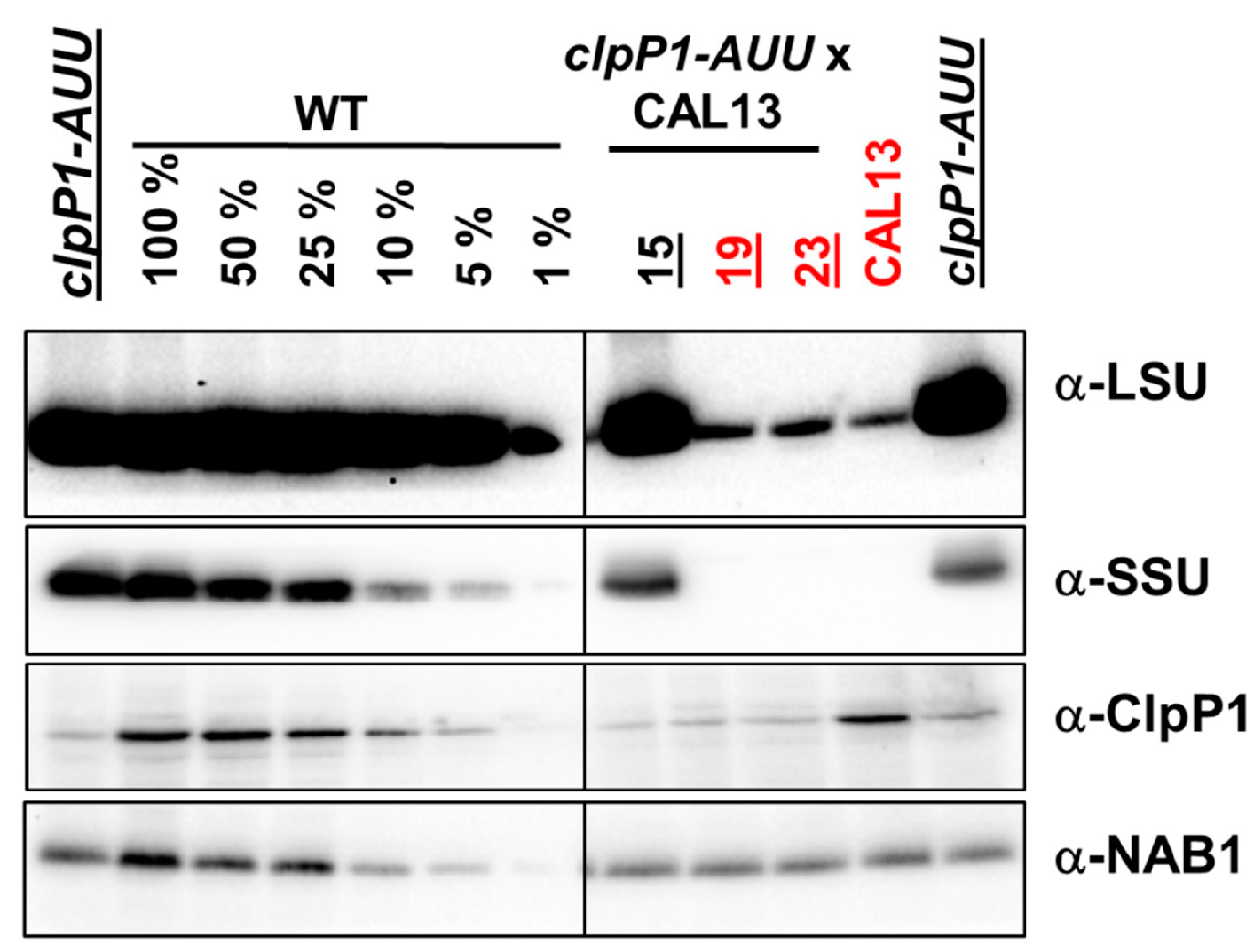
2.2. Double Mutants Combining Reduced ClpP Accumulation and Mutant LSU are not Viable
2.3. Effect of ClpP Attenuation in the Absence of LSU or SSU
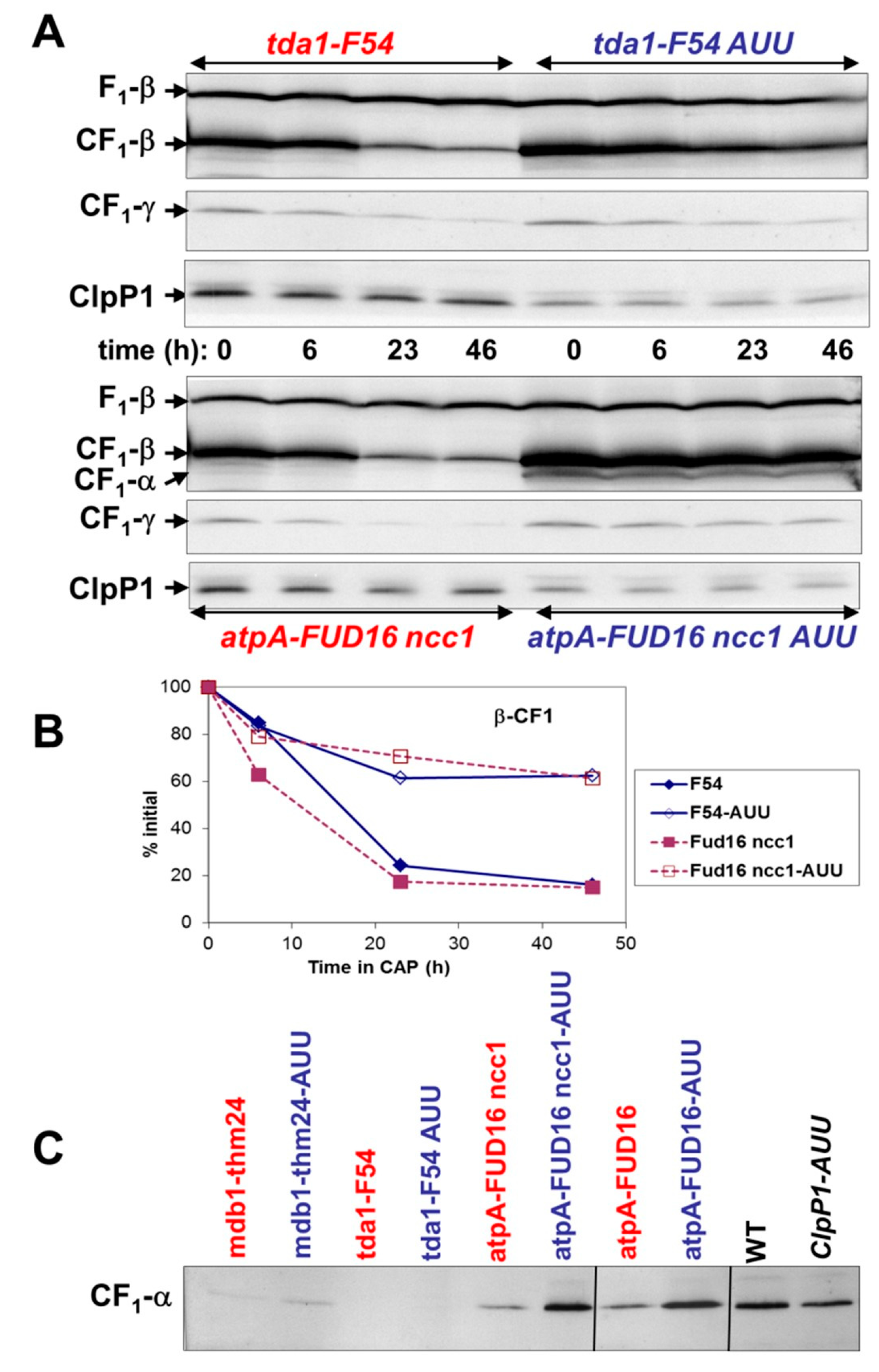
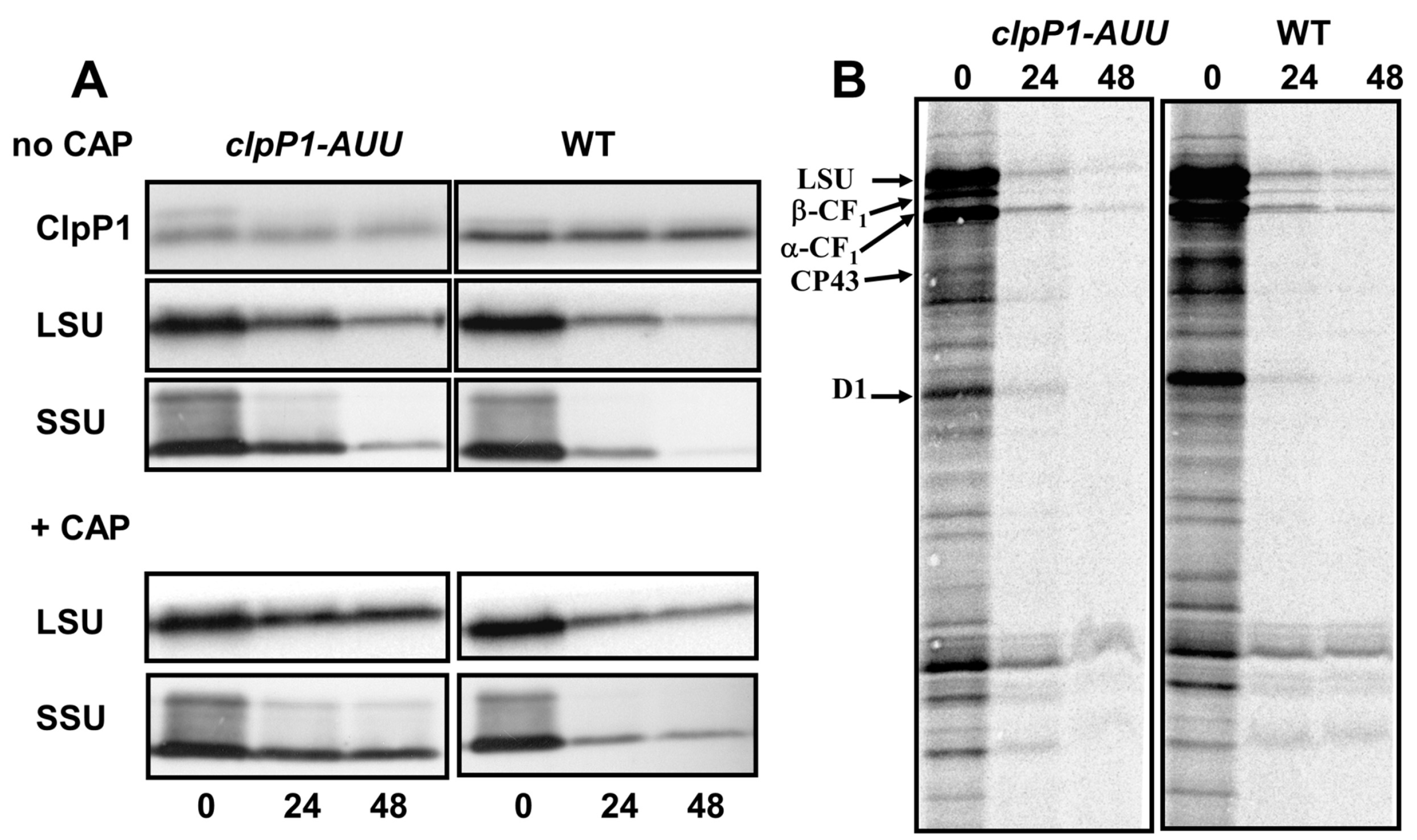
2.4. Effect of ClpP Attenuation on the Degradation of Unassembled Subunits in Other Photosynthetic Enzymes
2.5. RuBisCO Degradation during Sulfur and Nitrogen Starvation is ClpP Dependent
3. Discussion
3.1. Role of ClpP in the Degradation of Assembled RuBisCO
3.2. ClpP Levels Control the Accumulation of Unassembled Subunits of Photosynthetic Complexes.
3.3. ClpP is Necessary for the Proteolytic Disposal of Mutant LSU
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Chloroplast Transformation
4.3. Biochemical Analysis and Fluorescence Measurements.
4.4. Pulse Labeling and Chase
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Badger, M.R.; Bek, E.J. Multiple Rubisco forms in proteobacteria: Their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 2008, 59, 1525–1541. [Google Scholar] [CrossRef] [PubMed]
- Houtz, R.L.; Portis, A.R., Jr. The life of ribulose 1,5-bisphosphate carboxylase/oxygenase--posttranslational facts and mysteries. Arch. Biochem. Biophys. 2003, 414, 150–158. [Google Scholar] [CrossRef]
- Spreitzer, R.J. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 2003, 414, 141–149. [Google Scholar] [CrossRef]
- Bowes, G.; Ogren, W.L.; Hageman, R.H. Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 1971, 45, 716–722. [Google Scholar] [CrossRef]
- Ellis, R.J. The most abundant protein in the world. Trends Plant Sci. 1979, 4, 241–244. [Google Scholar] [CrossRef]
- Mastrobuoni, G.; Irgang, S.; Pietzke, M.; Assmus, H.E.; Wenzel, M.; Schulze, W.X.; Kempa, S. Proteome dynamics and early salt stress response of the photosynthetic organism Chlamydomonas reinhardtii. BMC Genom. 2012, 13, 215. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Stessman, D.J.; Spalding, M.H. The CO2 concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: How Chlamydomonas works against the gradient. Plant J. Cell Mol. Biol. 2015, 82, 429–448. [Google Scholar] [CrossRef] [PubMed]
- Mackinder, L.C.; Meyer, M.T.; Mettler-Altmann, T.; Chen, V.K.; Mitchell, M.C.; Caspari, O.; Freeman Rosenzweig, E.S.; Pallesen, L.; Reeves, G.; Itakura, A.; et al. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc. Natl. Acad. Sci. USA 2016, 113, 5958–5963. [Google Scholar] [CrossRef] [PubMed]
- Vitlin Gruber, A.; Feiz, L. Rubisco assembly in the chloroplast. Front. Mol. Biosci. 2018, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.H.; Hayer-Hartl, M. Complex chaperone dependence of rubisco biogenesis. Biochemistry 2018, 57, 3210–3216. [Google Scholar] [CrossRef] [PubMed]
- Hayer-Hartl, M. From chaperonins to rubisco assembly and metabolic repair. Protein Sci. A Publ. Protein Soc. 2017, 26, 2324–2333. [Google Scholar] [CrossRef] [PubMed]
- Hubbs, A.E.; Roy, H. Assembly of in vitro synthesized large subunits into ribulose- bisphosphate carboxylase/oxygenase. Formation and discharge of an L8- like species. J. Biol. Chem. 1993, 268, 13519–13525. [Google Scholar] [PubMed]
- Hall, L.N.; Rossini, L.; Cribb, L.; Langdale, J.A. GOLDEN 2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 1998, 10, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Wostrikoff, K.; Stern, D. Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 2007, 104, 6466–6471. [Google Scholar] [CrossRef] [PubMed]
- Feiz, L.; Williams-Carrier, R.; Wostrikoff, K.; Belcher, S.; Barkan, A.; Stern, D.B. Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell 2012, 24, 3435–3446. [Google Scholar] [CrossRef] [PubMed]
