CRISPR/Cas9-Mediated pds Knockout in Potato Reveals Network-Level Transcriptomic Reorganization Beyond Pigment Loss
Abstract
1. Introduction
2. Results
2.1. Establishment of a Visual CRISPR/Cas9 Editing System Targeting pds in Potato
2.2. On-Target Genotyping and Albino Phenotype
2.3. RNA-Seq Data Quality Assessment and Global Transcriptomic Overview
2.4. Functional Enrichment Revealed Distinct Transcriptomic Reprogramming Between Partial and Complete Albinism
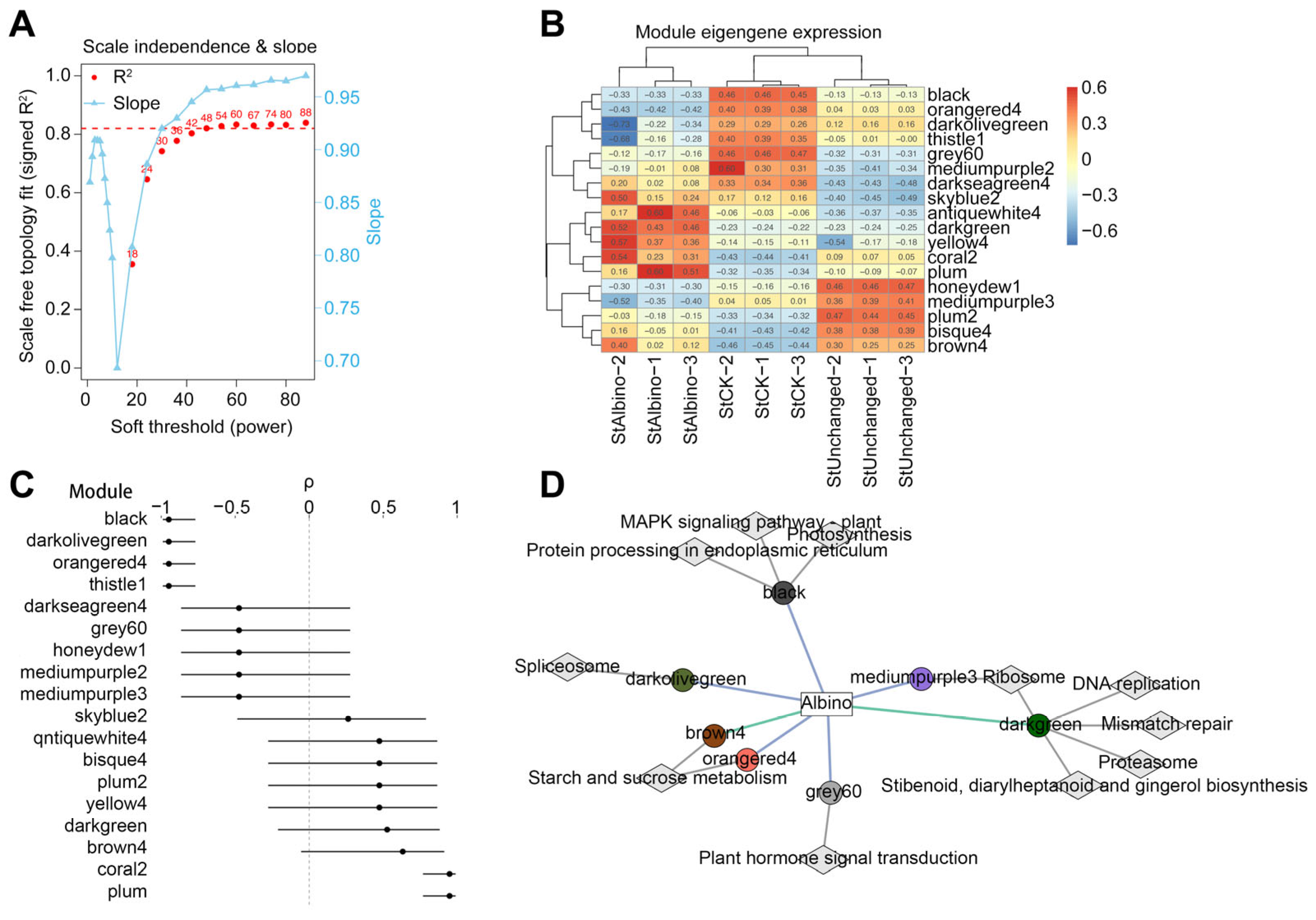
2.5. WGCNA Module Identification and Phenotypic Association
2.6. Functional Enrichment of Trait-Associated Modules Revealed Coordinated Metabolic and Signaling Shifts
3. Discussion
3.1. Albino Phenotype Is Associated with Broad Transcriptional and Metabolic Alterations
3.2. Passive Damage or Programmed Response in Albino Plants
3.3. Rethinking pds-Based Visual Screening Tools
4. Materials and Methods
4.1. Tissue Culture Conditions and CRISPR/Cas9 Knockout Vector Construction
4.2. Agrobacterium-Mediated Transformation of Potato
4.3. Identification of Positive Transformants and Phenotypic Assessment
4.4. TIDE Analysis of Editing Efficiency
4.5. Transcriptome Sequencing and Differential Expression
4.6. Co-Expression Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAO. FAO World Food and Agriculture-Statistical Yearbook 2024; FAO: Rome, Italy, 2024. [Google Scholar]
- Hu, X.; Jiang, H.; Liu, Z.; Gao, M.; Liu, G.; Tian, S.; Zeng, F. The global potato-processing industry: A review of production, products, quality and sustainability. Foods 2025, 14, 1758. [Google Scholar] [CrossRef]
- del Mar Martínez-Prada, M.; Curtin, S.J.; Gutiérrez-González, J.J. Potato improvement through genetic engineering. GM Crops Food 2021, 12, 479–496. [Google Scholar] [CrossRef]
- de Vries, M.E.; Adams, J.R.; Eggers, E.-j.; Ying, S.; Stockem, J.E.; Kacheyo, O.C.; van Dijk, L.C.; Khera, P.; Bachem, C.W.; Lindhout, P. Converting hybrid potato breeding science into practice. Plants 2023, 12, 230. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Z.; Tang, D.; Zhu, Y.; Wang, P.; Li, D.; Zhu, G.; Xiong, X.; Shang, Y.; Li, C. Genome design of hybrid potato. Cell 2021, 184, 3873–3883. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Challam, C.; Zinta, R.; Bhatia, N.; Dalamu, D.; Naik, S.; Poonia, A.K.; Singh, R.K.; Luthra, S.K. CRISPR/Cas genome editing in potato: Current status and future perspectives. Front. Genet. 2022, 13, 827808. [Google Scholar] [CrossRef] [PubMed]
