Genome-Wide Association Study Dissects the Genetic Architecture of Pericarp Traits in Fresh-Eating Maize
Abstract
1. Introduction
2. Results
2.1. Descriptive Statistics and Correlation Analysis of Pericarp-Related Traits
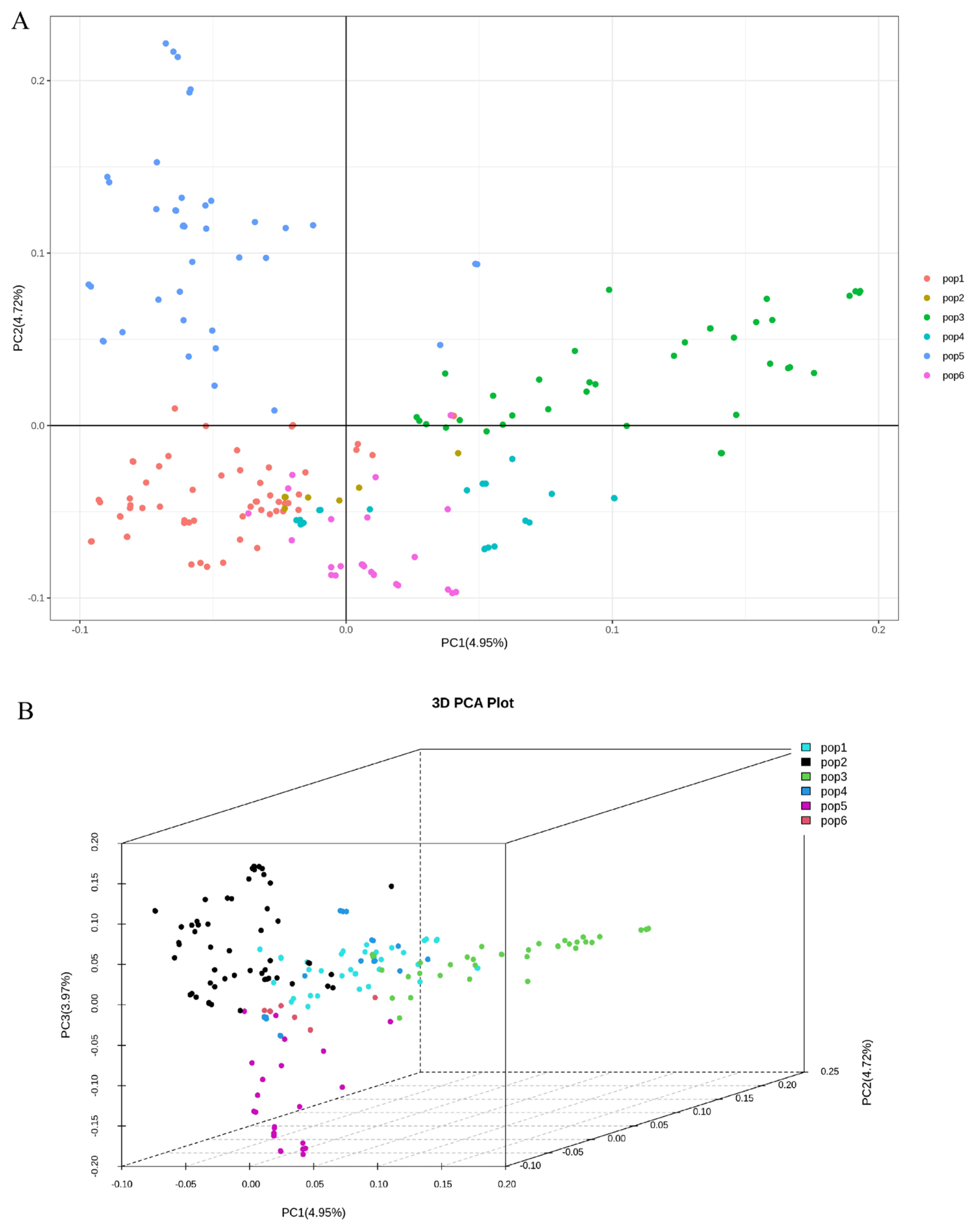
2.2. Genotyping and Principal Component Analysis of Natural Populations
2.3. Linkage Disequilibrium Analysis in Natural Populations
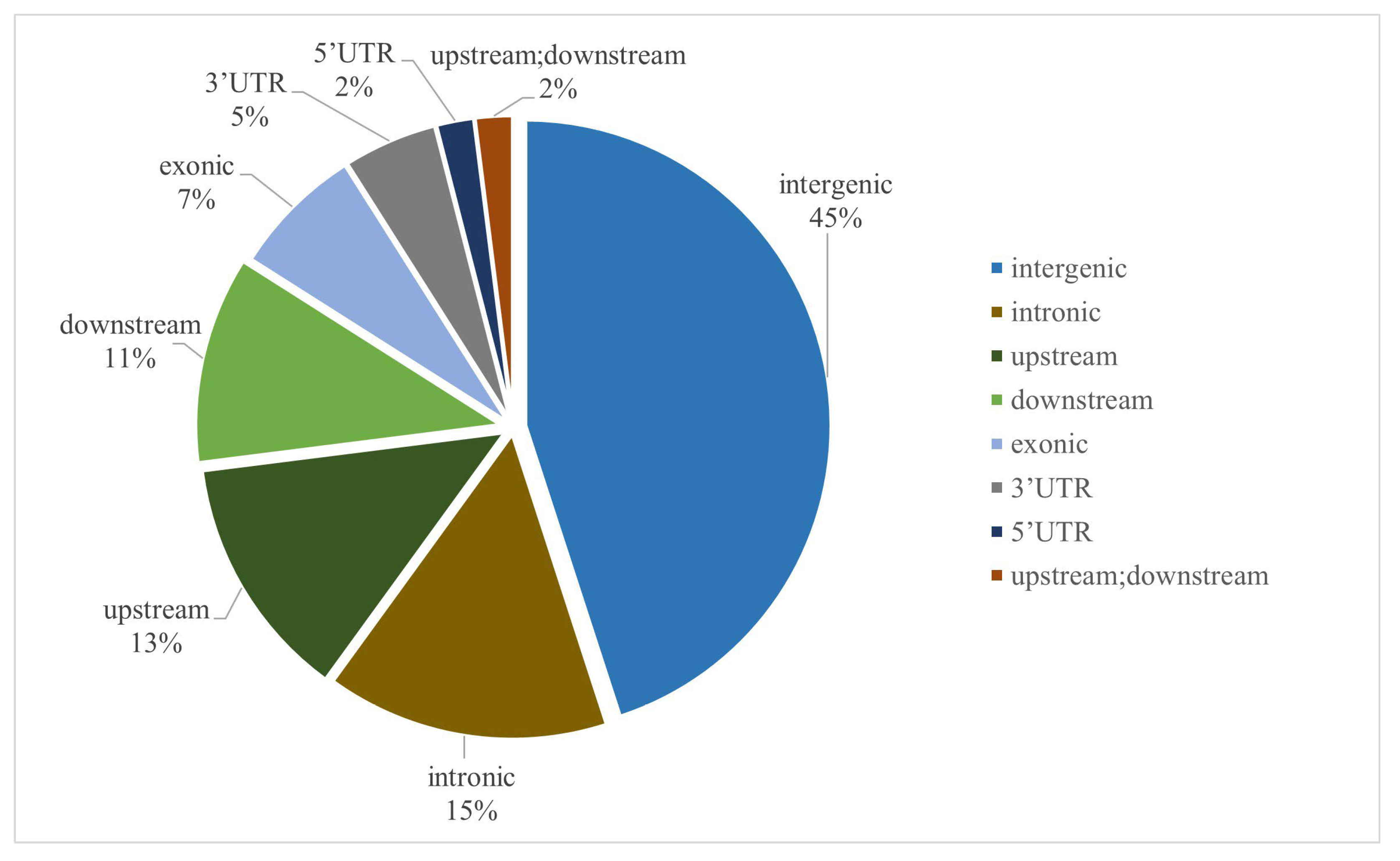
2.4. Genome-Wide Association Study and Candidate Gene Identification for Pericarp-Related Traits
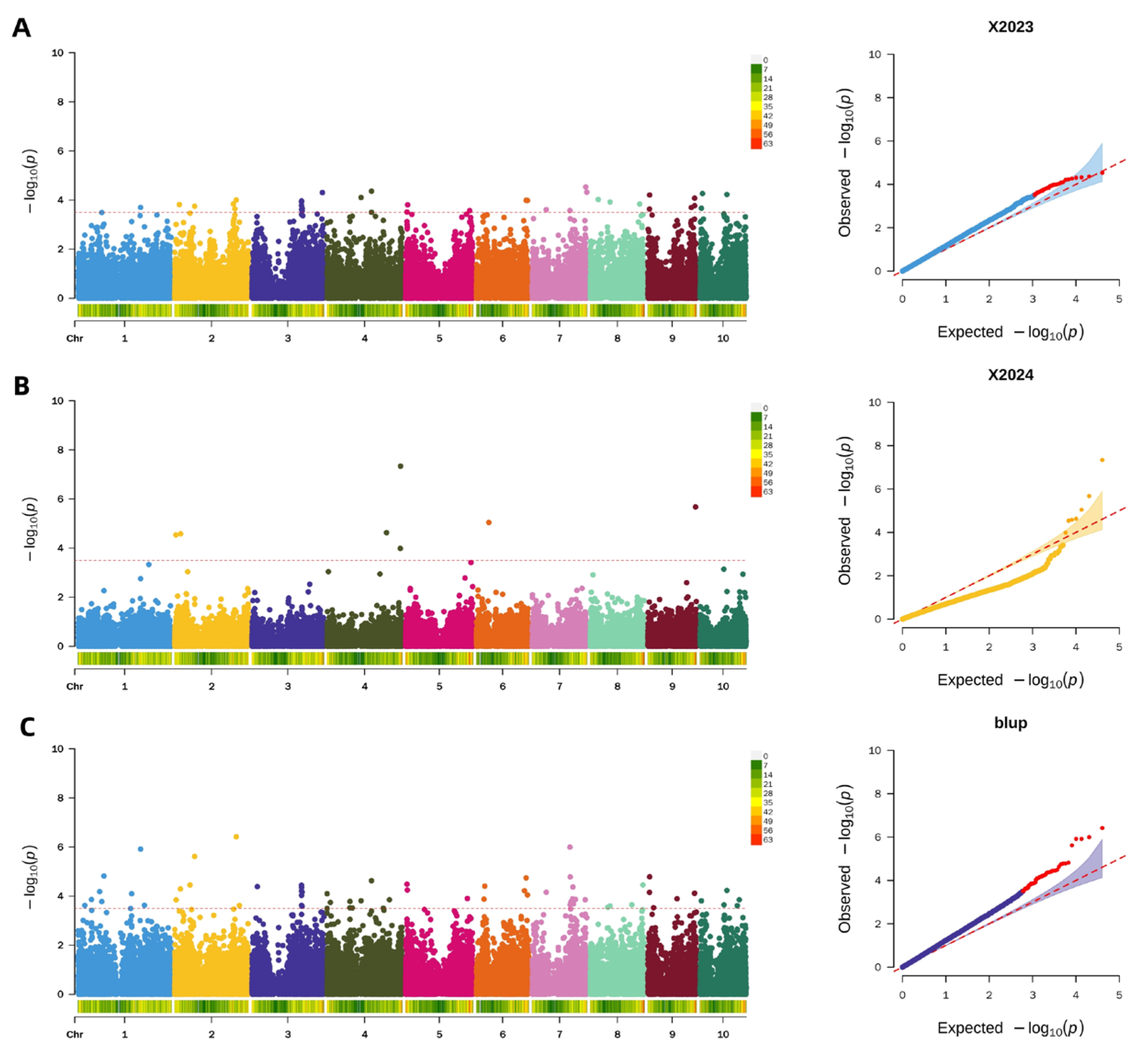