- Li, L.A.; Tabita, F.R. Maximum activity of recombinant ribulose 1,5-bisphosphate carboxylase/oxygenase of Anabaena sp. strain CA requires the product of the rbcX gene. J. Bacteriol. 1997, 179, 3793–3796. [Google Scholar] [CrossRef] [PubMed]
- Kolesinski, P.; Piechota, J.; Szczepaniak, A. Initial characteristics of RbcX proteins from Arabidopsis thaliana. Plant Mol. Biol. 2011, 77, 447–459. [Google Scholar] [CrossRef][Green Version]
- Liu, C.; Young, A.L.; Starling-Windhof, A.; Bracher, A.; Saschenbrecker, S.; Rao, B.V.; Rao, K.V.; Berninghausen, O.; Mielke, T.; Hartl, F.U.; et al. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 2010, 463, 197–202. [Google Scholar] [CrossRef]
- Hauser, T.; Popilka, L.; Hartl, F.U.; Hayer-Hartl, M. Role of auxiliary proteins in Rubisco biogenesis and function. Nat. Plants 2015, 1, 15065. [Google Scholar] [CrossRef]
- Aigner, H.; Wilson, R.H.; Bracher, A.; Calisse, L.; Bhat, J.Y.; Hartl, F.U.; Hayer-Hartl, M. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 2017, 358, 1272–1278. [Google Scholar] [CrossRef]
- Van de Loo, F.J.; Salvucci, M.E. Activation of ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) involves Rubisco activase Trp16. Biochemistry 1996, 35, 8143–8148. [Google Scholar] [CrossRef] [PubMed]
- Kuras, R.; Wollman, F.-A. The assembly of cytochrome b6f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 1994, 13, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.W.; Mishkind, M.L. Rapid degradation of unassembled ribulose 1,5-bisphosphate carboxylase small subunits in chloroplasts. Proc. Natl. Acad. Sci. USA 1983, 80, 2632–2636. [Google Scholar] [CrossRef] [PubMed]
- Khrebtukova, I.; Spreitzer, R.J. Elimination of the Chlamydomonas gene family that encodes the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc. Natl. Acad. Sci. USA 1996, 93, 13689–13693. [Google Scholar] [CrossRef] [PubMed]
- Rodermel, S.; Haley, J.; Jiang, C.Z.; Tsai, C.H.; Bogorad, L. A mechanism for intergenomic integration: Abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc. Natl. Acad. Sci. USA 1996, 93, 3881–3885. [Google Scholar] [CrossRef]
- Wostrikoff, K.; Clark, A.; Sato, S.; Clemente, T.; Stern, D. Ectopic expression of Rubisco subunits in maize mesophyll cells does not overcome barriers to cell type-specific accumulation. Plant Phys. 2012, 160, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Spreitzer, R.J.; Goldschmidt-Clermont, M.; Rahire, M.; Rochaix, J.D. Nonsense mutations in the Chlamydomonas chloroplast gene that codes for the large subunit of ribulosebisphosphate carboxylase/oxygenase. Proc. Natl. Acad. Sci. USA 1985, 82, 5460–5464. [Google Scholar] [CrossRef]
- Spreitzer, R.J.; Thow, G.; Zhu, G. Pseudoreversion substitution at large-subunit residue 54 influences the CO2/O2 specificity of chloroplast ribulose-bisphosphate carboxylase/oxygenase. Plant Phys. 1995, 109, 681–685. [Google Scholar] [CrossRef]
- Adam, Z.; Adamska, I.; Nakabayashi, K.; Ostersetzer, O.; Haussuhl, K.; Manuell, A.; Zheng, B.; Vallon, O.; Rodermel, S.R.; Shinozaki, K.; et al. Chloroplast and mitochondrial proteases in arabidopsis. a proposed nomenclature. Plant Phys. 2001, 125, 1912–1918. [Google Scholar] [CrossRef]
- Nishimura, K.; Kato, Y.; Sakamoto, W. Essentials of proteolytic machineries in chloroplasts. Mol. Plant 2017, 10, 4–19. [Google Scholar] [CrossRef]
