New Insight into HPts as Hubs in Poplar Cytokinin and Osmosensing Multistep Phosphorelays: Cytokinin Pathway Uses Specific HPts
Abstract
1. Introduction
2. Results
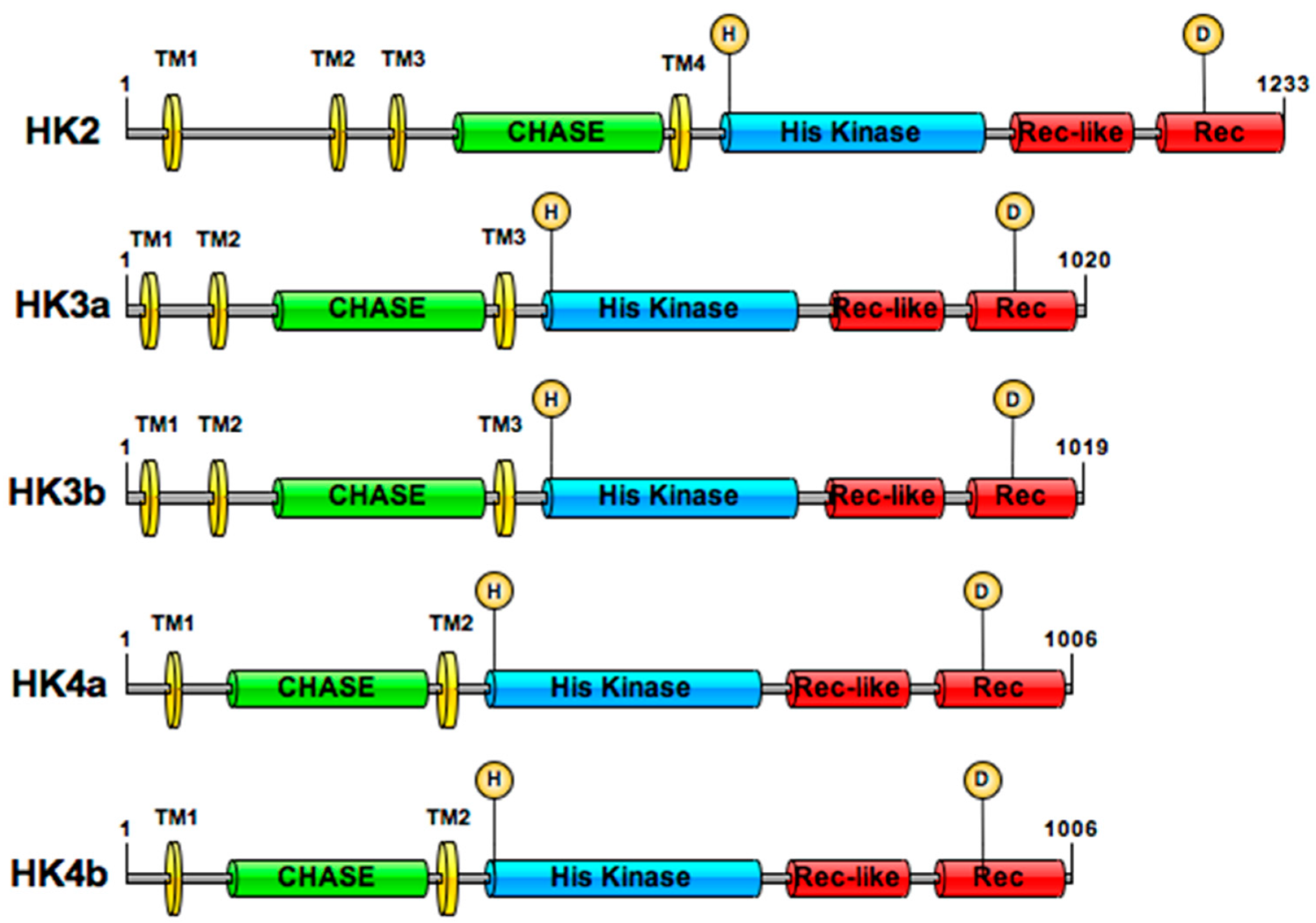
2.1. Isolation of Five HKs from the Poplar Dorskamp Genotype
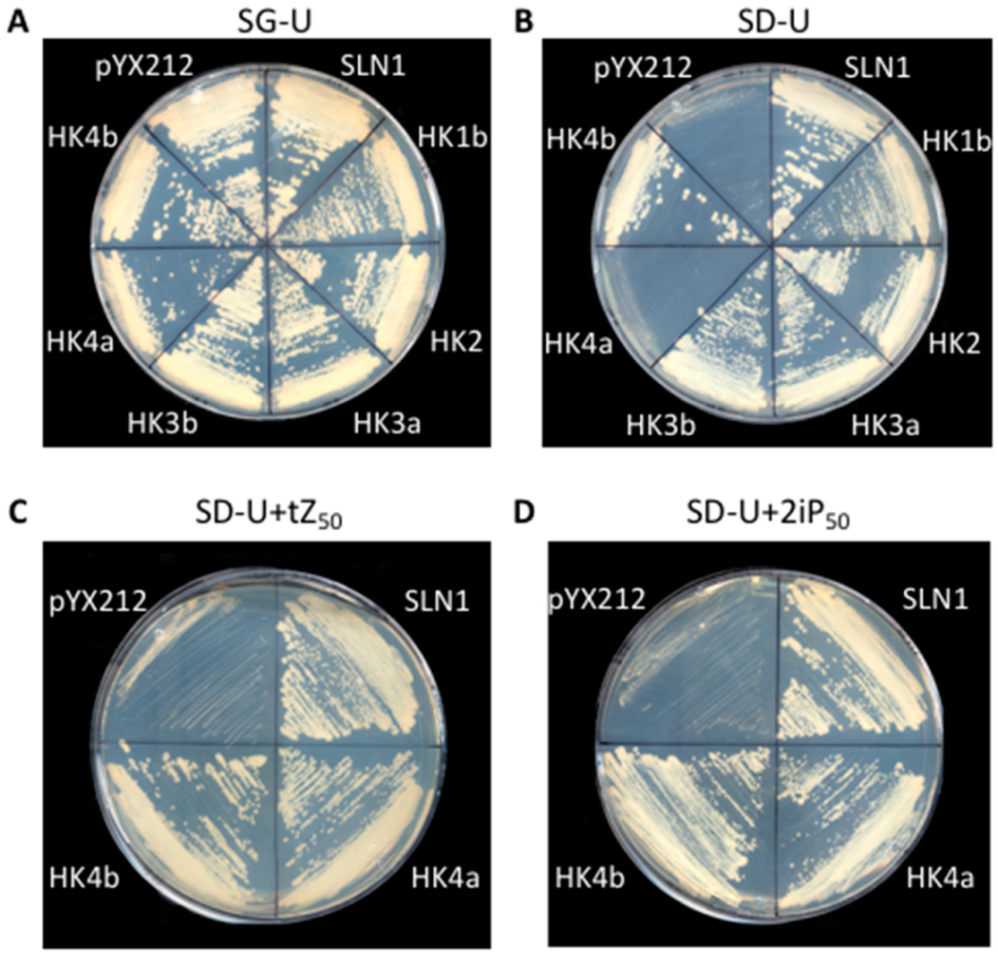
2.2. Isolated Poplar CK HK Sequences Are Functional HKs
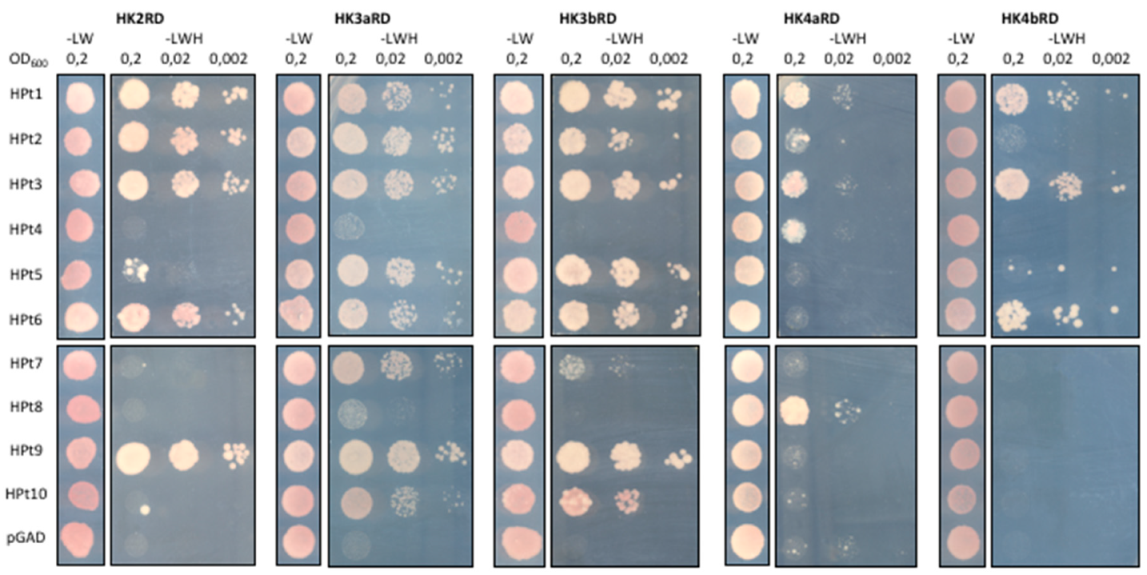
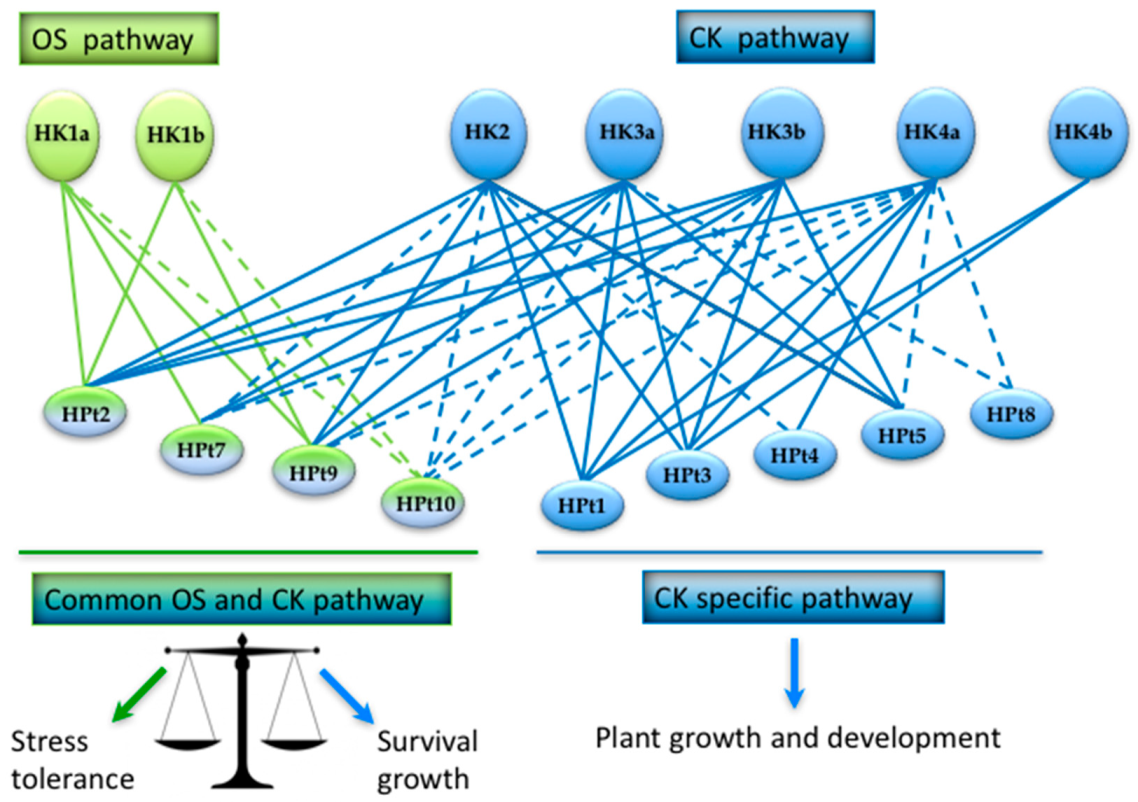
2.3. Identification of HPt Partners of CK HKs in Y2H Assay
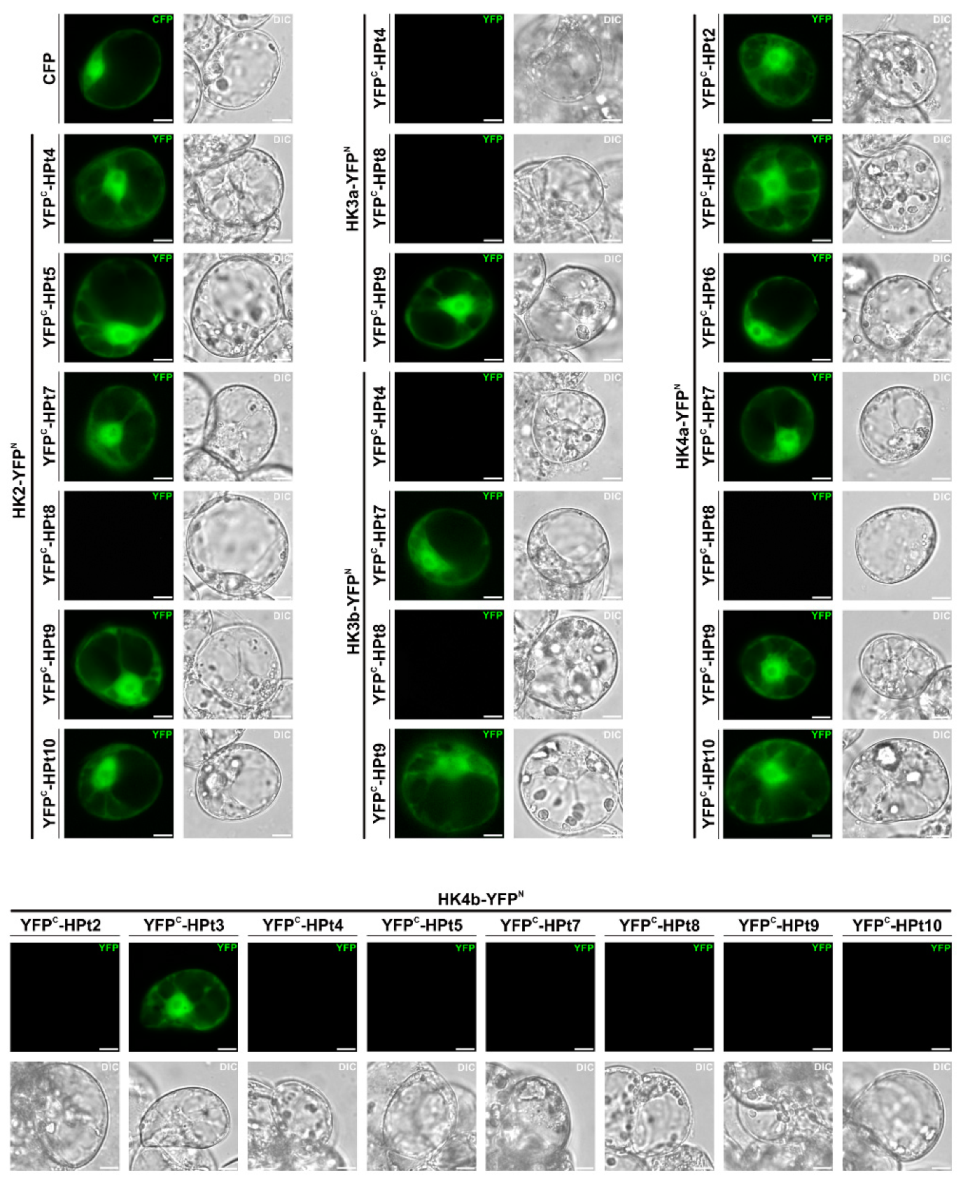
2.4. Validation of HPt Partners of CK HKs in Planta
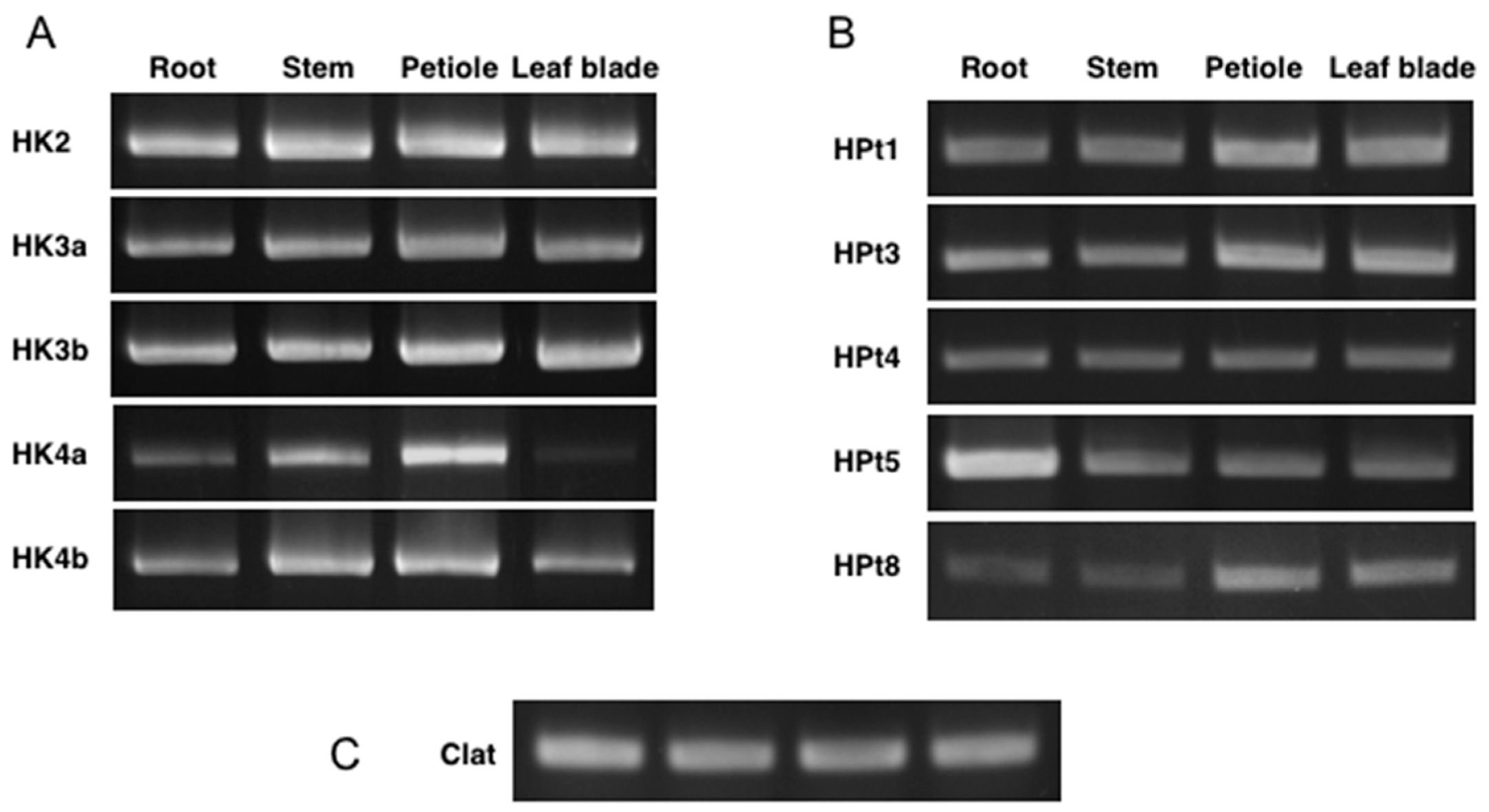
2.5. Expression of CK HK and HPt Genes
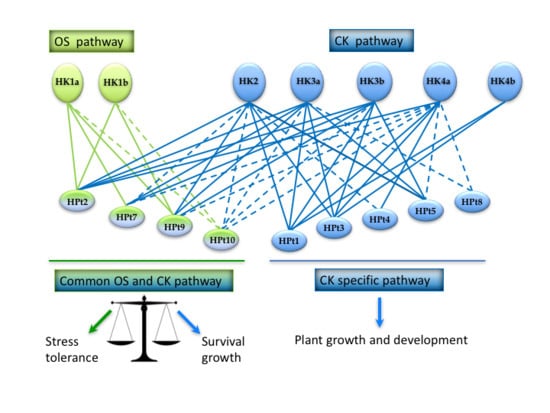
3. Discussion
4. Materials and Methods
4.1. Isolation of CK HK CDSs
4.2. Complementation Analysis of the sln1Δ sho1Δ Deletion Mutant MH179
4.3. Yeast Two-Hybrid Tests
4.4. BiFC Assays
4.5. Expression of HK and HPt Genes
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Sheen, J.; Müller, B. Cytokinin Signaling Networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012, 17, 172–179. [Google Scholar] [CrossRef]
- Zwack, P.J.; Rashotte, A.M. Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 2015, 66, 4863–4871. [Google Scholar] [CrossRef]
- Pavlù, J.; Novák, J.; Koukalová, V.; Luklová, M.; Brzobohaty, B.; Cerny, M. Cytokinin at the crossroads of abiotic stress signalling pathways. Int. J. Mol. Sci. 2018, 19, 2050. [Google Scholar] [CrossRef]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Shinozaki, K.; Yamagushi-Shinozaki, K. Role of cytokinin two-component system in ABA and osmotic stress signalings. Plant Signal Behav. 2010, 5, 148–150. [Google Scholar] [CrossRef]
- Nongpiur, R.; Soni, P.; Karan, R.; Singla-Pareek, S.L.; Pareek, A. Histidine kinases in plants: Cross talk between hormone and stress responses. Plant Signal Behav. 2012, 7, 1230–1237. [Google Scholar] [CrossRef]
