Genetic Diversity, Population Structure, and Marker-Trait Association for Drought Tolerance in US Rice Germplasm
Abstract
1. Introduction
2. Results
2.1. Assessment of Drought Tolerance Variability in US Rice Germplasm
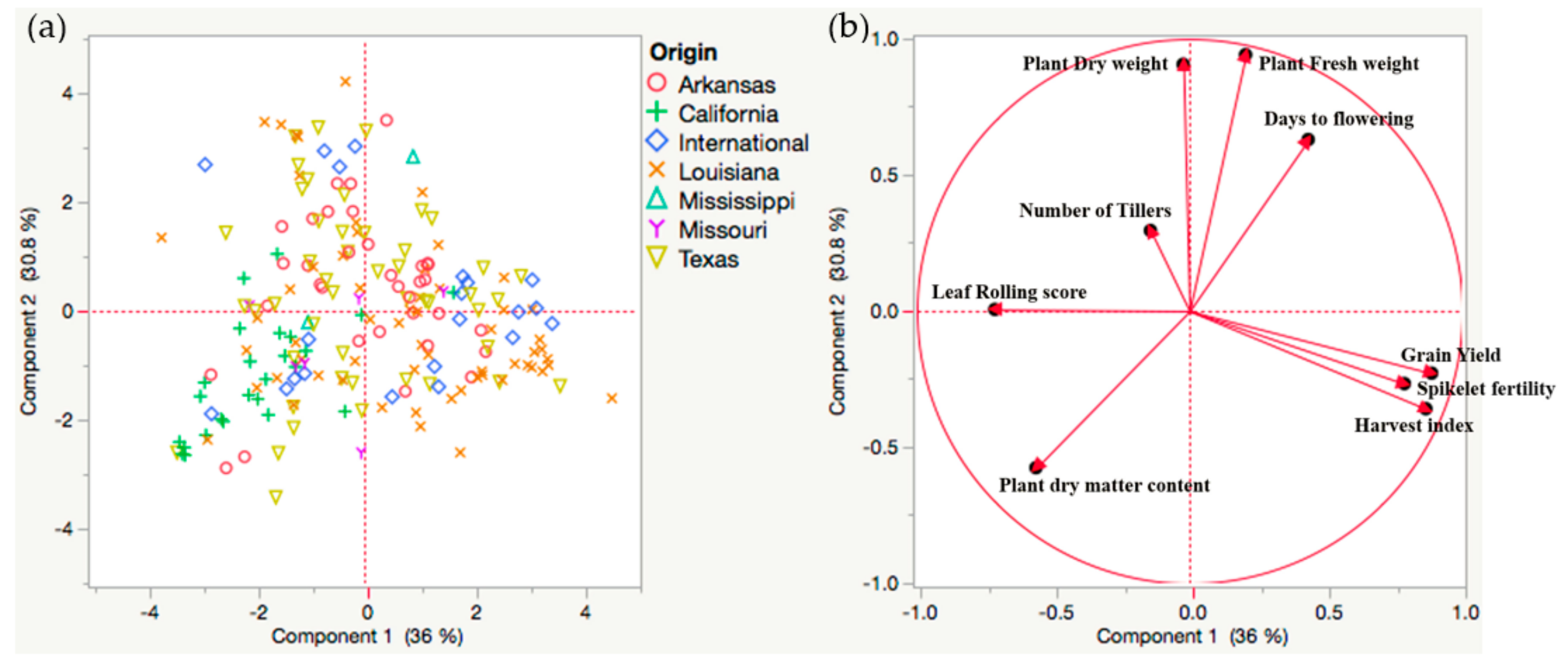
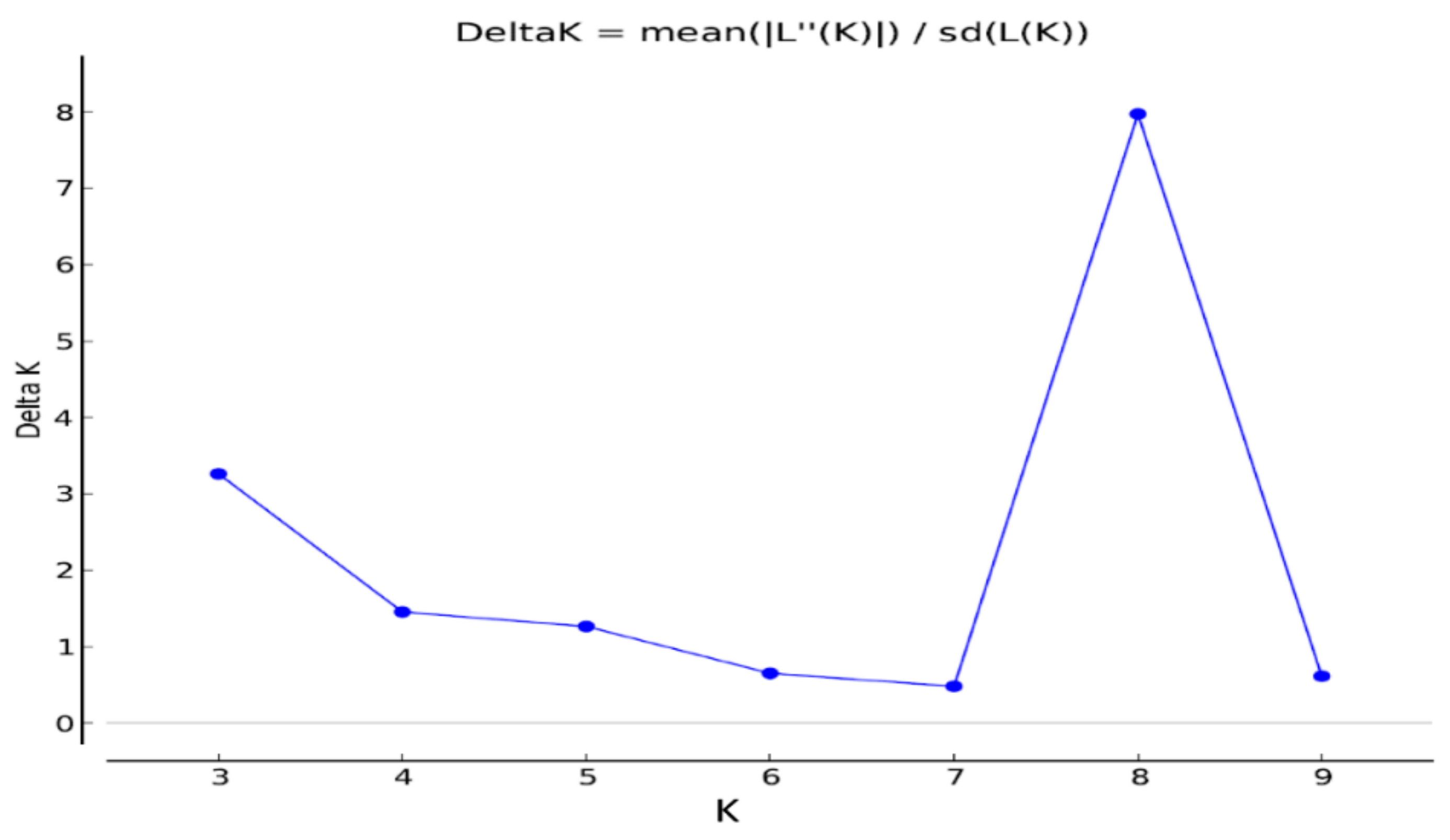
2.2. Genetic Variability in US Rice Germplasm
2.3. Marker Trait Associations
3. Discussion
4. Methods
4.1. Plant Materials and Drought Tolerance Screening
4.2. Genotyping
4.3. Statistical Analysis
4.4. Genetic Diversity and Population Structure Analysis
4.5. Marker-Trait Associations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Howitt, R.; McEwan, D.; Azuara, J.M.; Lund, J.; Sumner, D. Economic Analysis of the 2015 for California Agriculture; UC Davis Center for Watershed Science; California Department of Food and Agriculture University of California—Davis California: Davis, CA, USA, 2015. [Google Scholar]
- F.A.O. Corporate Document Repository. Sustainable Rice Production for Food Security. In Proceedings of the 20th Session of International Rice Commission, Bangkok, Thailand, 23–25 July 2002. [Google Scholar]
- Islam, M.Z.; Khalequzzaman, M.; Prince, M.F.R.K.; Siddique, M.A.; Rashid, E.S.M.H.; Ahmed, M.S.U.; Pittendrigh, B.R.; Ali, M.P. Diversity and population structure of red rice germplasm in Bangladesh. PLoS ONE 2018, 13, e0196096. [Google Scholar] [CrossRef] [PubMed]
- Kamoshita, A.; Babu, R.C.; Boopathi, M.; Fukai, S. Phenotypic and genotypic analysis of drought-resistance traits for development of rice cultivars adapted to rainfed environments. Field Crops Res. 2008, 109, 1–23. [Google Scholar] [CrossRef]
- Das, B.; Sengupta, S.; Parida, S.K.; Roy, B.; Gosh, M.; Prasad, M.; Ghose, T.K. Genetic diversity and population structure of rice landraces from Eastern and North-Eastern states of India. BMC Genet. 2013, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Anandan, A.; Anumalla, M.; Pradhan, S.K.; Ali, J. Population structure, genetic diversity, and trait association analysis in rice (Oryza sativa L.) germplasm for early seedling vigor (ESV) using trait linked SSR markers. PLoS ONE 2016, 11, e0152406. [Google Scholar] [CrossRef]