- Olinares, P.D.; Kim, J.; van Wijk, K.J. The Clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta 2011, 1807, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, S.; Chen, L.; Lemieux, C.; Otis, C.; Turmel, M.; Liu, X.-Q. The Chlamydomonas chloroplast ClpP gene contains translated large insertion sequences and its essential for cell growth. Mol. Gen. Genet. 1994, 244, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Majeran, W.; Friso, G.; van Wijk, K.J.; Vallon, O. The chloroplast ClpP complex in Chlamydomonas reinhardtii contains an unusual high molecular mass subunit with a large apical domain. FEBS J. 2005, 272, 5558–5571. [Google Scholar] [CrossRef] [PubMed]
- Peltier, J.B.; Ytterberg, J.; Liberles, D.A.; Roepstorff, P.; van Wijk, K.J. Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 16318–16327. [Google Scholar] [CrossRef]
- Derrien, B.; Majeran, W.; Effantin, G.; Ebenezer, J.; Friso, G.; van Wijk, K.J.; Steven, A.C.; Maurizi, M.R.; Vallon, O. The purification of the Chlamydomonas reinhardtii chloroplast ClpP complex: Additional subunits and structural features. Plant Mol. Biol. 2012, 80, 189–202. [Google Scholar] [CrossRef]
- Olinares, P.D.; Kim, J.; Davis, J.I.; van Wijk, K.J. Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in arabidopsis. Plant Cell 2011, 23, 2348–2361. [Google Scholar] [CrossRef]
- Cahoon, A.B.; Cunningham, K.A.; Stern, D.B. The plastid ClpP gene may not be essential for plant cell viability. Plant Cell Phys. 2003, 44, 93–95. [Google Scholar] [CrossRef][Green Version]
- Majeran, W.; Wollman, F.A.; Vallon, O. Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 2000, 12, 137–150. [Google Scholar] [CrossRef]
- Majeran, W.; Olive, J.; Drapier, D.; Vallon, O.; Wollman, F.A. The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Phys. 2001, 126, 421–433. [Google Scholar] [CrossRef]
- Ramundo, S.; Casero, D.; Muhlhaus, T.; Hemme, D.; Sommer, F.; Crevecoeur, M.; Rahire, M.; Schroda, M.; Rusch, J.; Goodenough, U.; et al. Conditional depletion of the chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell 2014, 26, 2201–2222. [Google Scholar] [CrossRef]
- Feller, U.; Anders, I.; Mae, T. Rubiscolytics: Fate of Rubisco after its enzymatic function in a cell is terminated. J. Exp. Bot. 2008, 59, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Hortensteiner, S.; Feller, U. Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 2002, 53, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Nakamura, S. Chloroplast protein turnover: The influence of extraplastidic processes, including autophagy. Int. J. Mol. Sci. 2018, 19, 828. [Google Scholar] [CrossRef] [PubMed]
- Lunde, C.; Zygadlo, A.; Simonsen, H.T.; Nielsen, P.L.; Blennow, A.; Haldrup, A. Sulfur starvation in rice: The effect on photosynthesis, carbohydrate metabolism, and oxidative stress protective pathways. Phys. Plant. 2008, 134, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Lopez Garcia de Lomana, A.; Schauble, S.; Valenzuela, J.; Imam, S.; Carter, W.; Bilgin, D.D.; Yohn, C.B.; Turkarslan, S.; Reiss, D.J.; Orellana, M.V.; et al. Transcriptional program for nitrogen starvation-induced lipid accumulation in Chlamydomonas reinhardtii. Biotechnol. Biofuels 2015, 8, 207. [Google Scholar] [CrossRef] [PubMed]
- De Mia, M.; Lemaire, S.D.; Choquet, Y.; Wollman, F.-A. Nitric oxide remodels the photosynthetic apparatus upon S-starvation in Chlamydomonas reinhardtii. Plant Phys. 2019, 179, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Derrien, B.; Gautier, A.; Houille-Vernes, L.; Boulouis, A.; Saint-Marcoux, D.; Malnoe, A.; Rappaport, F.; de Vitry, C.; Vallon, O.; et al. Nitric Oxide-Triggered Remodeling of Chloroplast Bioenergetics and Thylakoid Proteins upon Nitrogen Starvation in Chlamydomonas reinhardtii. Plant Cell 2014, 26, 353–372. [Google Scholar] [CrossRef]
- Giordano, M.; Pezzoni, V.; Hell, R. Strategies for the allocation of resources under sulfur limitation in the green alga Dunaliella salina. Plant Phys. 2000, 124, 857–864. [Google Scholar] [CrossRef]