- Cardi, T.; Murovec, J.; Bakhsh, A.; Boniecka, J.; Bruegmann, T.; Bull, S.E.; Eeckhaut, T.; Fladung, M.; Galovic, V.; Linkiewicz, A. CRISPR/Cas-mediated plant genome editing: Outstanding challenges a decade after implementation. Trends Plant Sci. 2023, 28, 1144–1165. [Google Scholar] [CrossRef]
- Lu, Q.S.M.; Tian, L. An efficient and specific CRISPR-Cas9 genome editing system targeting soybean phytoene desaturase genes. BMC Biotechnol. 2022, 22, 7. [Google Scholar] [CrossRef]
- Siddappa, S.; Sharma, N.; Salaria, N.; Thakur, K.; Pathania, S.; Singh, B.; Sharma, H.; Sood, S.; Bhardwaj, V.; Thakur, A.K. CRISPR/Cas9-mediated editing of phytoene desaturase (PDS) gene in an important staple crop, potato. 3 Biotech 2023, 13, 129. [Google Scholar] [CrossRef]
- Yang, S.H.; Kim, E.; Park, H.; Koo, Y. Selection of the high efficient sgRNA for CRISPR-Cas9 to edit herbicide related genes, PDS, ALS, and EPSPS in tomato. Appl. Biol. Chem. 2022, 65, 13. [Google Scholar] [CrossRef]
- Molina-Risco, M.; Ibarra, O.; Faion-Molina, M.; Kim, B.; Septiningsih, E.M.; Thomson, M.J. Optimizing Agrobacterium-mediated transformation and CRISPR-Cas9 gene editing in the tropical japonica rice variety presidio. Int. J. Mol. Sci. 2021, 22, 10909. [Google Scholar] [CrossRef] [PubMed]
- Ntui, V.O.; Tripathi, J.N.; Tripathi, L. Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 2020, 21, 100128. [Google Scholar] [CrossRef]
- Bánfalvi, Z.; Csákvári, E.; Villányi, V.; Kondrák, M. Generation of transgene-free PDS mutants in potato by Agrobacterium-mediated transformation. BMC Biotechnol. 2020, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B.J. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016, 67, 25–53. [Google Scholar] [CrossRef]
- Fernández, A.P.; Strand, Å. Retrograde signaling and plant stress: Plastid signals initiate cellular stress responses. Curr. Opin. Plant Biol. 2008, 11, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Leister, D. Piecing the puzzle together: The central role of reactive oxygen species and redox hubs in chloroplast retrograde signaling. Antioxid. Redox Signal. 2019, 30, 1206–1219. [Google Scholar] [CrossRef]
- Hooghvorst, I.; López-Cristoffanini, C.; Nogués, S. Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci. Rep. 2019, 9, 17077. [Google Scholar] [CrossRef]
- Kaur, N.; Alok, A.; Shivani, N.; Kaur, N.; Pandey, P.; Awasthi, P.; Tiwari, S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 2018, 18, 89–99. [Google Scholar] [CrossRef]
- Dietz, K.-J.; Wesemann, C.; Wegener, M.; Seidel, T. Toward an integrated understanding of retrograde control of photosynthesis. Antioxid. Redox Signal. 2019, 30, 1186–1205. [Google Scholar] [CrossRef]