- Riemann, M.; Dhakarey, R.; Hazman, M.; Miro, B.; Kohli, A.; Nick, P. Exploring jasmonates in the hormonal network of drought and salinity responses. Front. Plant Sci. 2015, 6, 1077. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Lata, C.; Chauhan, P.S.; Prasad, V.; Prasad, M. A functional genomic perspective on drought signalling and its crosstalk with phytohormone-mediated signalling pathways in plants. Curr. Genom. 2017, 18, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Herrera-Estrella, L.; Tran, L.-S.P. Do cytokinins and strigolactones crosstalk during drought adaptation? Trends Plant Sci. 2019, 24, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Herrera-Estrella, L.; Tran, L.-S.P. The Yin-Yang of cytokinin homeostasis and drought acclimation/adaptation. Trends Plant Sci. 2016, 21, 548–550. [Google Scholar] [CrossRef] [PubMed]
- Haswell, E.S.; Verslues, P.E. The ongoing search for the molecular basis of plant osmosensing. J. Gen. Physiol. 2015, 145, 389–394. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Urao, T.; Yakubov, B.; Satoh, R.; Yamaguchi-Shinozaki, K.; Seki, M.; Hirayama, T.; Shinozaki, K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 1999, 11, 1743–1754. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamagushi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abcissic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef]
- Wohlbach, D.J.; Quirino, B.F.; Sussman, M.R. Analysis of the Arabidopsis Histidine Kinase ATHK1 Reveals a Connection between Vegetative Osmotic Stress Sensing and Seed Maturation. Plant Cell 2008, 20, 1101–1117. [Google Scholar] [CrossRef]
- Heyl, A.; Riefler, M.; Romanov, G.A.; Schmülling, T. Properties, functions and evolution of cytokinin receptors. Eur. J. Cell Biol. 2012, 91, 246–256. [Google Scholar] [CrossRef]
- Dortay, H.; Mehnert, N.; Bürkle, L.; Schmülling, T.; Heyl, A. Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J. 2006, 273, 4631–4644. [Google Scholar] [CrossRef] [PubMed]
- Urao, T.; Miyata, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 2000, 478, 227–232. [Google Scholar] [CrossRef]