- Ferreira, R.M.; Teixeira, A.R. Sulfur starvation in Lemna leads to degradation of ribulose-bisphosphate carboxylase without plant death. J. Boil. Chem. 1992, 267, 7253–7257. [Google Scholar]
- Zhang, L.; Happe, T.; Melis, A. Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 2002, 214, 552–561. [Google Scholar] [CrossRef]
- Wykoff, D.D.; Davies, J.P.; Melis, A.; Grossman, A.R. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Phys. 1998, 117, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhao, L.; Sun, Y.L.; Cui, S.X.; Zhang, L.F.; Yang, B.; Wang, J.; Kuang, T.Y.; Huang, F. Proteomic analysis of hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. J. Proteome Res. 2010, 9, 3854–3866. [Google Scholar] [CrossRef] [PubMed]
- Melis, A.; Zhang, L.; Forestier, M.; Ghirardi, M.L.; Seibert, M. Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Phys. 2000, 122, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Hemschemeier, A.; Fouchard, S.; Cournac, L.; Peltier, G.; Happe, T. Hydrogen production by Chlamydomonas reinhardtii: An elaborate interplay of electron sources and sinks. Planta 2008, 227, 397–407. [Google Scholar] [CrossRef]
- Zhang, Z.; Shrager, J.; Jain, M.; Chang, C.W.; Vallon, O.; Grossman, A.R. Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot. Cell 2004, 3, 1331–1348. [Google Scholar] [CrossRef] [PubMed]
- Philipps, G.; Happe, T.; Hemschemeier, A. Nitrogen deprivation results in photosynthetic hydrogen production in Chlamydomonas reinhardtii. Planta 2012, 235, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.T.; Ullrich, N.; Joo, S.; Waffenschmidt, S.; Goodenough, U. Algal lipid bodies: Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 2009, 8, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.C.; Goodenough, U.W. Gametic differentiation in Chlamydomonas reinhardtii. I. Production of gametes and their fine structure. J. Cell Biol. 1975, 67, 587–605. [Google Scholar] [CrossRef] [PubMed]
- Bulte, L.; Wollman, F.A. Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6/f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur. J. Biochem. 1992, 204, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Plumley, F.G.; Schmidt, G.W. Nitrogen-dependent regulation of photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 1989, 86, 2678–2682. [Google Scholar] [CrossRef] [PubMed]
- Majeran, W. La Proteolyse Dans le Chloroplaste d eChlamydomonas Reinhardtii: Rôle et Organisation Structurale de la Protéase ClpP. Ph.D. Thesis, Institut National Agronomique ParisGrignon, Paris, France, 2002. [Google Scholar]
- Spreitzer, R.J.; Ogren, W.L. Rapid recovery of chloroplast mutations affecting ribulosebisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1983, 80, 6293–6297. [Google Scholar] [CrossRef] [PubMed]
- Thow, G.; Spreitzer, R.J. Missense mutations in the chloroplast rbcL gen that affect Rubisco holoenzyme assembly. Res. Photosynth. 1992, 3, 633–636. [Google Scholar]
- Hemmingsen, S.M.; Woolford, C.; van der Vies, S.M.; Tilly, K.; Dennis, D.T.; Georgopoulos, C.P.; Hendrix, R.W.; Ellis, R.J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 1988, 333, 330–334. [Google Scholar] [CrossRef]
- Roy, H.; Hubbs, A.; Cannon, S. Stability and Dissociation of the Large Subunit RuBisCO Binding Protein Complex in Vitro and in Organello. Plant Phys. 1988, 86, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Johnson, X.; Wostrikoff, K.; Finazzi, G.; Kuras, R.; Schwarz, C.; Bujaldon, S.; Nickelsen, J.; Stern, D.B.; Wollman, F.A.; Vallon, O. MRL1, a conserved pentatricopeptide repeat protein, is required for Stabilization of rbcL mRNA in chlamydomonas and arabidopsis. Plant Cell 2010, 22, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Johnson, X. Manipulating RuBisCO accumulation in the green alga, Chlamydomonas reinhardtii. Plant Mol. Biol. 2011, 76, 397–405. [Google Scholar] [CrossRef]