2.4.1. Genome-Wide Association Study for Pericarp Thickness
2.4.2. Screening of Candidate Genes for Pericarp Thickness
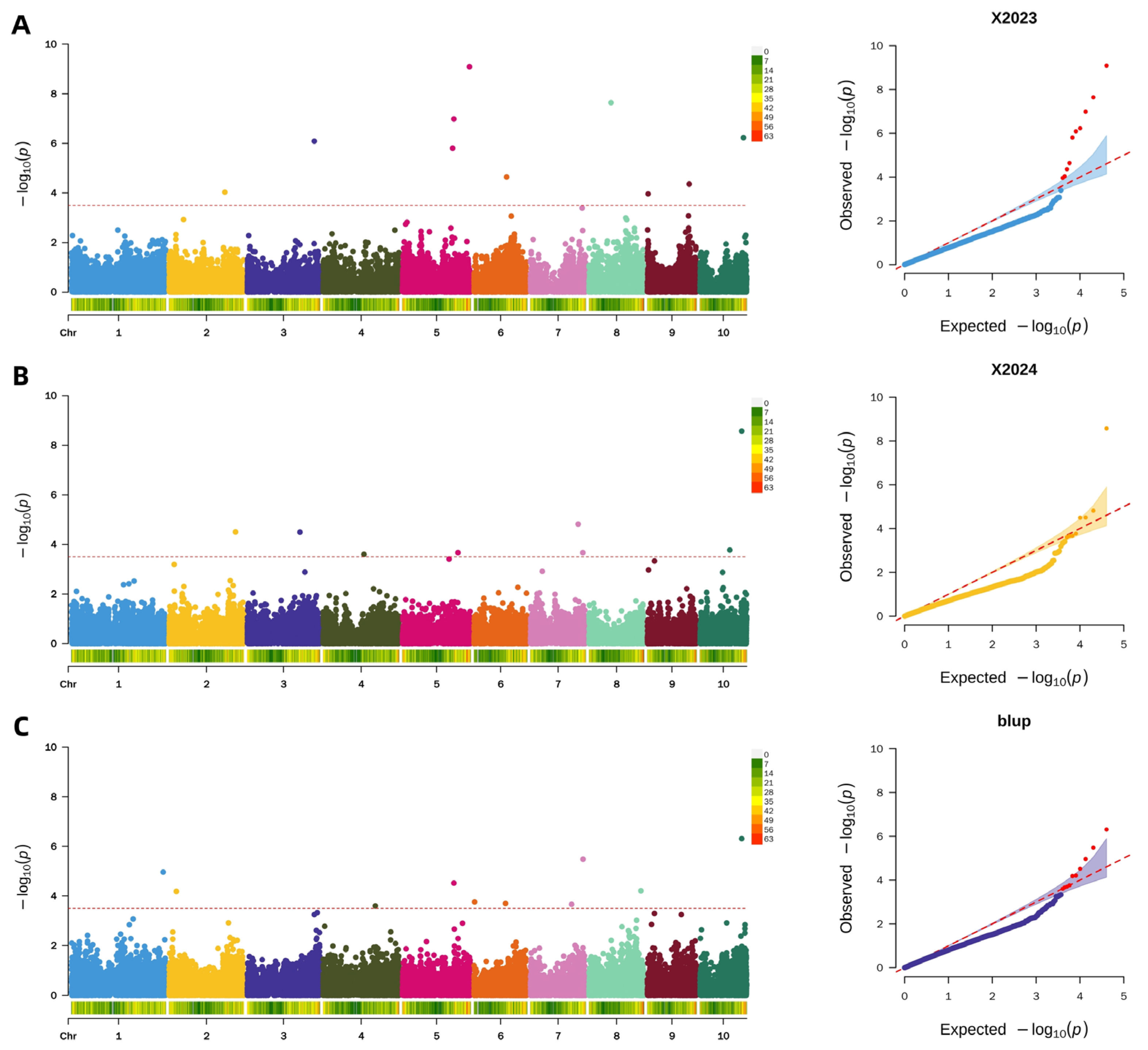
2.4.3. Genome-Wide Association Study of Pericarp Rupture Strength
2.4.4. Screening of Candidate Genes for Pericarp Rupture Strength
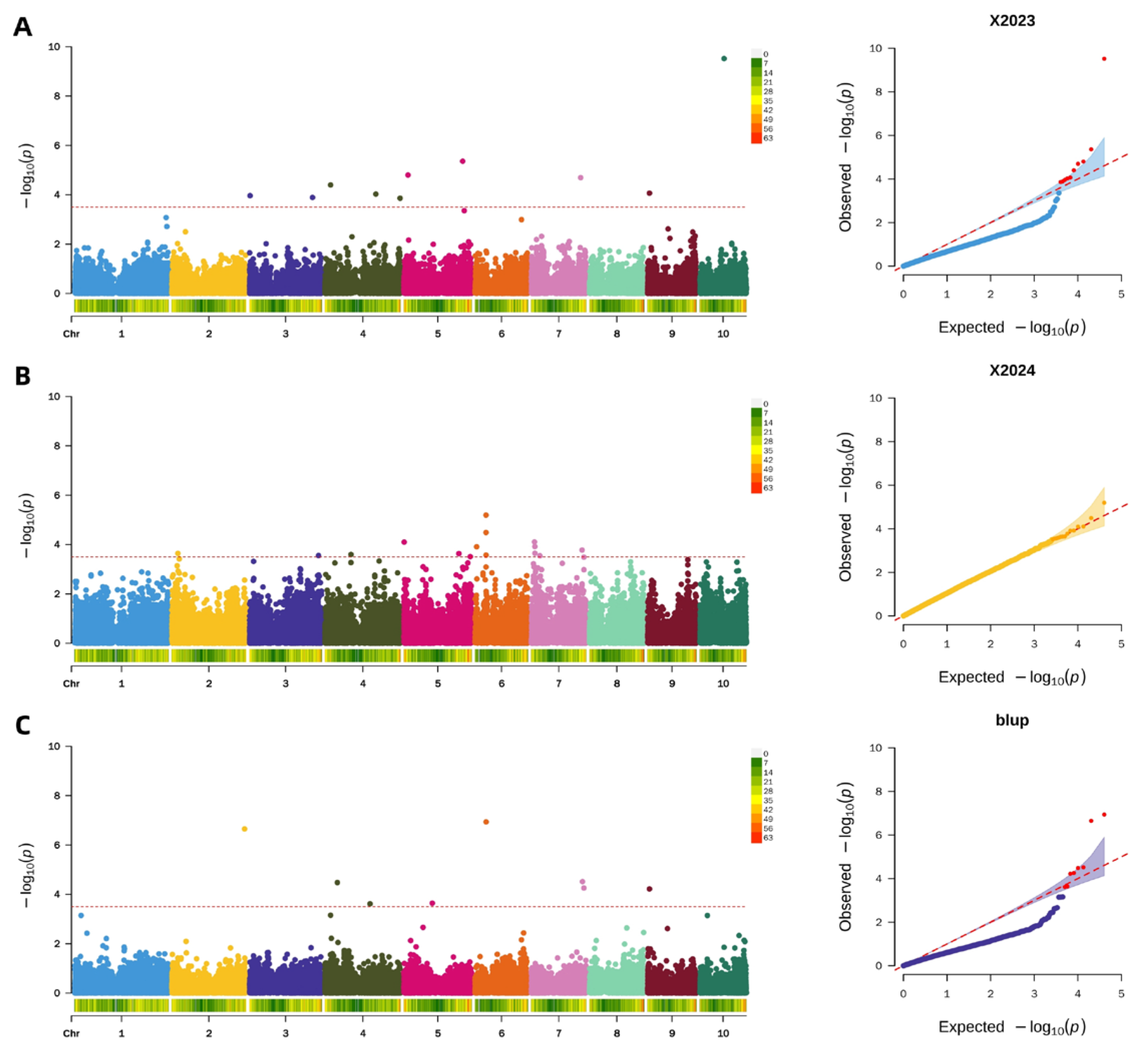
2.4.5. Genome-Wide Association Study of Pericarp Brittleness
2.4.6. Screening of Candidate Genes for Pericarp Brittleness
2.4.7. Overlapping Loci for Candidate Genes Associated with Pericarp-Related Traits
2.4.8. Functional Prioritization and Mechanistic Hypotheses for Candidate Genes
3. Discussion
3.1. Extensive Phenotypic Variation and Novel Genetic Loci Revealed by GWAS
3.2. Functional Analysis of ZmbZIP130 as a Key Candidate Gene
3.3. Genotype-by-Environment Interaction, Analytical Strategy, and Study Limitations
4. Materials and Methods
4.1. Plant Materials
4.2. Experimental Methods
4.2.1. Phenotypic Identification of Pericarp-Related Traits
4.2.2. Genotyping of the Natural Population
- Removal of paired-end reads containing > 10% N bases relative to read length.