- Sierra, J.; McQuinn, R.P.; Leon, P. The role of carotenoids as a source of retrograde signals: Impact on plant development and stress responses. J. Exp. Bot. 2022, 73, 7139–7154. [Google Scholar] [CrossRef]
- Li, M.; Kim, C. Chloroplast ROS and stress signaling. Plant Commun. 2022, 3, 100264. [Google Scholar] [CrossRef] [PubMed]
- Chi, W.; Sun, X.; Zhang, L. Intracellular signaling from plastid to nucleus. Annu. Rev. Plant Biol. 2013, 64, 559–582. [Google Scholar] [CrossRef]
- Moreno, J.C.; Mi, J.; Alagoz, Y.; Al-Babili, S. Plant apocarotenoids: From retrograde signaling to interspecific communication. Plant J. 2021, 105, 351–375. [Google Scholar] [CrossRef]
- Escobar-Tovar, L.; Sierra, J.; Hernández-Muñoz, A.; McQuinn, R.P.; Mathioni, S.; Cordoba, E.; Colas des Francs-Small, C.; Meyers, B.C.; Pogson, B.; León, P. Deconvoluting apocarotenoid-mediated retrograde signaling networks regulating plastid translation and leaf development. Plant J. 2021, 105, 1582–1599. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF transcription factor responses and tolerance to various abiotic stresses in plants: A comprehensive review. Int. J. Mol. Sci. 2024, 25, 893. [Google Scholar] [CrossRef]
- Kim, S.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulatory networks in plant responses to drought and cold stress. Plant Physiol. 2024, 195, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg Valero, K.C. Aligning functional network constraint to evolutionary outcomes. BMC Evol. Biol. 2020, 20, 58. [Google Scholar] [CrossRef] [PubMed]
- Gaudinier, A.; Brady, S.M. Mapping transcriptional networks in plants: Data-driven discovery of novel biological mechanisms. Annu. Rev. Plant Biol. 2016, 67, 575–594. [Google Scholar] [CrossRef]
- Walley, J.W.; Sartor, R.C.; Shen, Z.; Schmitz, R.J.; Wu, K.J.; Urich, M.A.; Nery, J.R.; Smith, L.G.; Schnable, J.C.; Ecker, J.R. Integration of omic networks in a developmental atlas of maize. Science 2016, 353, 814–818. [Google Scholar] [CrossRef]
- Zhao, W.; Chen, X.; Wang, J.; Cheng, Z.; Ma, X.; Zheng, Q.; Xu, Z.; Zhang, F. Emerging Mechanisms of Plant Responses to Abiotic Stress. Plants 2025, 14, 3445. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, H.; Wu, X.; Wang, J.; Li, H.; Zhang, R. Transcriptomic and physiological responses of contrasting maize genotypes to drought stress. Front. Plant Sci. 2022, 13, 928897. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Guo, C.; Ma, X.; Gao, F.; Guo, Y. Off-target effects in CRISPR/Cas9 gene editing. Front. Bioeng. Biotechnol. 2023, 11, 1143157. [Google Scholar] [CrossRef]