- Chefdor, F.; Bénédetti, H.; Depierreux, C.; Delmotte, F.; Morabito, D.; Carpin, S. Osmotic stress sensing in Populus: Components identification of a phosphorelay system. FEBS Lett. 2006, 580, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Héricourt, F.; Chefdor, F.; Djeghdir, I.; Larcher, M.; Lafontaine, F.; Courdavault, V.; Auguin, D.; Coste, F.; Depierreux, C.; Tanigawa, M.; et al. Functional divergence of poplar histidine-aspartate kinase HK1 paralogs in response to osmotic stress. Int. J. Mol. Sci. 2016, 17, 2061. [Google Scholar] [CrossRef]
- Héricourt, F.; Chefdor, F.; Bertheau, L.; Tanigawa, M.; Maeda, T.; Guirimand, G.; Courdavault, V.; Larcher, M.; Depierreux, C.; Bénédetti, H.; et al. Characterization of histidine-aspartate kinase HK1 and identification of Histidine Phosphotransfer proteins as potential partners in a Populus multistep phosphorelay. Physiol. Plant 2013, 149, 188–199. [Google Scholar] [CrossRef]
- Chefdor, F.; Héricourt, F.; Koudounas, K.; Carqueijeiro, I.; Courdavault, V.; Mascagni, F.; Bertheau, L.; Larcher, M.; Depierreux, C.; Lamblin, F.; et al. Highlighting type A RRs as potential regulators of the dkHK1 multi-step phosphorelay pathway in Populus. Plant Sci. 2018, 277, 68–78. [Google Scholar] [CrossRef]
- Bertheau, L.; Chefdor, F.; Guirimand, G.; Courdavault, V.; Depierreux, C.; Morabito, D.; Brignolas, F.; Héricourt, F.; Carpin, S. Identification of five B-type response regulators as members of a multistep phosphorelay system interacting with histidine-containing phosphotransfer partners of Populus osmosensor. BMC Plant Biol. 2012, 12, 241. [Google Scholar] [CrossRef]
- Bertheau, L.; Miranda, M.; Foureau, E.; Fernanda, L.; Hoyos, R.; Chefdor, F.; Héricourt, F.; Depierreux, C.; Morabito, D.; Papon, N.; et al. In planta validation of HK1 homodimerization and recruitment of preferential HPt downstream partners involved in poplar multistep phosphorelay systems. Plant Biosyst. 2013, 147, 991–995. [Google Scholar] [CrossRef]