- Removal of paired-end reads with >40% low-quality bases (Q ≤ 20).
4.2.3. Genome-Wide Association Study (GWAS)
4.2.4. GO Functional Annotation of Pericarp-Related Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, X.-Y.; Long, W.-J.; Chen, D.; Zhou, G.-Y.; Du, J.; Wu, S.-Y.; Cai, Q. Waxy allele diversity in waxy maize landraces of Yunnan Province, China. J. Integr. Agric. 2022, 21, 578–585. [Google Scholar] [CrossRef]
- Bao, F.; Zhang, P.; Yu, Q.; Cai, Y.; Chen, B.; Tan, H.; Han, H.; Hou, J.; Zhao, F. Response of fresh maize yield to nitrogen application rates and characteristics of nitrogen-efficient varieties. J. Integr. Agric. 2025, 24, 3803–3818. [Google Scholar] [CrossRef]
- Yang, H.; Cai, X.; Lu, D. Effects of Waterlogging at Flowering Stage on the Grain Yield and Starch Quality of Waxy Maize. Plants 2024, 13, 108. [Google Scholar] [CrossRef] [PubMed]
- Paranhos, J.; Foshee, W.; Coolong, T.; Heyes, B.; Salazar-Gutierrez, M.; Kesheimer, K.; da Silva, A.L. Characterization of Sweet Corn Production in Subtropical Environmental Conditions. Agriculture 2023, 13, 1156. [Google Scholar] [CrossRef]
- So, Y.-S. Pericarp thickness of Korean maize landraces. Plant Genet. Resour. Charact. Util. 2019, 17, 87–90. [Google Scholar] [CrossRef]
- Okagaki, R.J.; Weil, C.F. Analysis of Recombination Sites Within the Maize waxy Locus. Genetics 1997, 147, 815–821. [Google Scholar] [CrossRef]
- Juvik, J.A.; LaBonte, D.R. Single-kernel Analysis for the Presence of the sugary enhancer (se) Gene in Sweet Corn. HortScience 1988, 23, 384–386. [Google Scholar] [CrossRef]
- Du, Y.; Yang, B.; Lu, Y.; Zhao, L.; Zhang, T.; Liu, J.; Pei, Y.; Cai, D.; Zhang, H.; Zhang, Z.; et al. Shrunken2 (Sh2) and Brittle2 (Bt2) play a crucial role in the developmental processes of the aleurone layers in maize. Plant Physiol. 2025, 199, kiaf464. [Google Scholar] [CrossRef]
- Sharma, S.P.; Zuo, T.; Peterson, T. Transposon-induced inversions activate gene expression in the maize pericarp. Genetics 2021, 218, iyab062. [Google Scholar] [CrossRef]
- Zhang, J.; Yue, Y.; Hu, M.; Yi, F.; Chen, J.; Lai, J.; Xin, B. Dynamic transcriptome landscape of maize pericarp development. Plant J. Cell Mol. Biol. 2024, 117, 1574–1591. [Google Scholar] [CrossRef]
- Sahito, J.H.; Zhang, H.; Gishkori, Z.G.; Ma, C.; Wang, Z.; Ding, D.; Zhang, X.; Tang, J. Advancements and Prospects of Genome-Wide Association Studies (GWAS) in Maize. Int. J. Mol. Sci. 2024, 25, 1918. [Google Scholar] [CrossRef]
- Tang, R.; Zhuang, Z.; Bian, J.; Ren, Z.; Ta, W.; Peng, Y. GWAS and Meta-QTL Analysis of Kernel Quality-Related Traits in Maize. Plants 2024, 13, 2730. [Google Scholar] [CrossRef] [PubMed]
- Son, J.-H.; Hong, S.; Kim, J.W.; Go, J.; Choi, J.; Lee, S.-B.; Ha, J.Y.; Go, Y.-S.; Bae, H.-H.; Jung, T.-W.; et al. Kernel type-based entries are efficient to develop a core collection of maize (Zea mays L.). Appl. Biol. Chem. 2025, 68, 7. [Google Scholar] [CrossRef]
- Li, C.; Jia, Y.; Zhou, R.; Liu, L.; Cao, M.; Zhou, Y.; Wang, Z.; Di, H. GWAS and RNA-seq analysis uncover candidate genes associated with alkaline stress tolerance in maize (Zea mays L.) seedlings. Front. Plant Sci. 2022, 13, 963874. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Jia, L.; Zhong, Z.; Zhuang, Z.; Jin, B.; Ji, X.; Bai, M.; Peng, Y. Integrated GWAS and Transcriptome Analysis Reveal the Genetic and Molecular Basis of Low Nitrogen Tolerance in Maize Seedlings. Plants 2025, 14, 2689. [Google Scholar] [CrossRef]
- Cao, X.; Guo, Z.; Wang, P.; Lu, S.; Li, W.; Ma, Z.; Mao, J.; Chen, B. MdbZIP44–MdCPRF2-like–Mdα-GP2 regulate starch and sugar metabolism in apple under nitrogen supply. Hortic. Res. 2024, 11, uhae072. [Google Scholar] [CrossRef]