- Tabanao, D.A.; Pocsedio, A.E.; Yabes, J.C.; Nino, M.C.; Millas, R.A.; Sevilla, N.R.L.; Yulong, Z.; Yu, J. Genetic diversity and population structure in a rice drought stress panel. Plant Genet. Resour. 2014, 13, 195–205. [Google Scholar] [CrossRef]
- Xu, Y.; Beachell, H.; McCouch, S.R. A marker-based approach to broadening the genetic base of rice in the USA. Crop Sci. 2004, 44, 1947–1959. [Google Scholar] [CrossRef]
- Lu, H.; Redus, M.A.; Coburn, J.R.; Rutger, N.; McCouch, S.R.; Tai, T.H. Population structure and breeding patterns of 145 U.S. rice cultivars based on SSR marker analysis. Crop Sci. 2005, 45, 66–76. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Ndjiondjop, M.; Cisse, F.; Futakuchi, K.; Lorieux, M.; Manneh, B.; Bocco, R.; Fatondji, B. Effect of drought on rice (Oryza spp.) genotypes according to their drought tolerance level. In Innovation and Partnerships to Realize Africa’s Rice Potential; Seck, P.A., General, D., Eds.; Second Africa Rice Congress: Bamako, Mali, 2010. [Google Scholar]
- Sandhu, N.; Kumar, A. Bridging the rice yield gaps under drought: QTLs, genes and their use in breeding programs. Agronomy 2017, 7, 27. [Google Scholar] [CrossRef]
- Bhattarai, U.; Subudhi, P. Genetic analysis of yield and agronomic traits under reproductive-stage drought stress in rice using a high-resolution linkage map. Gene 2018, 669, 69–76. [Google Scholar] [CrossRef]
- Prince, S.J.; Beena, R.; Gomez, S.M.; Senthivel, S.; Babu, R.C. Mapping consistent rice (Oryza sativa L.) yield QTLs under drought stress in target rainfed environments. Rice 2015, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, U.; Subudhi, P. Identification of drought responsive QTLs during vegetative growth stage of rice using a saturated GBS-based SNP linkage map. Euphytica 2018, 214, 38. [Google Scholar] [CrossRef]
- Vikram, P.; Swamy, B.P.M.; Dixit, S.; Ahmed, H.U.; Cruz, M.T.S.; Singh, A.K.; Kumar, A. qDTY1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet. 2011, 12, 89. [Google Scholar] [CrossRef] [PubMed]
- Swamy, B.P.M.; Shamsudin, N.A.A.; Rahman, S.N.A.; Mauleon, R.; Ratnam, W.; Cruz, M.T.S.; Kumar, A. Association mapping of yield and yield related traits under reproductive stage drought stress in rice (Oryza sativa L.). Rice 2017, 10, 21. [Google Scholar] [CrossRef]
