In-Depth Exploration of the Coloration Mechanism of Iris dichotoma Pall. via Transcriptomic and Metabolomic Analyses
Abstract
1. Introduction
2. Results
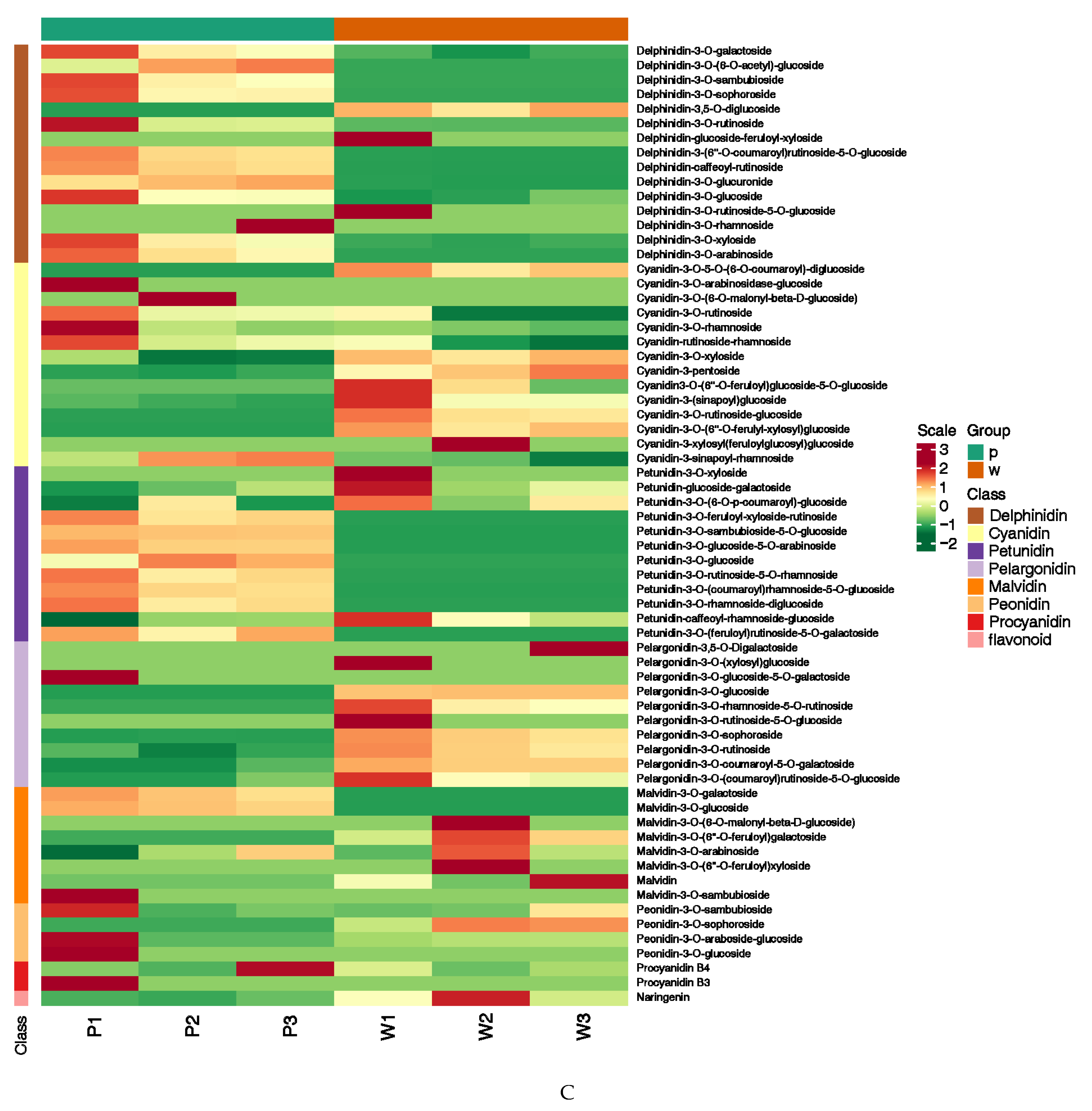
2.1. Determination of Targeted Anthocyanin Metabolites in Flowers of Different Colors
2.2. Transcriptome Analysis of Petals
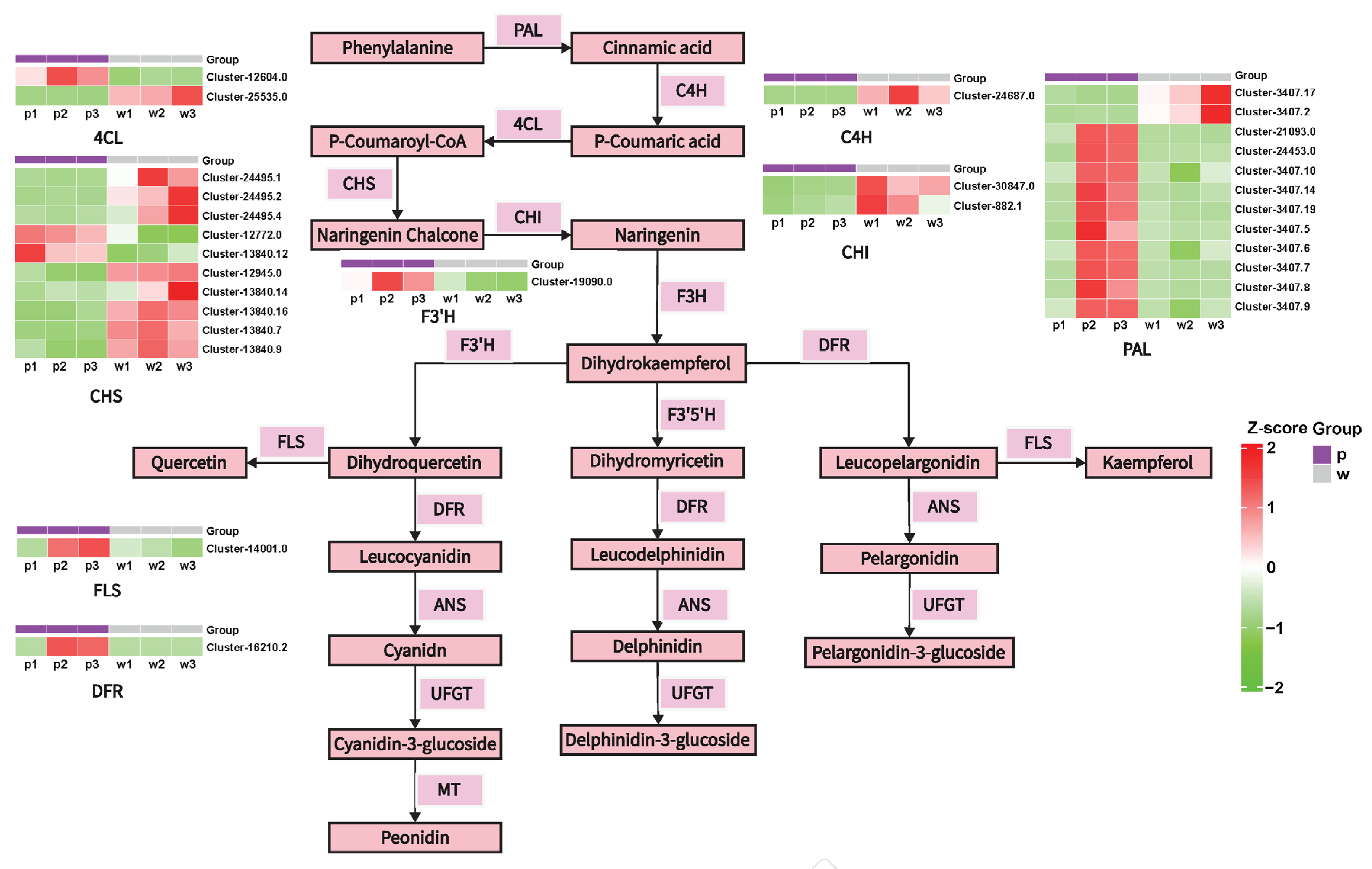
2.3. Differential Expression of Anthocyanin Structural Genes
2.4. Analysis of Key Transcription Factors Regulating Anthocyanin Synthesis
3. Discussion
4. Methods
4.1. Plant Materials
4.2. Extraction, Separation, Identification, and Quantification of Anthocyanins
4.3. RNA-Seq
4.4. Weighted Gene Co-Expression Network Analysis
4.5. qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, G.; Liu, H.; Shi, G.; Xu, N.; Niu, Z.; Wang, L.; Zhao, R.; Wang, L.; Fan, L. Multi-omics analysis of Iris sanguinea with distinctive flower colors provides insights into petal coloration. Hortic. Plant J. 2024, 11, 1274–1290. [Google Scholar] [CrossRef]
- Ichino, T.; Fuji, K.; Ueda, H.; Takahashi, H.; Koumoto, Y.; Takagi, J.; Tamura, K.; Sasaki, R.; Aoki, K.; Shimada, T.; et al. GFS9/TT9 contributes to intracellular membrane trafficking and flavonoid accumulation in Arabidopsis thaliana. Plant J. 2014, 80, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Yang, R.; Wang, Q.; Gao, L.; Li, S.; Li, L.; Chen, M.; Jiang, S.; Liu, D.; Li, Y.; et al. Multi-omics analysis uncovers novel gene regulatory networks of flower coloration in Lagerstroemia indica. Ind. Crops Prod. 2024, 216, 118710. [Google Scholar] [CrossRef]
- Peng, Q.-Z.; Zhu, Y.; Liu, Z.; Du, C.; Li, K.-G.; Xie, D.-Y. An integrated approach to demonstrating the ANR pathway of proanthocyanidin biosynthesis in plants. Planta 2012, 236, 901–918. [Google Scholar] [CrossRef]
- Fukui, Y.; Tanaka, Y.; Kusumi, T.; Iwashita, T.; Nomoto, K. A rationale for the shift in colour towards blue in transgenic carnation flowers expressing the flavonoid 3′,5′-hydroxylase gene. Phytochemistry 2003, 63, 15–23. [Google Scholar] [CrossRef]