- Bertheau, L.; Djeghdir, I.; Foureau, E.; Chefdor, F.; Glévarec, G.; Oudin, A.; Depierreux, C.; Morabito, D.; Brignolas, F.; Courdavault, V.; et al. Insights into B-type RR members as signaling partners actiong downstream of HPt partners of HK1 in the osmotic stress response in Populus. Plant Physiol. Biochem. 2015, 94, 244–252. [Google Scholar] [CrossRef]
- Pils, B.; Heyl, A. Unraveling the evolution of cytokinin signaling. Plant Physiol. 2009, 151, 782–791. [Google Scholar] [CrossRef]
- Immanen, J.; Nieminen, K.; Silva, H.D.; Rojas, F.R.; Meisel, L.A.; Silva, H.; Albert, V.A.; Hvidsten, T.R.; Helariutta, Y. Characterization of cytokinin signaling and homeostasis gene families in two hardwood tree species: Populus trichocarpa and Prunus persica. BMC Genom. 2013, 14, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, K.; Immanen, J.; Laxell, M.; Kauppinen, L.; Tarkowski, P.; Dolezal, K.; Tähtiharju, S.; Elo, A.; Decourteix, M.; Ljung, K.; et al. Cytokinin signalling regulates cambial development in poplar. Proc. Natl. Acad. Sci. USA 2008, 150, 20032–20037. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.; et al. The Arabidopsis Histidine Phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Watanabe, Y.; Levya-Gonzalez, M.A.; Ha, C.V.; Fujita, Y.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Herrera-Estrella, L.; et al. Arabidopsis AHP2, AHP3 and AHP5 histidine phosphotranfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. USA 2013, 110, 4840–4845. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.E.; Kieber, J.J. Signaling via Histidine-containing Phosphotransfer proteins in Arabidopsis. Plant Signal Behav. 2007, 2, 287–289. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Daudu, D.; Allion, E.; Liesecke, F.; Papon, N.; Courdavault, V.; Dugé de Bernonville, T.; Mélin, C.; Oudin, A.; Clastre, M.; Lanoue, A.; et al. CHASE-containing histidine kinase receptors in apple tree: From a common receptor structure to divergent cytokinin binding properties and specific functions. Front. Plant Sci. 2017, 8, 1614. [Google Scholar] [CrossRef]