- Osakabe, K.; Wada, N.; Miyaji, T.; Murakami, E.; Marui, K.; Ueta, R.; Hashimoto, R.; Abe-Hara, C.; Kong, B.; Yano, K. Genome editing in plants using CRISPR type ID nuclease. Commun. Biol. 2020, 3, 648. [Google Scholar] [CrossRef]
- Rurek, M.; Smolibowski, M. Variability of plant transcriptomic responses under stress acclimation: A review from high throughput studies. Acta Biochim. Pol. 2024, 71, 13585. [Google Scholar] [CrossRef]
- Gan, W.C.; Ling, A.P. CRISPR/Cas9 in plant biotechnology: Applications and challenges. BioTechnologia 2022, 103, 81. [Google Scholar] [CrossRef]
- Singer, S.D.; Laurie, J.D.; Bilichak, A.; Kumar, S.; Singh, J. Genetic variation and unintended risk in the context of old and new breeding techniques. Crit. Rev. Plant Sci. 2021, 40, 68–108. [Google Scholar] [CrossRef]
- Yu, T.; Ma, X.; Zhang, J.; Cao, S.; Li, W.; Yang, G.; He, C. Progress in Transcriptomics and Metabolomics in Plant Responses to Abiotic Stresses. Curr. Issues Mol. Biol. 2025, 47, 421. [Google Scholar] [CrossRef]
- Zaman, Q.U.; Raza, A.; Lozano-Juste, J.; Chao, L.; Jones, M.G.; Wang, H.-F.; Varshney, R.K. Engineering plants using diverse CRISPR-associated proteins and deregulation of genome-edited crops. Trends Biotechnol. 2024, 42, 560–574. [Google Scholar] [CrossRef] [PubMed]
- Din, A.; Wani, M.A.; Jin, C.; Nazki, I.T.; Ma, J.; Li, F. Post-genomic era of CRISPR/Cas technology in ornamental plants: Advantages, limitations, and prospects. Ornam. Plant Res. 2025, 5, e010. [Google Scholar] [CrossRef]
- Aziz, M.A.; Masmoudi, K. Molecular breakthroughs in modern plant breeding techniques. Hortic. Plant J. 2025, 11, 15–41. [Google Scholar] [CrossRef]
- Razavi, Z.; Soltani, M.; Souri, M.; van Wijnen, A.J. CRISPR innovations in tissue engineering and gene editing. Life Sci. 2024, 358, 123120. [Google Scholar] [CrossRef]
- Vaia, G.; Pavese, V.; Moglia, A.; Cristofori, V.; Silvestri, C. Knockout of phytoene desaturase gene using CRISPR/Cas9 in highbush blueberry. Front. Plant Sci. 2022, 13, 1074541. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, P.; Zhang, J.; Yang, D.; Lu, L.; Hao, Z.; Ma, Y.; Shi, J.; Chen, J. Highly Efficient Homozygous CRISPR/Cas9 Gene Editing Based on Single-Cell-Originated Somatic Embryogenesis in Liriodendron tulipifera. Plants 2025, 14, 472. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Huang, S.; Wang, J.; Sun, D.; Wang, M.; Lin, S.; Liu, J.; Xu, K.; Liu, B.; Ma, H. Review and Validation of Plant Gene Function Research Methods Bypassing Tissue Culture. Agronomy 2025, 15, 603. [Google Scholar] [CrossRef]