- Seitz, C.; Vitten, M.; Steinbach, P.; Hartl, S.; Hirsche, J.; Rathje, W.; Treutter, D.; Forkmann, G. Redirection of anthocyanin synthesis in Osteospermum hybrida by a two-enzyme manipulation strategy. Phytochemistry 2007, 68, 824–833. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F.; Chandler, S. Recent Progress of Flower Colour Modification by Biotechnology. Int. J. Mol. Sci. 2009, 10, 5350–5369. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F.; Kalc, G.; Senior, M.; Dyson, B.; Nakamura, N.; Katsumoto, Y.; Chandler, S. Flower color modification by engineering of the flavonoid biosynthetic pathway: Practical perspectives. Biosci. Biotechnol. Biochem. 2010, 74, 1760–1769. [Google Scholar] [CrossRef]
- Xu, W.-J.; Yu, F.-Y.; Jia, Q.-X.; Luo, G.-J.; Bi, X.-Y. ‘Sweet Princess’: A New Summer Ornamental Iris Cultivar. HortScience 2017, 52, 1832–1833. [Google Scholar] [CrossRef]
- Faraco, M.; Spelt, C.; Bliek, M.; Verweij, W.; Hoshino, A.; Espen, L.; Prinsi, B.; Jaarsma, R.; Tarhan, E.; de Boer, A.H.; et al. Hyperacidification of Vacuoles by the Combined Action of Two Different P-ATPases in the Tonoplast Determines Flower Color. Cell Rep. 2014, 6, 32–43. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Lu, C.; Wang, Z.; Deng, C.; Gao, K.; Li, J.; Fang, Z.; Liu, H.; Hong, Y.; et al. Flavonoid extracts from chrysanthemum with appropriate anthocyanins turn blue when exposed to iron ions. Hortic. Plant J. 2024, 10, 837–852. [Google Scholar] [CrossRef]
- Sundaramoorthy, J.; Park, G.T.; Lee, J.-D.; Kim, J.H.; Seo, H.S.; Song, J.T. A P3A-Type ATPase and an R2R3-MYB Transcription Factor Are Involved in Vacuolar Acidification and Flower Coloration in Soybean. Front. Plant Sci. 2020, 11, 580085. [Google Scholar] [CrossRef] [PubMed]
- Tindal, R.A.; Jeffery, D.W.; Muhlack, R.A. Nonlinearity and anthocyanin colour expression: A mathematical analysis of anthocyanin association kinetics and equilibria. Food Res. Int. 2024, 183, 114195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Y.; Ye, J.; Fan, Q.; Lin, X.; Gou, Z.; Jiang, S. Dietary supplementation of bilberry anthocyanin on growth performance, intestinal mucosal barrier and cecal microbes of chickens challenged with Salmonella Typhimurium. J. Anim. Sci. Biotechnol. 2023, 14, 15. [Google Scholar] [CrossRef]