- Dixit, S.; Huang, B.E.; Cruz, M.T.S.; Maturan, P.T.; Ontoy, J.C.E.; Kumar, A. QTLs for tolerance of drought and breeding for tolerance of abiotic and biotic stress: An integrated approach. PLoS ONE 2014, 9, e109574. [Google Scholar] [CrossRef]
- Kumar, A.; Dixit, S.; Ram, T.; Yadaw, R.B.; Mishra, K.K.; Mandal, N.P. Breeding high-yielding drought-tolerant rice: Genetic variations and conventional and molecular approaches. J. Exp. Bot. 2014, 65, 6265–6278. [Google Scholar] [CrossRef]
- Singh, B.; Reddy, K.R.; Redona, E.D.; Walker, T. Screening of rice cultivars for morpho-physiological responses to early-season soil moisture stress. Rice Sci. 2017, 24, 322–335. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Gregorio, G.; Subudhi, P.K. Genetic variation in southern USA rice genotypes for seedling salinity tolerance. Front. Plant Sci. 2015, 6, 374. [Google Scholar] [CrossRef]
- Nounjan, N.; Chansongkrow, P.; Charoensawan, V.; Siangliw, J.L.; Toojinda, T.; Chadchawan, S.; Theerakulpisut, P. High performance of photosynthesis and osmotic adjustment are associated with salt tolerance ability in rice carrying drought tolerance QTL; physiological and co-expression network analysis. Front. Plant Sci. 2018, 9, 1135. [Google Scholar] [CrossRef]
- Zhang, P.; Li, J.; Li, X.; Liu, X.; Zhao, X.; Lu, Y. Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoS ONE 2011, 6, e2756. [Google Scholar]
- Courtois, B.; Frouin, J.; Greco, R.; Bruschi, G.; Droc, G.; Hamelin, C.; Ruiz, M.; Clement, G.; Evrard, J.C.; Coppenole, S.V.; et al. Genetic diversity and population structure in a European collection of rice. Crop Sci. 2012, 52, 1163–1675. [Google Scholar] [CrossRef]
- Nachimuthu, V.V.; Muthurajan, R.; Duraialaguraja, S.; Sivakami, R.; Pandin, B.A.; Ponniah, G.; Gunasekaran, K.; Swaminathan, M.; Suji, K.K.; Sabariappan, R. Analysis of population structure and genetic diversity in rice germplasm using SSR markers: An initiative towards association mapping of agronomic traits in Oryza sativa. Rice 2015, 8, 30. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Barik, S.R.; Sahoo, A.; Mohapatra, S.; Nayak, D.K.; Mahender, A.; Mehar, J.; Anandan, A.; Pandit, E. Population structure, genetic diversity and molecular marker-trait association analysis for high temperature stress tolerance in rice. PLoS ONE 2016, 11, e0160027. [Google Scholar] [CrossRef]