- Grefen, C.; Harter, K. Plant two-component systems: Principles, functions, complexity and cross talk. Planta 2004, 219, 733–742. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. The perception of cytokinin: A story 50 years in the making. Plant Physiol. 2010, 154, 487–492. [Google Scholar] [CrossRef]
- Pekárová, B.; Klumpler, T.; Třísková, O.; Horák, J.; Jansen, S.; Dopitová, R.; Borkovcová, P.; Papoušková, V.; Nejedlá, E.; Sklenář, V.; et al. Structure and binding specificity of the receiver domain of sensor histidine kinase CKI1 from Arabidopsis thaliana. Plant J. 2011, 67, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Wurgler-Murphy, S.M.; Saito, H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 1994, 369, 42–245. [Google Scholar] [CrossRef] [PubMed]
- Posas, F.; Wurgley-Murphy, S.M.; Maeda, T.; Witten, E.A.; Thai, T.C.; Saito, H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 1996, 86, 865–875. [Google Scholar] [CrossRef]
- Persinova, M.; Grochova, M.; Konecny, T.; Plackova, L.; Harustiakova, D.; Kakimoto, T.; Heisler, M.G.; Novak, O.; Hejatko, J. Cytokinin signalling regulates organ identity via the AHK4 receptor in Arabidopsis. Development 2018, 145, dev163907. [Google Scholar]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Jaworek, P.; Kopecny, D.; Zalabak, D.; Sebela, M.; Kouril, S.; Hluska, T.; Koncitikova, K.; Tarkowski, P. Occurence and biosynthesis of cytokinins in poplar. Planta 2019, 250, 229–244. [Google Scholar] [CrossRef]
- Jaworek, P.; Tarkowski, P.; Hluska, T.; Kouril, S.; Vrobel, O.; Nisler, J.; Kopecny, D. Characterization of five CHASE-containing histidine kinase receptors from Populus × canadensis cv. Robusta sensing isoprenoid and aromatic cytokinins. Planta 2020, 251, 1. [Google Scholar] [CrossRef]
- Cutcliffe, J.W.; Hellmann, E.; Heyl, A.; Rashotte, A.M. CRFs form protein-protein interactions with each other and with members of the cytokinin signalling pathway in Arabidopsis via the CRF domain. J. Exp. Bot. 2011, 62, 4995–5002. [Google Scholar] [CrossRef]