- Dent, R.M.; Sharifi, M.N.; Malnoe, A.; Haglund, C.; Calderon, R.H.; Wakao, S.; Niyogi, K.K. Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. Cell Mol. Biol. 2015, 82, 337–351. [Google Scholar] [CrossRef]
- Drapier, D.; Rimbault, B.; Vallon, O.; Wollman, F.A.; Choquet, Y. Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J. 2007, 26, 3581–3591. [Google Scholar] [CrossRef] [PubMed]
- Minai, L.; Wostrikoff, K.; Wollman, F.A.; Choquet, Y. Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 2006, 18, 159–175. [Google Scholar] [CrossRef] [PubMed]
- Choquet, Y.; Zito, F.; Wostrikoff, K.; Wollman, F.A. Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 2003, 15, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Rochaix, J.D.; Kuchka, M.; Mayfield, S.; Schirmer-Rahire, M.; Girard-Bascou, J.; Bennoun, P. Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 1989, 8, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, C.; Wollman, F.A. The chloroplast ATP synthase in Chlamydomonas reinhardtii. II. Biochemical studies on its biogenesis using mutants defective in photophosphorylation. J. Biol. Chem. 1989, 264, 10235–10242. [Google Scholar] [PubMed]
- Drapier, D.; Girard-Bascou, J.; Wollman, F.-A. Evidence for nuclear control of the expression of the atpA and atpB chloroplast genes in Chlamydomonas. Plant Cell 1992, 4, 283–295. [Google Scholar] [CrossRef][Green Version]
- Ketchner, S.L.; Drapier, D.; Olive, J.; Gaudriault, S.; Girard-Bascou, J.; Wollman, F.A. Chloroplasts can accommodate inclusion bodies. Evidence from a mutant of Chlamydomonas reinhardtii defective in the assembly of the chloroplast ATP synthase. J. Biol. Chem. 1995, 270, 15299–15306. [Google Scholar] [CrossRef]
- Boulouis, A.; Drapier, D.; Razafimanantsoa, H.; Wostrikoff, K.; Tourasse, N.J.; Pascal, K.; Girard-Bascou, J.; Vallon, O.; Wollman, F.A.; Choquet, Y. Spontaneous dominant mutations in Chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell 2015, 27, 984–1001. [Google Scholar] [CrossRef] [PubMed]
- Ravina, C.G.; Chang, C.I.; Tsakraklides, G.P.; McDermott, J.P.; Vega, J.M.; Leustek, T.; Gotor, C.; Davies, J.P. The sac mutants of Chlamydomonas reinhardtii reveal transcriptional and posttranscriptional control of cysteine biosynthesis. Plant Phys. 2002, 130, 2076–2084. [Google Scholar] [CrossRef]
- Martinez, D.E.; Costa, M.L.; Guiamet, J.J. Senescence-associated degradation of chloroplast proteins inside and outside the organelle. Plant Biol. 2008, 10 (Suppl. 1), 15–22. [Google Scholar] [CrossRef]
- Poret, M.; Chandrasekar, B.; van der Hoorn, R.A.L.; Avice, J.C. Characterization of senescence-associated protease activities involved in the efficient protein remobilization during leaf senescence of winter oilseed rape. Plant Sci. Int. J. Exp. Plant Biol. 2016, 246, 139–153. [Google Scholar] [CrossRef]
- Bushnell, T.; Bushnell, D.; Jagendorf, A.T. A purified zinc ptotease of pea chloroplasts, EP1, degrades the large subunit of ribulose-1,5-biphosphate carboxylase/oxygenase. Plant Phys. 1993, 103, 585–591. [Google Scholar] [CrossRef]
- Kato, Y.; Murakami, S.; Yamamoto, Y.; Chatani, H.; Kondo, Y.; Nakano, T.; Yokota, A.; Sato, F. The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta 2004, 220, 97–104. [Google Scholar] [CrossRef]
- Murakami, S.; Kondo, Y.; Nakano, T.; Sato, F. Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells [In Process Citation]. FEBS Lett. 2000, 468, 15–18. [Google Scholar] [CrossRef]