- Alappat, B.; Alappat, J. Anthocyanin Pigments: Beyond Aesthetics. Molecules 2020, 25, 5500. [Google Scholar] [CrossRef]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N.; et al. Allelic Variation of MYB10 Is the Major Force Controlling Natural Variation in Skin and Flesh Color in Strawberry (Fragaria spp.). Fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef]
- Del Rio, D.; Rodriguez-Mateos, A.; Spencer, J.P.E.; Tognolini, M.; Borges, G.; Crozier, A. Dietary (Poly)phenolics in Human Health: Structures, Bioavailability, and Evidence of Protective Effects Against Chronic Diseases. Antioxid. Redox Signal. 2013, 18, 1818–1892. [Google Scholar] [CrossRef]
- Huang, X.; Liu, L.; Qiang, X.; Meng, Y.; Li, Z.; Huang, F. Integrated Metabolomic and Transcriptomic Profiles Provide Insights into the Mechanisms of Anthocyanin and Carotenoid Biosynthesis in Petals of Medicago sativa ssp. sativa and Medicago sativa ssp. falcata. Plants 2024, 13, 700. [Google Scholar] [CrossRef]
- Ming, H.; Wang, Q.; Wu, Y.; Liu, H.; Zheng, L.; Zhang, G. Transcriptome analysis reveals the mechanism of anthocyanidins biosynthesis during grains development in purple corn (Zea mays L.). J. Plant Physiol. 2021, 257, 153328. [Google Scholar] [CrossRef]
- Xu, P.; Li, X.; Fan, J.; Tian, S.; Cao, M.; Lin, A.; Gao, Q.; Xiao, K.; Wang, C.; Kuang, H.; et al. An arginine-to-histidine mutation in flavanone-3-hydroxylase results in pink strawberry fruits. Plant Physiol. 2023, 193, 1849–1865. [Google Scholar] [CrossRef]
- Bharathy, P.; Thanikachalam, P.V. Pharmacological relevance of anthocyanin derivative: A review. Pharmacol. Res.-Mod. Chin. Med. 2025, 14, 100565. [Google Scholar] [CrossRef]
- Cai, T.; Ge-Zhang, S.; Song, M. Anthocyanins in metabolites of purple corn. Front. Plant Sci. 2023, 14, 1154535. [Google Scholar] [CrossRef] [PubMed]