- Giomi, G.M.; Sampietro, D.A.; Velazco, J.G.; Iglesias, J.; Fernández, M.; Oviedo, M.S.; Presello, D.A. Map overlapping of QTL for resistance to Fusarium ear rot and associated traits in maize. Euphytica 2021, 217, 81. [Google Scholar] [CrossRef]
- Gong, G.; Jia, H.; Tang, Y.; Pei, H.; Zhai, L.; Huang, J. Genetic analysis and QTL mapping for pericarp thickness in maize (Zea mays L.). BMC Plant Biol. 2024, 24, 338. [Google Scholar] [CrossRef]
- Cao, L.; Lu, X.; Zhang, P.; Wang, G.; Wei, L.; Wang, T. Systematic Analysis of Differentially Expressed Maize ZmbZIP Genes between Drought and Rewatering Transcriptome Reveals bZIP Family Members Involved in Abiotic Stress Responses. Int. J. Mol. Sci. 2019, 20, 4103. [Google Scholar] [CrossRef]
- Wang, B.; Zheng, J.; Liu, Y.; Wang, J.; Wang, G. Cloning and characterization of the stress-induced bZIP gene ZmbZIP60 from maize. Mol. Biol. Rep. 2012, 39, 6319–6327. [Google Scholar] [CrossRef]
- Ma, F.; Zhou, H.; Xu, Y.; Huang, D.; Wu, B.; Xing, W.; Chen, D.; Xu, B.; Song, S. Comprehensive analysis of bZIP transcription factors in passion fruit. iScience 2023, 26, 106556. [Google Scholar] [CrossRef]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, Y.; Lou, L.; Wang, Y.; Dai, B.; Wang, Y.; Kebbeh, M.; Huan, C.; Shen, S.; Liu, Y.; et al. Transcriptomic analysis reveals the role of abscisic acid and ethylene in regulating starch-source biosynthesis associated with soft nose disorder in ‘Keitt’ mango fruit during postharvest. Postharvest Biol. Technol. 2024, 209, 112698. [Google Scholar] [CrossRef]
- Wu, C.; Shan, W.; Liang, S.; Zhu, L.; Guo, Y.; Chen, J.; Lu, W.; Li, Q.; Su, X.; Kuang, J. MaMPK2 enhances MabZIP93-mediated transcriptional activation of cell wall modifying genes during banana fruit ripening. Plant Mol. Biol. 2019, 101, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-F.; Zhang, Y.-L.; Shan, W.; Cai, Y.-J.; Liang, S.-M.; Chen, J.-Y.; Lu, W.-J.; Kuang, J.-F. Identification of Two Transcriptional Activators MabZIP4/5 in Controlling Aroma Biosynthetic Genes during Banana Ripening. J. Agric. Food Chem. 2018, 66, 6142–6150. [Google Scholar] [CrossRef]
- Liu, X.; Bulley, S.M.; Varkonyi-Gasic, E.; Zhong, C.; Li, D. Kiwifruit bZIP transcription factor AcePosF21 elicits ascorbic acid biosynthesis during cold stress. Plant Physiol. 2023, 192, 982–999. [Google Scholar] [CrossRef]
- Jin, Z.; Xu, W.; Liu, A. Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 2014, 239, 299–312. [Google Scholar] [CrossRef]
- Mohamed, H.N.; Abd-Elfattah, E.F.; Abd-El-Monem, A.; Abd El-Raheem, A.E.-R.M. Saddlepoint p-values for a class of location-scale tests under randomized block design. Sci. Rep. 2024, 14, 3092. [Google Scholar] [CrossRef]
- Lis, A.; Staniewski, B.; Ziajka, J. A comparison of butter texture measurements with the AP 4/2 penetrometer and TA.XT. Plus texture analyzer. Int. J. Food Prop. 2021, 24, 1744–1757. [Google Scholar] [CrossRef]
- Chen, X.; Guo, L.; Chen, P.; Xu, Y.; Hao, H.; Du, X. Investigation of the high-amylose maize starch gelatinization behaviours in glycerol-water systems. J. Cereal Sci. 2017, 77, 135–140. [Google Scholar] [CrossRef]
- Farcuh, M.; Copes, B.; Le-Navenec, G.; Marroquin, J.; Jaunet, T.; Chi-Ham, C.; Cantu, D.; Bradford, K.J.; Van Deynze, A. Texture diversity in melon (Cucumis melo L.): Sensory and physical assessments. Postharvest Biol. Technol. 2020, 159, 111024. [Google Scholar] [CrossRef]