- Shivrain, V.K.; Burgos, N.R.; Agrama, H.A.; Rauth, A.L.; Lu, B.; Sales, M.A.; Boyett, V.; Gealy, D.R.; Moldenhauer, K. Genetic diversity of weedy red rice (Oryza sativa) in Arkansas, USA. Weed Res. 2010, 50, 289–302. [Google Scholar] [CrossRef]
- Vikram, P.; Swamy, B.P.M.; Dixit, S.; Trinidad, J.; Cruz, M.T.S.; Maturan, P.C.; Amante, M.; Kumar, A. Linkages and interactions analysis of major effect drought grain yield QTLs in rice. PLoS ONE 2016, 11, e0151532. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Ashikar, M.; Yamanouchi, U.; Sasaki, T.; Yano, M. Identification and characterization of a quantitative trait locus, Hd9, controlling heading date in rice. Breed Sci. 2002, 52, 35–41. [Google Scholar] [CrossRef][Green Version]
- Ramchander, S.; Raveendran, M.; Robin, S. Mapping QTLs for physiological traits associated with drought tolerance in rice (Oryza sativa L.). J. Investig Genom. 2016, 3, 00052. [Google Scholar]
- Venuprasad, R.; Bool, M.E.; Quiatchon, L.; Atlin, G.N. A QTL for rice grain yield in aerobic environments with large effects in three genetic backgrounds. Theor. Appl. Genet. 2012, 124, 323–332. [Google Scholar] [CrossRef]
- International Rice Research Institute (IRRI). Standard Evaluation System for Rice (SES); IRRI: Los Baños, Philippines, 2002; Available online: http://www.knowledgebank.irri.org/images/docs/rice-standard-evaluation-system.pdf (accessed on 10 January 2017).
- Chen, D.H.; Ronald, P.C. A rapid DNA minipreparation method suitable for AFLP and other applications. Plant Mol. Biol. Rep. 1999, 17, 53–57. [Google Scholar] [CrossRef]
- SAS Institute Inc. Base SAS 9.3 Procedures Guide; SAS Institute Inc: Cary, NC, USA, 2011. [Google Scholar]
- SAS Institute Inc. Using JMP11; SAS Institute Inc: Cary, NC, USA, 2013. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Vonholdt, B.M. Structure harvester: A website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemound-collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/darwin (accessed on 15 August 2017).