- Nakano, T.; Murakami, S.; Shoji, T.; Yoshida, S.; Yamada, Y.; Sato, F. A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell 1997, 9, 1673–1682. [Google Scholar] [PubMed]
- James, M.; Poret, M.; Masclaux-Daubresse, C.; Marmagne, A.; Coquet, L.; Jouenne, T.; Chan, P.; Trouverie, J.; Etienne, P. SAG12, a major cysteine protease involved in nitrogen mobilization during senescence for seed production in Arabidopsis thaliana. Plant Cell Phys. 2018, 50, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Izumi, M.; Wada, S.; Makino, A. Roles of autophagy in chloroplast recycling. Biochim. Biophys. Acta 2013. [Google Scholar] [CrossRef] [PubMed]
- Chiba, A.; Ishida, H.; Nishizawa, N.K.; Makino, A.; Mae, T. Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant cell Phys. 2003, 44, 914–921. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Yoshimoto, K.; Izumi, M.; Reisen, D.; Yano, Y.; Makino, A.; Ohsumi, Y.; Hanson, M.R.; Mae, T. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Phys. 2008, 148, 142–155. [Google Scholar] [CrossRef]
- Goodenough, U.; Blaby, I.; Casero, D.; Gallaher, S.D.; Goodson, C.; Johnson, S.; Lee, J.H.; Merchant, S.S.; Pellegrini, M.; Roth, R.; et al. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot. Cell 2014, 13, 591–613. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.P.; Horst, I.; Duong, G.H.; Tomsett, E.V.; Litvinenko, A.C.; Howe, C.J.; Smith, A.G. Triacylglyceride production and autophagous responses in Chlamydomonas reinhardtii depend on resource allocation and carbon source. Eukaryot. Cell 2014, 13, 392–400. [Google Scholar] [CrossRef]
- Heredia-Martinez, L.G.; Andres-Garrido, A.; Martinez-Force, E.; Perez-Perez, M.E.; Crespo, J.L. Chloroplast damage induced by the inhibition of fatty acid synthesis triggers autophagy in Chlamydomonas. Plant Phys. 2018. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Ito, M.; Kiyosue, T.; Shinozaki, K.; Watanabe, A. Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Phys. 1999, 40, 504–514. [Google Scholar] [CrossRef][Green Version]
- Nishimura, K.; van Wijk, K.J. Organization, function and substrates of the essential Clp protease system in plastids. Biochim. Biophys. Acta 2015, 1847, 915–930. [Google Scholar] [CrossRef] [PubMed]
- Saroussi, S.; Sanz-Luque, E.; Kim, R.G.; Grossman, A.R. Nutrient scavenging and energy management: Acclimation responses in nitrogen and sulfur deprived Chlamydomonas. Curr. Opin. Plant Biol. 2017, 39, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Cakmak, T.; Angun, P.; Demiray, Y.E.; Ozkan, A.D.; Elibol, Z.; Tekinay, T. Differential effects of nitrogen and sulfur deprivation on growth and biodiesel feedstock production of Chlamydomonas reinhardtii. Biotechnol. Bioeng. 2012, 109, 1947–1957. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Cui, X.; Wahid, F.; Xia, F.; Zhong, C.; Jia, S. Analysis of the Physiological and Molecular Responses of Dunaliella salina to Macronutrient Deprivation. PLoS ONE 2016, 11, e0152226. [Google Scholar] [CrossRef] [PubMed]
- Nagy, V.; Vidal-Meireles, A.; Podmaniczki, A.; Szentmihalyi, K.; Rakhely, G.; Zsigmond, L.; Kovacs, L.; Toth, S.Z. The mechanism of photosystem-II inactivation during sulphur deprivation-induced H2 production in Chlamydomonas reinhardtii. Plant J. Cell Mol. Biol. 2018, 94, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Desimone, M.; Henke, A.; Wagner, E. Oxidative Stress Induces Partial Degradation of the Large Subunit of Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase in Isolated Chloroplasts of Barley. Plant Phys. 1996, 111, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Ishida, H.; Makino, A.; Mae, T. Fe2+-catalyzed site-specific cleavage of the large subunit of ribulose 1,5-bisphosphate carboxylase close to the active site. J. Biol. Chem. 2002, 277, 12382–12387. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.; Garcia-Murria, M.J.; Marin-Navarro, J. Redox modulation of Rubisco conformation and activity through its cysteine residues. J. Exp. Bot. 2008, 59, 1605–1614. [Google Scholar] [CrossRef]