- Wen, J.; Shen, Y.; Xing, Y.; Wang, Z.; Han, S.; Li, S.; Yang, C.; Hao, D.; Zhang, Y. QTL Mapping of Fusarium Ear Rot Resistance in Maize. Plant Dis. 2020, 105, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, J.; Shi, X.; Chen, L.; Qin, J.; Zhang, M.; Yang, C.; Song, Q.; Yan, L. Development of SNP marker panels for genotyping by target sequencing (GBTS) and its application in soybean. Mol. Breed. 2023, 43, 26. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Du, X.; Wang, Y.; Yang, X.; Cheng, X.; Huang, C.; Li, T.; Li, T.; Chen, C.; Zhao, J.; et al. GenoBaits® WheatplusEE: A targeted capture sequencing panel for quick and accurate identification of wheat–Thinopyrum derivatives. Theor. Appl. Genet. 2024, 137, 36. [Google Scholar] [CrossRef]
- Duan, S.; Wang, D.; Kang, Q.; Yan, H.; Cui, J.; Zhang, M.; Liu, D.; Yang, S.; Zhu, Y.; Niu, H.; et al. The development of liquid-phase chip by target sequencing and their application in watermelon molecular breeding. Hortic. Plant J. 2025, 11, 2109–2120. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Andorf, C.M.; Ross-Ibarra, J.; Seetharam, A.S.; Hufford, M.B.; Woodhouse, M.R. A unified VCF dataset from nearly 1,500 diverse maize accessions and resources to explore the genomic landscape of maize. G3|Genes|Genomes|Genet. 2025, 15, jkae281. [Google Scholar] [CrossRef]
- Lin, M.; Qiao, P.; Matschi, S.; Vasquez, M.; Ramstein, G.P.; Bourgault, R.; Mohammadi, M.; Scanlon, M.J.; Molina, I.; Smith, L.G.; et al. Integrating GWAS and TWAS to elucidate the genetic architecture of maize leaf cuticular conductance. Plant Physiol. 2022, 189, 2144–2158. [Google Scholar] [CrossRef]
- Jamann, T.M.; Sood, S.; Wisser, R.J.; Holland, J.B. High-Throughput Resequencing of Maize Landraces at Genomic Regions Associated with Flowering Time. PLoS ONE 2017, 12, e0168910. [Google Scholar] [CrossRef]
- Jagtap, A.B.; Vikal, Y.; Johal, G.S. Genome-Wide Development and Validation of Cost-Effective KASP Marker Assays for Genetic Dissection of Heat Stress Tolerance in Maize. Int. J. Mol. Sci. 2020, 21, 7386. [Google Scholar] [CrossRef]
- Zorić, M.; Gunjača, J.; Galić, V.; Jukić, G.; Varnica, I.; Šimić, D. Best Linear Unbiased Predictions of Environmental Effects on Grain Yield in Maize Variety Trials of Different Maturity Groups. Agronomy 2022, 12, 922. [Google Scholar] [CrossRef]
- Liu, H.-M.; Liu, Z.-F.; Zheng, J.-P.; Yang, D.; Hu, S.-Z.; Yan, S.-H.; He, X.-W. coPLINK: A complementary tool to PLINK. PLoS ONE 2020, 15, e0239144. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Song, L.; Gu, W.; Guan, Y.; Wang, H.; Shi, B.; Zhou, Z.; Zheng, H.; Jiang, Y.; Yao, Y. Genome-Wide Comparative Analysis of Genetic Diversity of Regular and Specialty Maize Inbred Lines Through Genotyping by Target Sequencing (GBTS). Plant Mol. Biol. Rep. 2022, 40, 221–231. [Google Scholar] [CrossRef]
- Rio, S.; Moreau, L.; Charcosset, A.; Mary-Huard, T. Accounting for Group-Specific Allele Effects and Admixture in Genomic Predictions: Theory and Experimental Evaluation in Maize. Genetics 2020, 216, 27–41. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- He, W.; Xu, L.; Wang, J.; Yue, Z.; Jing, Y.; Tai, S.; Yang, J.; Fang, X. VCF2PCACluster: A simple, fast and memory-efficient tool for principal component analysis of tens of millions of SNPs. BMC Bioinform. 2024, 25, 173. [Google Scholar] [CrossRef]
- Vos, P.G.; Paulo, M.J.; Voorrips, R.E.; Visser, R.G.F.; van Eck, H.J.; van Eeuwijk, F.A. Evaluation of LD decay and various LD-decay estimators in simulated and SNP-array data of tetraploid potato. Theor. Appl. Genet. 2017, 130, 123–135. [Google Scholar] [CrossRef]