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]

| Traits | Mean | Range | R-square a |
|---|---|---|---|
| Days to heading | 74.9 | 62–88 | 0.94 |
| Number of tillers | 4.0 | 2.5–7 | 0.69 |
| Leaf rolling score b | 6.2 | 3–9 | 0.73 |
| Shoot fresh weight (g/plant) | 86.4 | 29.2–230.4 | 0.88 |
| Shoot dry weight (g/plant) | 38.8 | 25–93.5 | 0.91 |
| Shoot dry matter content (%) | 47.9 | 13.9–90.0 | 0.76 |
| Spikelet fertility (%) | 30.4 | 0.1–90.0 | 0.88 |
| Grain yield (g/plant) | 5.7 | 0.1–46.3 | 0.87 |
| Harvest index | 0.15 | 0.001–0.62 | 0.88 |
| DTH | NT | LRS | SFW | SDW | SDMC | SF | GY | HI | |
|---|---|---|---|---|---|---|---|---|---|
| DTH | 1.00 | −0.06 | −0.19 ** | 0.55 ** | 0.53 ** | −0.39 ** | 0.19 ** | 0.23 ** | 0.14 * |
| NT | 1.00 | −0.07 | 0.29 ** | 0.25 ** | −0.25 ** | −0.11 | −0.13 * | −0.17 ** | |
| LRS | 1.00 | −0.17 ** | 0.07 | 0.52 ** | −0.37 ** | −0.45 ** | −0.46 ** | ||
| SFW | 1.00 | 0.84 ** | −0.66 ** | −0.07 | 0.03 | −0.11 | |||
| SDW | 1.00 | −0.30 ** | −0.21 ** | −0.13 * | −0.26 ** | ||||
| SDMC | 1.00 | −0.21 ** | −0.28 ** | −0.22 ** | |||||
| SF | 1.00 | 0.68 ** | 0.70 ** | ||||||
| GY | 1.00 | 0.93 ** | |||||||
| HI | 1.00 |
| Group a | Count b | DTH | NT | LRS | SFW | SDW | SDMC | SF | GY | HI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 (S) | 36 | 83.0 | 3.8 | 6.8 | 121.4 | 52.8 | 44.7 | 15.3 | 2.5 | 0.06 |
| 2 (MS) | 37 | 70.3 | 4.0 | 6.9 | 77.3 | 33.7 | 47.5 | 15.1 | 3.0 | 0.08 |
| 3 (T) | 22 | 70.1 | 4.0 | 6.8 | 57.2 | 32.0 | 57.7 | 46.0 | 6.3 | 0.22 |
| 4 (MT) | 17 | 72.4 | 6.0 | 6.0 | 97.0 | 42.5 | 47.1 | 25.4 | 4.0 | 0.11 |
| 5 (HS) | 17 | 66.5 | 3.3 | 8.0 | 39.4 | 28.7 | 73.7 | 13.1 | 1.5 | 0.05 |
| 6 (HT) | 68 | 77.2 | 3.7 | 4.9 | 91.6 | 37.9 | 42.2 | 54.4 | 10.3 | 0.28 |
| Group (Level of Tolerance) | List of Rice Genotypes |
|---|---|
| Group 1 (Susceptible) | Starbonnet-1, Rexark-1, Starbonnet-2, Bluebonnet, Toro, Nova, Glutinous selection, FL378, Melrose, Arkansas fortune, Prelude, Rexark Rogue-9262, RD, Nova-66, Stormproof, Carolina Gold, Rexark-2, Lady wright, Sierra, Zenith-2, Epagri, C-4, Tokalon, Texas Patna, TP-49, Moroberekan, Lacrosse, Salvo, Delitus-120, Delitus, Rexark Rogue-9214, Nira-43, Nira, Cheriviruppu, Pokkali |
| Group 2 (Moderately Susceptible) | Bond, CL162, Tebonnet, S-201, Calrose, Gulfrose, Early Colusa, Vista, M-202, Cheniere, M-102, Jackson, Azucena, LA-0702086, R-52, Sabine, M-301, Calrose-2, Conway, M-201, Catahoula, Bluebelle, Vegold, Bluebelle-2, Pacos, Caloro, MS-1995-15, Gold Zenith, Brazos, Smooth Zenith, Newrex, Kamrose, Colusa, Family-24, Nato, Calady, Skybonnet |