- Marin-Navarro, J.; Moreno, J. Cysteines 449 and 459 modulate the reduction-oxidation conformational changes of ribulose 1.5-bisphosphate carboxylase/oxygenase and the translocation of the enzyme to membranes during stress. Plant Cell Environ. 2006, 29, 898–908. [Google Scholar] [CrossRef]
- Lubben, T.H.; Donaldson, G.K.; Viitanen, P.V.; Gatenby, A.A. Several proteins imported into chloroplasts form stable complexes with the GroEL-related chloroplast molecular chaperone. Plant Cell 1989, 1, 1223–1230. [Google Scholar] [CrossRef]
- Tsugeki, R.; Nishimura, M. Interaction of homologues of Hsp70 and Cpn60 with ferredoxin-NADP+ reductase upon its import into chloroplasts. FEBS Lett. 1993, 320, 198–202. [Google Scholar] [CrossRef]
- Madueno, F.; Napier, J.A.; Gray, J.C. Newly imported Rieske Iron-Sulfur protein associates with both Cpn60 and Hsp70 in the chloroplast stroma. Plant Cell. 1993, 5, 1865–1876. [Google Scholar] [CrossRef] [PubMed]
- Choquet, Y.; Wostrikoff, K.; Rimbault, B.; Zito, F.; Girard-Bascou, J.; Drapier, D.; Wollman, F.A. Assembly-controlled regulation of chloroplast gene translation. Biochem. Soc. Trans. 2001, 29, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Wostrikoff, K.; Girard-Bascou, J.; Wollman, F.A.; Choquet, Y. Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 2004, 23, 2696–2705. [Google Scholar] [CrossRef]
- Choquet, Y.; Stern, D.B.; Wostrikoff, K.; Kuras, R.; Girard-Bascou, J.; Wollman, F.A. Translation of cytochrome f is autoregulated through the 5’ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 1998, 95, 4380–4385. [Google Scholar] [CrossRef] [PubMed]
- Schottler, M.A.; Toth, S.Z.; Boulouis, A.; Kahlau, S. Photosynthetic complex stoichiometry dynamics in higher plants: Biogenesis, function, and turnover of ATP synthase and the cytochrome b6f complex. J. Exp. Bot. 2015, 66, 2373–2400. [Google Scholar] [CrossRef]
- Lemaire, C.; Wollman, F.A.; Bennoun, P. Restoration of phototrophic growth in a mutant of Chlamydomonas reinhardtii in which the chloroplast atpB gene of the ATP synthase has a deletion: An example of mitochondria-dependent photosynthesis. Proc. Natl. Acad. Sci. USA 1988, 85, 1344–1348. [Google Scholar] [CrossRef]
- De Vitry, C.; Finazzi, G.; Baymann, F.; Kallas, T. Analysis of the nucleus-encoded and chloroplast-targeted rieske protein by classic and site-directed mutagenesis of Chlamydomonas. Plant Cell 1999, 11, 2031–2044. [Google Scholar] [CrossRef]
- Malnoe, A.; Wollman, F.A.; de Vitry, C.; Rappaport, F. Photosynthetic growth despite a broken Q-cycle. Nat. Commun. 2011, 2, 301. [Google Scholar] [CrossRef]
- Spreitzer, R.J.; Salvucci, M.E. Rubisco: Structure, regulatory interactions, and possibilities for a better enzyme. Ann. Rev. Plant Biol. 2002, 53, 449–475. [Google Scholar] [CrossRef]
- Avni, A.; Edelman, M.; Rachailovich, I.; Aviv, D.; Fluhr, R. A point mutation in the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase affects holoenzyme assembly in Nicotiana tabacum. EMBO J. 1989, 8, 1915–1918. [Google Scholar] [CrossRef] [PubMed]
- Brinker, A.; Pfeifer, G.; Kerner, M.J.; Naylor, D.J.; Hartl, F.U.; Hayer-Hartl, M. Dual function of protein confinement in chaperonin-assisted protein folding. Cell 2001, 107, 223–233. [Google Scholar] [CrossRef]