- Mira-Rodado, V.; Veerabagu, M.; Witthöft, J.; Teply, J.; Harter, K.; Desikan, R. Identification of two-component system elements downstream of AHK5 in the stomatal closure response of Arabidopsis thaliana. Plant Sig. Behav. 2012, 7, 1–10. [Google Scholar] [CrossRef]
- Yamada, H.; Koizumi, N.; Nakamichi, N.; Kiba, T.; Yamashino, T.; Mizuno, T. Rapide response of Arabidopsis T87 cultured celles to cytokinin through His-to-Asp phosphorelay signal transduction. Biosci. Biotechnol. Biochem. 2004, 68, 1966–1976. [Google Scholar] [CrossRef]
- Jung, K.W.; Oh, S.-I.; Kim, Y.Y.; Yoo, K.S.; Cui, M.H.; Shin, J.S. Arabidopsis histidine-containing phosphotransfer factor 4 (AHP4) negatively regulates secondary wall thickening of the anther endothecium during flowering. Mol. Cells 2008, 25, 294–300. [Google Scholar] [PubMed]
- Ueguchi, C.; Koizumi, H.; Suzuki, T.; Mizuno, T. Novel Family of Sensor Histidine Kinase Genes in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Debellé, F.; Gamas, P.; Frugier, F.; Brault, M. Diversification of cytokinin phosphotransfer signalling genes in Medicago truncatula and other legume genomes. BMC Genom. 2019, 20, 373. [Google Scholar] [CrossRef] [PubMed]
- Pekárová, B.; Szmitkowska, A.; Dopitová, R.; Degtjarik, O.; Zídek, L.; Hejátko, J. Structural aspects of multistep phosphorelay-mediated signalling in plants. Mol. Plant 2016, 9, 71–85. [Google Scholar] [CrossRef]
- Sharan, A.; Soni, P.; Nongpiur, R.C.; Singla-Pareek, S.L.; Pareek, A. Mapping the “Two-component system” network in rice. Sci. Rep. 2017, 7, 9287. [Google Scholar] [CrossRef]
- Zdarska, M.; Cuyacot, A.R.; Tarr, P.T.; Yamoune, A.; Szmitkowska, A.; Hrdinová, V.; Gelová, Z.; Meyerowitz, E.M.; Hejátko, J. ETR1 intergrates responses to ethylene and cytokinins into a single multistep phosphorelay pathway to control root growth. Mol. Plant 2019, 12, 1338–1352. [Google Scholar] [CrossRef]