- Wallace, T.C. Anthocyanins in Cardiovascular Disease. Adv. Nutr. 2011, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, G.; Liu, T.; Deng, S. The MYB transcription factor RtAN2 promotes anthocyanin accumulation over proanthocyanidin during the fruit ripening of rose myrtle berries. Ind. Crops Prod. 2025, 224, 120433. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Y.; Wang, W.; Wang, N.; Yan, R.; Li, S.; Zhang, T.; Liu, J.; Zeng, X.; Zhao, S.; et al. Analysis of WD40 genes in kiwifruit reveals the key role of the light-induced AcTTG1-AcMYB75-AcbHLH2 complex in anthocyanin accumulation. Int. J. Biol. Macromol. 2025, 297, 139758. [Google Scholar] [CrossRef]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant Flavonoids—Biosynthesis, Transport and Involvement in Stress Responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef]
- Singh, A.K.; Ma, Y.-J.; Duan, H.-R.; Zhang, F.; Li, Y.; Yang, H.-S.; Tian, F.-P.; Zhou, X.-H.; Wang, C.-M.; Ma, R. Transcriptomic analysis of Lycium ruthenicum Murr. during fruit ripening provides insight into structural and regulatory genes in the anthocyanin biosynthetic pathway. PLoS ONE 2018, 13, e0208627. [Google Scholar] [CrossRef]
- Villegas, D.; Handford, M.; Alcalde, J.A.; Perez-Donoso, A. Exogenous application of pectin-derived oligosaccharides to grape berries modifies anthocyanin accumulation, composition, and gene expression. Plant Physiol. Biochem. 2016, 104, 125–133. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, B.; Wu, T.; Yang, Y.; Fan, L.; Wen, M.; Sui, J. Transcriptomic profiling of two Pak Choi varieties with contrasting anthocyanin contents provides an insight into structural and regulatory genes in the anthocyanin biosynthetic pathway. BMC Genom. 2017, 18, 288. [Google Scholar] [CrossRef]
- Husain, A.; Chanana, H.; Khan, S.A.; Dhanalekshmi, U.M.; Ali, M.; Alghamdi, A.A.; Ahmad, A. Chemistry and Pharmacological Actions of Delphinidin, a Dietary Purple Pigment in Anthocyanidin and Anthocyanin Forms. Front. Nutr. 2022, 9, 746881. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wei, J.; Wu, Y.; Chen, S.; Yu, C.; Zhu, Y.; Lin, Z.; Lv, H.; Chen, Y. Comprehensive study of non-volatile and volatile metabolites in five water lily species and varieties (Nymphaea spp.) using widely targeted metabolomics. Beverage Plant Res. 2024, 4, e012. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, M.; Zheng, X.; Shu, Q.; Li, H.; Zhong, P.; Zhang, H.; Xu, Y.; Wang, L.; Wang, L. Relationship between the Composition of Flavonoids and Flower Colors Variation in Tropical Water Lily (Nymphaea) Cultivars. PLoS ONE 2012, 7, e34335. [Google Scholar] [CrossRef]