- Mansoor, S.; Karunathilake, E.; Tuan, T.T.; Chung, Y.S. Genomics, Phenomics, and Machine Learning in Shaping the Future of Plant Research: Advancements and Challenges. Hortic. Plant J. 2024, 11, 486–503. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Schindele, P.; Merker, L.; Schreiber, T.; Prange, A.; Tissier, A.; Puchta, H. Enhancing gene editing and gene targeting efficiencies in Arabidopsis thaliana by using an intron-containing version of ttLbCas12a. Plant Biotechnol. J. 2022, 21, 457. [Google Scholar] [CrossRef]
- Brinkman, E.K.; van Steensel, B. Rapid quantitative evaluation of CRISPR genome editing by TIDE and TIDER. In CRISPR Gene Editing: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2019; pp. 29–44. [Google Scholar]
- Shen, Y.; Li, R.; Tian, F.; Chen, Z.; Lu, N.; Bai, Y.; Ge, Q.; Lu, Z. Impact of RNA integrity and blood sample storage conditions on the gene expression analysis. OncoTargets Ther. 2018, 11, 3573–3581. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kolde, R. Package ‘pheatmap’; R package, 1.0.12, 2015. Available online: https://cran.ms.unimelb.edu.au/web/packages/pheatmap/pheatmap.pdf (accessed on 10 November 2025).
- Mai, H.; Zhang, Y.; Li, D.; Leung, H.C.-M.; Luo, R.; Wong, C.-K.; Ting, H.-F.; Lam, T.-W. AC-DIAMOND v1: Accelerating large-scale DNA–protein alignment. Bioinformatics 2018, 34, 3744–3746. [Google Scholar] [CrossRef]
- Mulder, N.; Apweiler, R. InterPro and InterProScan: Tools for protein sequence classification and comparison. In Comparative genomics; Springer: Berlin/Heidelberg, Germany, 2007; pp. 59–70. [Google Scholar]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Lai, X.; Xiang, Y.; Liu, S.; Zhang, Y.; Zhang, Y.; Chen, Z.; Liu, S.; Yan, L. CRISPR/Cas9-Mediated pds Knockout in Potato Reveals Network-Level Transcriptomic Reorganization Beyond Pigment Loss. Plants 2026, 15, 96. https://doi.org/10.3390/plants15010096
Lai X, Xiang Y, Liu S, Zhang Y, Zhang Y, Chen Z, Liu S, Yan L. CRISPR/Cas9-Mediated pds Knockout in Potato Reveals Network-Level Transcriptomic Reorganization Beyond Pigment Loss. Plants. 2026; 15(1):96. https://doi.org/10.3390/plants15010096
Chicago/Turabian StyleLai, Xianjun, Yuxin Xiang, Siqi Liu, Yandan Zhang, Yizheng Zhang, Zihan Chen, Shifeng Liu, and Lang Yan. 2026. "CRISPR/Cas9-Mediated pds Knockout in Potato Reveals Network-Level Transcriptomic Reorganization Beyond Pigment Loss" Plants 15, no. 1: 96. https://doi.org/10.3390/plants15010096
APA StyleLai, X., Xiang, Y., Liu, S., Zhang, Y., Zhang, Y., Chen, Z., Liu, S., & Yan, L. (2026). CRISPR/Cas9-Mediated pds Knockout in Potato Reveals Network-Level Transcriptomic Reorganization Beyond Pigment Loss. Plants, 15(1), 96. https://doi.org/10.3390/plants15010096