| Group 3 (Tolerant) | Newbonnet, Cypress, Jazzman-2, Jodon, R-50, Pin Kaeo, N-22, Trenasse, Presidio, Kokubelle, Lafitte, Mermentau, Dixieblle, Palmyra, Rico-1, Early Wataribur, Maybelle, Della-2, Chengri, Kalia-2, Djogolon, Caffey |
| Group 4 (Moderately Tolerant) | Early Prolific, MS-1996-9, CL261, Hybrid Mix, Lebonnet, Lotus, Damodar, Rexona, S-301, CL111, M-204, CL131, R27, Neches, Lavaoa, Bellemont, Jefferson |
| Group 5 (Highly Susceptible) | Alan, Terso, Tauri Mai, M-103, Carlpearl, Maxwell, Nipponbare, M-401, Belle Patna, Earlirose, M302, Cocodrie, Millie, Texmont, Gody, Rossmont, Adair |
| Group 6 (Highly Tolerant) | Zenith, Mars, Arkose selection, Saturn Rouge, Della, Hill Long Grain, Nortai, Cody, Jasmine-85, Evangeline, Dawn, Asahi, Rey, Acadia, CR5272, Saturn, SLO16, Northrose, Bengal, Dellamti, Katy, Taggert, FL478, Lacarus, CL152, MO R-500, Arkose, Gold Nato, Earl, LAH10, LA0802140, CL181, Wells, Templeton, TCCP-266, CL161, Glutinous Zenith, Hill medium, Magnolia, R54, Century Rogue, Toro-2, Short Century, Century Patna, SP14, Orion, CSR-11, Jupiter, Mercury, Dellrose, Geumgangbyeo, CL142, Madison, R-609, Roy J, Neptune, Lacassine, Pirogue, Dellmont, Jazzman, Leah, IRRI147, Ecrevisse, PSVRC, Dular, Jes, Kalia, LA110 |
| Source of Variation | DF a | SS b | MSS c | Estimated Variance | % variance | P-valuec |
|---|---|---|---|---|---|---|
| Among Population | 7 | 790.77 | 112.96 | 6.34 | 42 | <0.0001 |
| Within Population | 124 | 1095.75 | 8.84 | 8.84 | 58 | <0.0001 |
| Total | 131 | 1886.52 | 15.18 | 100 |
| Traits | Marker | Chr. | Pos. (Mb) | GLM a (Q) Model | MLM b (Q+K) Model | ||||
|---|---|---|---|---|---|---|---|---|---|
| F-value | P-value c | R-square d | F-value | P-value c | R-square d | ||||
| Days to heading | RM246 | 1 | 27.3 | 6.46 | 0.01 | 0.03 | |||
| RM22 | 3 | 1.5 | 8.12 | <0.01 | 0.03 | 7.61 | 0.01 | 0.04 | |
| RM3471 | 4 | 6.3 | 10.25 | <0.01 | 0.05 | 4.87 | 0.03 | 0.03 | |
| No. of Tillers | RM168 | 3 | 28.1 | 4.80 | 0.03 | 0.03 | 4.16 | 0.04 | 0.03 |
| Leaf rolling score | RM129 | 1 | 19.0 | 9.23 | <0.01 | 0.05 | 4.26 | 0.04 | 0.02 |
| RM168 | 3 | 28.1 | 7.69 | 0.01 | 0.04 | ||||
| RM570 | 3 | 35.6 | 8.35 | <0.01 | 0.04 | ||||
| RM351 | 7 | 23.9 | 10.53 | <0.01 | 0.06 | 5.25 | 0.02 | 0.04 | |
| RM152 | 8 | 0.7 | 10.60 | <0.01 | 0.05 | ||||
| RM256 | 8 | 24.3 | 4.54 | <0.01 | 0.02 | 5.51 | 0.02 | 0.03 | |
| RM216 | 10 | 5.4 | 11.27 | <0.01 | 0.05 | 4.38 | 0.04 | 0.02 | |
| RM7195 | 12 | 9.9 | 4.68 | 0.03 | 0.03 | ||||
| Shoot fresh weight | RM302 | 1 | 33 | 10.27 | <0.01 | 0.05 | 10.1 | <0.01 | 0.05 |
| RM431 | 1 | 38.9 | 6.08 | 0.01 | 0.03 | 6.81 | 0.01 | 0.04 | |