- Suzuki, K.; Nakanishi, H.; Bower, J.; Yoder, D.W.; Osteryoung, K.W.; Miyagishima, S.Y. Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in Arabidopsis thaliana. BMC Plant Biol. 2009, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.M.; Haglund, C.M.; Chin, B.L.; Kobayashi, M.C.; Niyogi, K.K. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of chlamydomonas reinhardtii. Plant Phys. 2005, 137, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.H. The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use; Harris, E.H., Ed.; Academic Press: San Diego, CA, USA, 1989; pp. 1–780. [Google Scholar]
- Boynton, J.E.; Gillham, N.W.; Harris, E.H.; Hosler, J.P.; Johnson, A.M.; Jones, A.R.; Randolph-Anderson, B.L.; Robertson, D.; Klein, T.M.; Shark, K.B. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 1988, 240, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
- Piccioni, R.G.; Bennoun, P.; Chua, N.H. A nuclear mutant of Chlamydomonas reinhardtii defective in photosynthetic photophosphorylation. Characterization of the algal coupling factor ATPase. Eur. J. Biochem. 1981, 117, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; Cramer, W.A.; von Jagow, G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994, 217, 220–230. [Google Scholar] [CrossRef]
- Smart, E.J.; Selman, B.R. Isolation and characterization of a Chlamydomonas reinhardtii mutant lacking the gamma-subunit of chloroplast coupling factor 1 (CF1). Mol. Cell Biol. 1991, 11, 5053–5058. [Google Scholar] [CrossRef]
| Mutation | Affected Genes | Mutation | Notes | Reference |
|---|---|---|---|---|
| rbcL-G54D (31-4E) rbcL-W451Opal (18-5B) | rbcL | G54D W451-Opal | Unstable LSU | Thow and Spreitzer, 1992 Spreitzer et al., 1985 |
| RBCS-T60 | RBCS1 (Cre02.g120100) | deletion RBCS1, RBCS2 and STT7 (?) | No SSu, leading to inhibition of LSU translation (CES process) | Khrebtukova and Spreitzer, 1996 |
| CAL13 (CAL005_01_13) | RBCS2 (Cre02.g120150) | deletion chromosome_2:6,908,477- 6.943.599 | Dent et al., 2015 | |
| clpP1-AUU | clpP1 | start (AUG)-->AUU | 25% accumulation of ClpP complex | Majeran et al., 2000 |
| Mrl1-5 | MRL1 (Cre06.g298300) | ? | No rbcL mRNA accumulation | Johnson, 2011 |
| Tbc1-F34 | TBC1 (?) | ? | no PscbC (CP43) translation | Rochaix, et al., 1989 |
| Tdal-F54 | TDA1 (Cre08.g358350) | W780-Opal | No atpA translation initiation leading to decreased β-CF1 translation (CES process) | Lemaire and Wollman, 1989, Eberhard, et al., 2011 |
| Mdb1-thm24 | MDB1 (Cre14.g614550) | unknown | No atpB mRNA accumulation and inhibition of α-CF1 translation (CES) | Drapier et al., 1992, unpublished |
| atpA-FUD16 | atpA | I184N and N186Y | aggregation of α-CF1 and β-CF1 in the stroma, forming inclusion bodies | Ketchner et al., 1995 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majeran, W.; Wostrikoff, K.; Wollman, F.-A.; Vallon, O. Role of ClpP in the Biogenesis and Degradation of RuBisCO and ATP Synthase in Chlamydomonas reinhardtii. Plants 2019, 8, 191. https://doi.org/10.3390/plants8070191
Majeran W, Wostrikoff K, Wollman F-A, Vallon O. Role of ClpP in the Biogenesis and Degradation of RuBisCO and ATP Synthase in Chlamydomonas reinhardtii. Plants. 2019; 8(7):191. https://doi.org/10.3390/plants8070191
Chicago/Turabian StyleMajeran, Wojciech, Katia Wostrikoff, Francis-André Wollman, and Olivier Vallon. 2019. "Role of ClpP in the Biogenesis and Degradation of RuBisCO and ATP Synthase in Chlamydomonas reinhardtii" Plants 8, no. 7: 191. https://doi.org/10.3390/plants8070191
APA StyleMajeran, W., Wostrikoff, K., Wollman, F.-A., & Vallon, O. (2019). Role of ClpP in the Biogenesis and Degradation of RuBisCO and ATP Synthase in Chlamydomonas reinhardtii. Plants, 8(7), 191. https://doi.org/10.3390/plants8070191