- Dang, D.; Guan, Y.; Zheng, H.; Zhang, X.; Zhang, A.; Wang, H.; Ruan, Y.; Qin, L. Genome-Wide Association Study and Genomic Prediction on Plant Architecture Traits in Sweet Corn and Waxy Corn. Plants 2023, 12, 303. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Qiu, C.-W.; Wu, Y.; Yao, L.; Cao, F.; Chen, Z.-H.; Wu, F. Integrated genome-wide association and RNA sequencing analyses reveal key long noncoding RNAs and target genes for drought tolerance in wheat. Plant Physiol. Biochem. 2025, 227, 110190. [Google Scholar] [CrossRef]
- Morgounov, A.; Li, H.; Shepelev, S.; Ali, M.; Flis, P.; Koksel, H.; Savin, T.; Shamanin, V. Genetic Characterization of Spring Wheat Germplasm for Macro-, Microelements and Trace Metals. Plants 2022, 11, 2173. [Google Scholar] [CrossRef]
- Daba, S.D.; Tyagi, P.; Brown-Guedira, G.; Mohammadi, M. Genome-Wide Association Studies to Identify Loci and Candidate Genes Controlling Kernel Weight and Length in a Historical United States Wheat Population. Front. Plant Sci. 2018, 9, 1045. [Google Scholar] [CrossRef]
- Lu, Y.; Rosenfeld, R.; Simon, I.; Nau, G.J.; Bar-Joseph, Z. A probabilistic generative model for GO enrichment analysis. Nucleic Acids Res. 2008, 36, e109. [Google Scholar] [CrossRef]
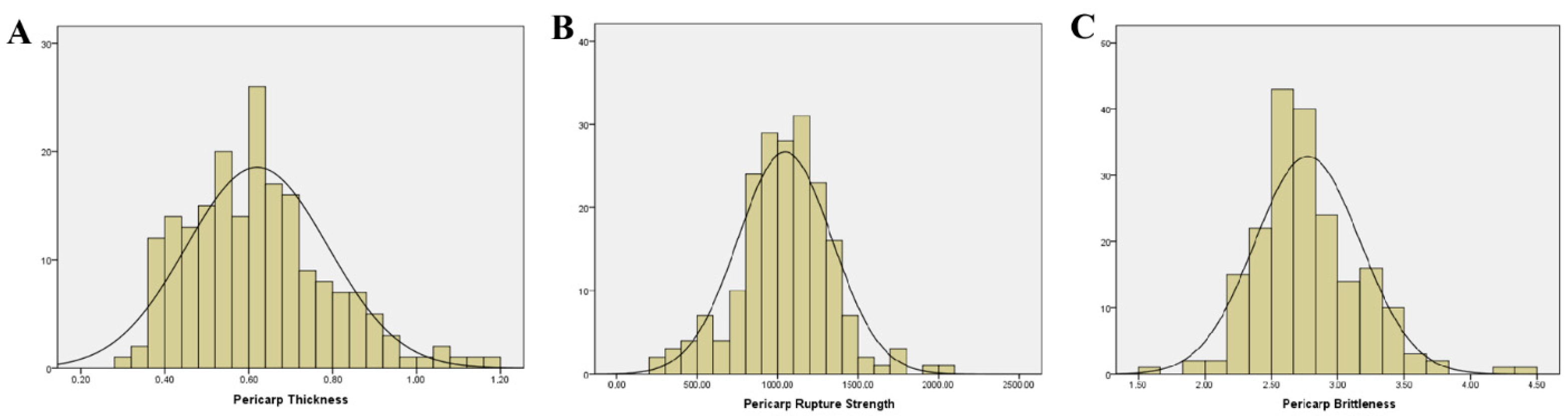

| Trait | Mean | Standard Deviation | Variation Rang | Coefficient of Variation (%) | Heritability (%) |
|---|---|---|---|---|---|
| Pericarp Thickness (mm) | 0.62 | 0.17 | 0.30–1.16 | 27.42 | 20.02 |
| Pericarp Rupture Strength (g) | 1045.54 | 293.43 | 213.13–2087.07 | 28.06 | 17.81 |
| Pericarp Brittleness (g.sec) | 2.77 | 0.40 | 1.60–4.50 | 14.44 | 26.57 |
| Pericarp Thickness | Pericarp Rupture Strength | Pericarp Brittleness | |
|---|---|---|---|
| Pericarp Thickness | 1 | ||
| Pericarp Rupture Strength | 0.140 * | 1 | |
| Pericarp Brittleness | 0.125 | −0.152 * | 1 |
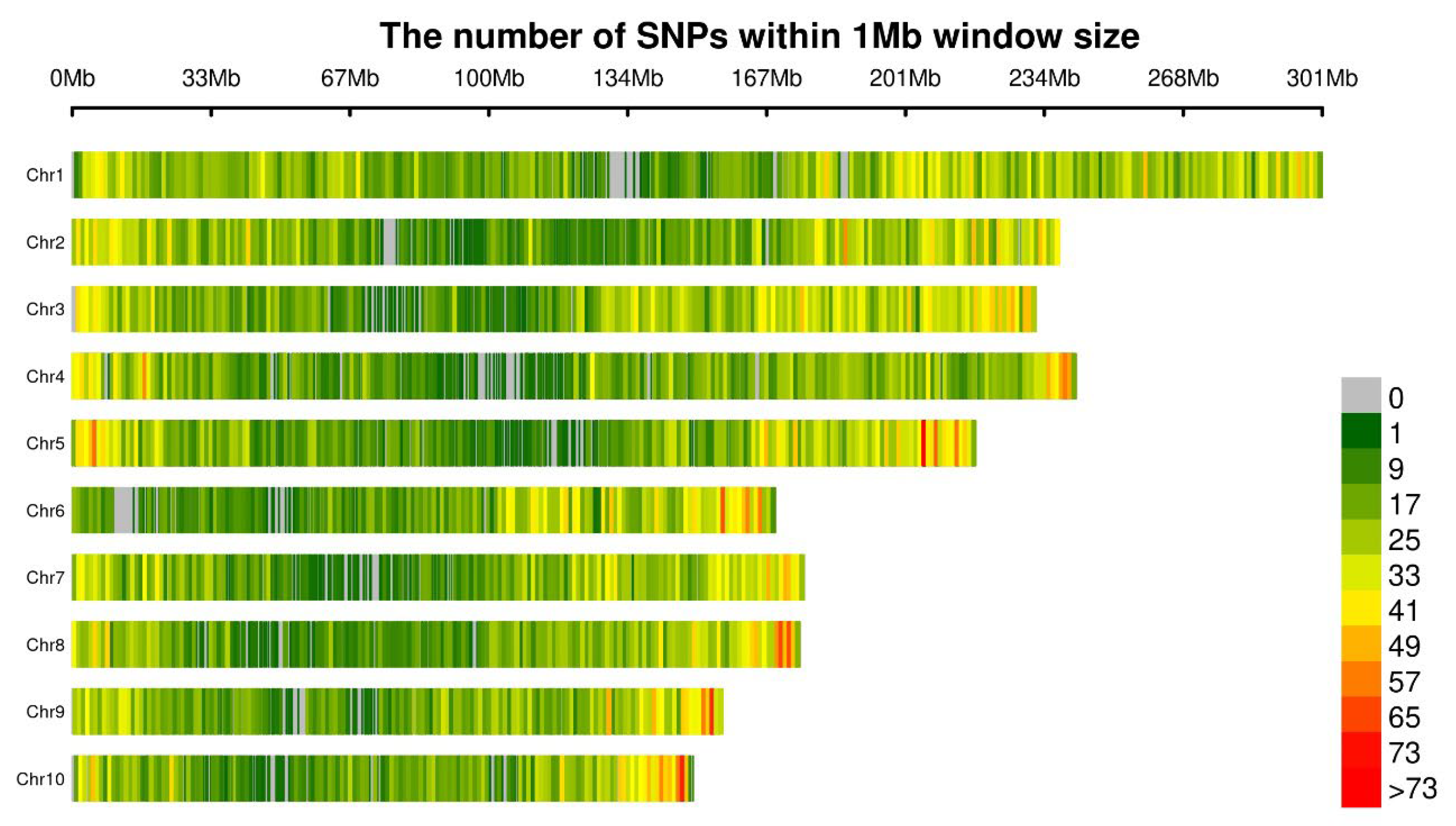
| Chromosome | SNPs | Density (Per Mbp) |
|---|---|---|
| Chr1 | 40,542 | 134.47 |
| Chr2 | 30,861 | 129.72 |
| Chr3 | 33,280 | 143.32 |
| Chr4 | 26,913 | 111.16 |
| Chr5 | 28,314 | 129.88 |
| Chr6 | 21,007 | 124.01 |
| Chr7 | 22,127 | 125.15 |
| Chr8 | 21,622 | 123.27 |
| Chr9 | 20,633 | 131.42 |
| Chr10 | 19,254 | 128.7 |
| Average | 26,455.3 | 128.43 |
| Chromosome | Position | Reference | Alternate | Candidate Genes | Annotations |
|---|---|---|---|---|---|
| 1 | 202,205,975 | G | A | Zm00001eb037610 | NC domain-containing protein-related |
| Zm00001eb037620 | Uncharacterized | ||||
| Zm00001eb037630 | photosystemI2 | ||||
| Zm00001eb037640 | Activator of 90 kDa heat shock protein ATPase | ||||
| 202,206,517 | G | A | Zm00001eb037630 | photosystemI2 | |
| Zm00001eb037640 | Activator of 90 kDa heat shock protein ATPase | ||||
| 2 | 64,578,153 | G | A | Zm00001eb084700 | Uncharacterized |
| Zm00001eb084710 | isopentenyl transferase2 | ||||
| 198,486,886 | C | T | Zm00001eb102550 | pyruvate dehydrogenase3 | |
| 3 | 160,120,677 | C | T | Zm00001eb142850 | L-type lectin-domain containing receptor kinase IX.1 |
| Zm00001eb142860 | Uncharacterized | ||||
| 160,157,364 | A | G | Zm00001eb142850 | L-type lectin-domain containing receptor kinase IX.1 | |
| Zm00001eb142860 | Uncharacterized | ||||
| 160,346,446 | C | A | Zm00001eb142890 | Uncharacterized | |
| Zm00001eb142900 | Uncharacterized | ||||
| Zm00001eb142910 | Uncharacterized | ||||
| Zm00001eb142920 | Uncharacterized | ||||
| 160,368,430 | T | C | Zm00001eb142890 | Uncharacterized | |
| Zm00001eb142900 | Uncharacterized | ||||
| Zm00001eb142910 | Uncharacterized | ||||
| Zm00001eb142920 | Uncharacterized | ||||
| 161,998,093 | A | T | Zm00001eb143120 | nucleobase: cation symporter10 | |
| Zm00001eb143130 | nucleobase: cation symporter10 | ||||
| Zm00001eb143150 | uncharacterized | ||||