- Albert, N.W. Subspecialization of R2R3-MYB Repressors for Anthocyanin and Proanthocyanidin Regulation in Forage Legumes. Front. Plant Sci. 2015, 6, 01165. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Jiang, L.; Yue, M.; Liu, Y.; Zhang, N.; Lin, Y.; Zhang, Y.; Wang, Y.; Li, M.; Luo, Y.; Zhang, Y.; et al. A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × ananassa). Plant Biotechnol. J. 2023, 21, 1140–1158. [Google Scholar] [CrossRef]
- Liu, H.; Lou, Q.; Ma, J.; Su, B.; Gao, Z.; Liu, Y. Cloning and Functional Characterization of Dihydroflavonol 4-Reductase Gene Involved in Anthocyanidin Biosynthesis of Grape Hyacinth. Int. J. Mol. Sci. 2019, 20, 4743. [Google Scholar] [CrossRef]
- Rajput, R.; Naik, J.; Stracke, R.; Pandey, A. Interplay between R2R3 MYB-type activators and repressors regulates proanthocyanidin biosynthesis in banana (Musa acuminata). New Phytol. 2022, 236, 1108–1127. [Google Scholar] [CrossRef]
- Yue, M.; Jiang, L.; Zhang, N.; Zhang, L.; Liu, Y.; Lin, Y.; Zhang, Y.; Luo, Y.; Zhang, Y.; Wang, Y.; et al. Regulation of flavonoids in strawberry fruits by FaMYB5/FaMYB10 dominated MYB-bHLH-WD40 ternary complexes. Front. Plant Sci. 2023, 14, 1145670. [Google Scholar] [CrossRef]
- Li, S. Transcriptional control of flavonoid biosynthesis. Plant Signal. Behav. 2014, 9, 27522. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Gao, R.; Han, T.; Zhang, J.; Wang, Y.; Kimani, S.; Wang, L.; Gao, X. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Commun. Biol. 2020, 3, 396. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, Z.; Li, J.; Zhang, H.; Peng, Y.; Li, Z. Uncovering Hierarchical Regulation among MYB-bHLH-WD40 Proteins and Manipulating Anthocyanin Pigmentation in Rice. Int. J. Mol. Sci. 2022, 23, 8203. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Jiang, Q.; Shui, Z.; An, P.; Shi, S.; Liu, T.; Zhang, H.; Huang, S.; Jing, B.; Xiao, E.; et al. HaMYBA-HabHLH1 regulatory complex and HaMYBF fine-tune red flower coloration in the corolla of sunflower (Helianthus annuus L.). Plant Sci. 2024, 338, 111901. [Google Scholar] [CrossRef]