| RM3471 | 4 | 6.3 | 5.50 | 0.02 | 0.03 | 4.32 | 0.04 | 0.02 | |
| RM289 | 5 | 7.8 | 4.72 | 0.03 | 0.02 | 5.62 | 0.02 | 0.03 | |
| RM5371 | 6 | 25.8 | 6.06 | 0.01 | 0.03 | 4.22 | 0.04 | 0.02 | |
| RM1376 | 8 | 3.2 | 5.25 | 0.02 | 0.03 | ||||
| Shoot dry weight | RM129 | 1 | 19.0 | 5.98 | 0.02 | 0.03 | |||
| RM302 | 1 | 33.0 | 20.95 | <0.01 | 0.09 | 5.74 | 0.02 | 0.04 | |
| RM14980 | 3 | 13.9 | 7.17 | 0.01 | 0.03 | ||||
| RM570 | 3 | 35.6 | 7.58 | 0.01 | 0.03 | ||||
| RM3471 | 4 | 6.3 | 11.72 | <0.01 | 0.06 | 8.43 | <0.01 | 0.05 | |
| RM289 | 5 | 7.8 | 6.39 | 0.01 | 0.03 | 6.82 | 0.01 | 0.04 | |
| RM5371 | 6 | 25.8 | 7.54 | 0.01 | 0.03 | ||||
| RM461 | 6 | 30.1 | 9.77 | <0.01 | 0.08 | 8.45 | <0.01 | 0.09 | |
| RM351 | 7 | 23.9 | 8.37 | <0.01 | 0.04 | ||||
| RM8207 | 10 | 9.8 | 7.48 | 0.01 | 0.07 | ||||
| Shoot dry matter content | RM315 | 1 | 36.7 | 6.60 | 0.01 | 0.03 | 6.27 | 0.01 | 0.04 |
| Spikelet fertility | RM431 | 1 | 38.9 | 6.09 | 0.01 | 0.03 | 4.44 | 0.04 | 0.03 |
| RM168 | 3 | 28.1 | 5.23 | 0.02 | 0.03 | ||||
| RM570 | 3 | 35.6 | 11.76 | <0.01 | 0.06 | 7.25 | 0.01 | 0.04 | |
| RM6054 | 5 | 22.8 | 8.95 | <0.01 | 0.04 | 7.17 | 0.01 | 0.04 | |
| RM351 | 7 | 23.9 | 6.68 | 0.01 | 0.04 | 5.02 | 0.03 | 0.03 | |
| RM216 | 10 | 5.4 | 5.29 | 0.02 | 0.03 | ||||
| Grain yield | RM523 | 3 | 1.3 | 5.38 | 0.02 | 0.03 | |||
| RM517 | 3 | 6.2 | 5.50 | 0.02 | 0.03 | ||||
| RM570 | 3 | 35.6 | 5.80 | 0.02 | 0.03 | 4.12 | 0.04 | 0.03 | |
| RM351 | 7 | 23.9 | 6.56 | 0.01 | 0.04 | ||||
| RM256 | 8 | 24.3 | 5.47 | 0.02 | 0.03 | ||||
| Harvest Index | RM523 | 3 | 1.3 | 5.41 | 0.02 | 0.03 | |||
| RM570 | 3 | 35.6 | 5.27 | 0.02 | 0.03 | 4.04 | 0.04 | 0.03 | |
| RM351 | 7 | 23.9 | 10.51 | <0.01 | 0.06 | 5.23 | 0.02 | 0.07 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhattarai, U.; Subudhi, P.K. Genetic Diversity, Population Structure, and Marker-Trait Association for Drought Tolerance in US Rice Germplasm. Plants 2019, 8, 530. https://doi.org/10.3390/plants8120530
Bhattarai U, Subudhi PK. Genetic Diversity, Population Structure, and Marker-Trait Association for Drought Tolerance in US Rice Germplasm. Plants. 2019; 8(12):530. https://doi.org/10.3390/plants8120530
Chicago/Turabian StyleBhattarai, Uttam, and Prasanta K. Subudhi. 2019. "Genetic Diversity, Population Structure, and Marker-Trait Association for Drought Tolerance in US Rice Germplasm" Plants 8, no. 12: 530. https://doi.org/10.3390/plants8120530
APA StyleBhattarai, U., & Subudhi, P. K. (2019). Genetic Diversity, Population Structure, and Marker-Trait Association for Drought Tolerance in US Rice Germplasm. Plants, 8(12), 530. https://doi.org/10.3390/plants8120530