- Hejátko, J.; Ryu, H.; Kim, G.-T.; Dobesová, R.; Choi, S.; Choi, S.M.; Soucek, P.; Horák, J.; Pekárová, B.; Palme, K.; et al. The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell 2009, 21, 2008–2021. [Google Scholar] [CrossRef]
- Deng, Y.; Dong, H.; Mu, J.; Ren, B.; Zheng, B.; Ji, Z.; Yang, W.-C.; Liang, Y.; Zuo, J. Arabidopsis histidine kinase CKI1 acts upstream of HISTIDINE PHOSPHOTRANSFER PROTEINS to regulate female gametophyte development and vegetative growth. Plant Cell 2010, 22, 1232–1248. [Google Scholar] [CrossRef]
- Desikan, R.; Horák, J.; Chaban, C.; Mira-Rodado, V.; Witthöft, J.; Elgass, K.; Grefen, C.; Cheung, M.-K.; Meixner, A.J.; Hooley, R.; et al. The Histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS ONE 2008, 3, e2491. [Google Scholar] [CrossRef]
- Caesar, K.; Thamm, A.M.K.; Witthöft, J.; Elgass, K.; Huppenberger, P.; Grefen, C.; Horak, J.; Harter, K. Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J. Exp. Bot. 2011, 62, 5571–5580. [Google Scholar] [CrossRef]
- Grefen, C.; Städele, K.; Ruzicka, K.; Obrdlik, P.; Harter, K.; Horak, J. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 2008, 1, 308–320. [Google Scholar] [CrossRef]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztányi, Z.; El-Gebali, S.; Fraser, M. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed]
- Manivasakam, P.; Weber, S.C.; McElver, J.; Schiestl, R.H. Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 1995, 23, 2799–2800. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. Transforming yeast with DNA. Method Mol. Cell Biol. 1995, 5, 255–269. [Google Scholar]
- Waadt, R.; Schmidt, L.; Lohse, M.; Hashimoto, K.; Bock, R.; Kudla, J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008, 56, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Guirimand, G.; Courdavault, V.; Lanoue, A.; Mahroug, S.; Guihur, A.; Blanc, N.; Giglioli-Guivarc’h, N.; St-Pierre, B.; Burlat, V. Strictosidine activation in Apocynaceae: Towards a “nuclear time bomb”? BMC Plant Biol. 2010, 10, 82. [Google Scholar] [CrossRef]