- Ma, S.; Zhou, H.; Ren, T.; Yu, E.-R.; Feng, B.; Wang, J.; Zhang, C.; Zhou, C.; Li, Y. Integrated transcriptome and metabolome analysis revealed that HaMYB1 modulates anthocyanin accumulation to deepen sunflower flower color. Plant Cell Rep. 2024, 43, 74. [Google Scholar] [CrossRef]
- He, F.; Mu, L.; Yan, G.-L.; Liang, N.-N.; Pan, Q.-H.; Wang, J.; Reeves, M.J.; Duan, C.-Q. Biosynthesis of Anthocyanins and Their Regulation in Colored Grapes. Molecules 2010, 15, 9057–9091. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory Mechanisms of Anthocyanin Biosynthesis in Apple and Pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef]
- Lou, Q.; Liu, Y.; Qi, Y.; Jiao, S.; Tian, F.; Jiang, L.; Wang, Y. Transcriptome sequencing and metabolite analysis reveal the role of delphinidin metabolism in flower colour in grape hyacinth. J. Exp. Bot. 2014, 65, 3157–3164. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Qiang, X.; Huang, F.; Huang, X.; Liu, L. In-Depth Exploration of the Coloration Mechanism of Iris dichotoma Pall. via Transcriptomic and Metabolomic Analyses. Plants 2025, 14, 1387. https://doi.org/10.3390/plants14091387
Yu Y, Qiang X, Huang F, Huang X, Liu L. In-Depth Exploration of the Coloration Mechanism of Iris dichotoma Pall. via Transcriptomic and Metabolomic Analyses. Plants. 2025; 14(9):1387. https://doi.org/10.3390/plants14091387
Chicago/Turabian StyleYu, Yalin, Xiaojing Qiang, Fan Huang, Xiuzheng Huang, and Lei Liu. 2025. "In-Depth Exploration of the Coloration Mechanism of Iris dichotoma Pall. via Transcriptomic and Metabolomic Analyses" Plants 14, no. 9: 1387. https://doi.org/10.3390/plants14091387
APA StyleYu, Y., Qiang, X., Huang, F., Huang, X., & Liu, L. (2025). In-Depth Exploration of the Coloration Mechanism of Iris dichotoma Pall. via Transcriptomic and Metabolomic Analyses. Plants, 14(9), 1387. https://doi.org/10.3390/plants14091387