| Zm00001eb143160 | uncharacterized | ||||
| 6 | 160,083,262 | T | C | Zm00001eb288790 | uncharacterized |
| Zm00001eb288800 | auxin import carrier4 | ||||
| 164,570,952 | G | T | Zm00001eb290360 | protein transporter | |
| Zm00001eb290370 | uncharacterized | ||||
| Zm00001eb290380 | uncharacterized | ||||
| Zm00001eb290390 | uncharacterized | ||||
| 7 | 45,582,095 | G | A | Zm00001eb306680 | protein binding protein |
| 8 | 64,175,962 | A | G | Zm00001eb342510 | mov34/MPN/PAD-1 family protein pseudogene |
| 9 | 5,510,195 | G | C | Zm00001eb372240 | uncharacterized |
| 5,991,255 | C | A | Zm00001eb372390 | uncharacterized | |
| Zm00001eb372400 | carboxyesterase2 | ||||
| 150,496,166 | A | T | Zm00001eb399220 | glutamate decarboxylase4 | |
| Zm00001eb399230 | Pentatricopeptide repeat-containing protein | ||||
| Zm00001eb399240 | leunig-related14 | ||||
| 10 | 5,365,786 | G | A | Zm00001eb406820 | dihydrodipicolinate decarboxylase1 |
| Zm00001eb406830 | DNA-3-methyladenine glycosylase4 | ||||
| Zm00001eb406840 | uncharacterized | ||||
| Zm00001eb406850 | uncharacterized | ||||
| Zm00001eb406860 | uncharacterized | ||||
| Zm00001eb406870 | uncharacterized | ||||
| 87,284,261 | T | G | Zm00001eb417200 | glycogen synthase kinase7 | |
| 120,473,230 | A | G | Zm00001eb417210 | uncharacterized |
| Chromosome | Position | Reference | Alternate | Candidate Genes | Annotations |
|---|---|---|---|---|---|
| 5 | 163,983,546 | C | G | Zm00001eb241080 | Os02g0478550-like protein |
| Zm00001eb241090 | ribosomal protein S27b | ||||
| Zm00001eb241100 | DUF679 domain membrane protein 7 | ||||
| Zm00001eb241110 | RS21-C6 protein | ||||
| 10 | 133,474,083 | C | T | Zm00001eb426710 | uncharacterized |
| Zm00001eb426720 | pentatricopeptide repeat protein 513 | ||||
| Zm00001eb426730 | integral membrane protein like protein |
| Chromosome | Position | Reference | Alternate | Candidate Genes | Annotations |
|---|---|---|---|---|---|
| 9 | 6,307,872 | G | A | Zm00001eb372460 | uncharacterized |
| Zm00001eb372470 | intramolecular lyase activity | ||||
| Zm00001eb372480 | osmotin-like protein | ||||
| Zm00001eb372490 | TCP-transcription factor 9 |
| Candidate Genes | Overlapping | Name | Annotations |
|---|---|---|---|
| Zm00001eb314860 | PRS BLUP + PT BLUP | ZmbZIP130 | bZIP130 transcription factor |
| Zm00001eb314870 | PRS BLUP + PT BLUP | non-coding RNA | |
| Zm00001eb314880 | PRS BLUP + PT BLUP | uncharacterized | |
| Zm00001eb415550 | 2023 PB + PT BLUP | ZmCCR4 | serine/threonine protein kinase CCR4 |
| Zm00001eb415560 | 2023 PB + PT BLUP | ZmCCR4 | serine/threonine protein kinase CCR4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Jin, Y.; Gao, S.; He, H.; Zhao, T.; Yue, Y.; Yang, X.; Wang, X. Genome-Wide Association Study Dissects the Genetic Architecture of Pericarp Traits in Fresh-Eating Maize. Plants 2026, 15, 74. https://doi.org/10.3390/plants15010074
Jin Y, Gao S, He H, Zhao T, Yue Y, Yang X, Wang X. Genome-Wide Association Study Dissects the Genetic Architecture of Pericarp Traits in Fresh-Eating Maize. Plants. 2026; 15(1):74. https://doi.org/10.3390/plants15010074
Chicago/Turabian StyleJin, Yukun, Song Gao, Huan He, Tong Zhao, Yaohai Yue, Xiangyu Yang, and Xinqi Wang. 2026. "Genome-Wide Association Study Dissects the Genetic Architecture of Pericarp Traits in Fresh-Eating Maize" Plants 15, no. 1: 74. https://doi.org/10.3390/plants15010074
APA StyleJin, Y., Gao, S., He, H., Zhao, T., Yue, Y., Yang, X., & Wang, X. (2026). Genome-Wide Association Study Dissects the Genetic Architecture of Pericarp Traits in Fresh-Eating Maize. Plants, 15(1), 74. https://doi.org/10.3390/plants15010074