- Guirimand, G.; Burlat, V.; Oudin, A.; Lanoue, A.; St-Pierre, B.; Courdavault, V. Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Rep. 2009, 28, 1215–1234. [Google Scholar] [CrossRef]
- Foureau, E.; Carqueijeiro, I.; Dugé de Bernonville, T.; Melin, C.; Lafontaine, F.; Besseau, S.; Lanoue, A.; Papon, N.; Oudin, A.; Glévarec, G.; et al. Prequels to Synthetic Biology: From Candidate Gene Identification and Validation to Enzyme Subcellular Localization in Plant and Yeast Cells. Methods Enzymol. 2016, 576, 167–206. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Héricourt, F.; Larcher, M.; Chefdor, F.; Koudounas, K.; Carqueijeiro, I.; Lemos Cruz, P.; Courdavault, V.; Tanigawa, M.; Maeda, T.; Depierreux, C.; et al. New Insight into HPts as Hubs in Poplar Cytokinin and Osmosensing Multistep Phosphorelays: Cytokinin Pathway Uses Specific HPts. Plants 2019, 8, 591. https://doi.org/10.3390/plants8120591
Héricourt F, Larcher M, Chefdor F, Koudounas K, Carqueijeiro I, Lemos Cruz P, Courdavault V, Tanigawa M, Maeda T, Depierreux C, et al. New Insight into HPts as Hubs in Poplar Cytokinin and Osmosensing Multistep Phosphorelays: Cytokinin Pathway Uses Specific HPts. Plants. 2019; 8(12):591. https://doi.org/10.3390/plants8120591
Chicago/Turabian StyleHéricourt, François, Mélanie Larcher, Françoise Chefdor, Konstantinos Koudounas, Inês Carqueijeiro, Pamela Lemos Cruz, Vincent Courdavault, Mirai Tanigawa, Tatsuya Maeda, Christiane Depierreux, and et al. 2019. "New Insight into HPts as Hubs in Poplar Cytokinin and Osmosensing Multistep Phosphorelays: Cytokinin Pathway Uses Specific HPts" Plants 8, no. 12: 591. https://doi.org/10.3390/plants8120591
APA StyleHéricourt, F., Larcher, M., Chefdor, F., Koudounas, K., Carqueijeiro, I., Lemos Cruz, P., Courdavault, V., Tanigawa, M., Maeda, T., Depierreux, C., Lamblin, F., Glévarec, G., & Carpin, S. (2019). New Insight into HPts as Hubs in Poplar Cytokinin and Osmosensing Multistep Phosphorelays: Cytokinin Pathway Uses Specific HPts. Plants, 8(12), 591. https://doi.org/10.3390/plants8120591