Validation of QTLs for Resistance to Pre-Harvest Sprouting in a Panel of European Wheat Cultivars
Abstract
1. Introduction
2. Results
2.1. Phenotypic Evaluation of PHS Resistance
2.2. Genotyping
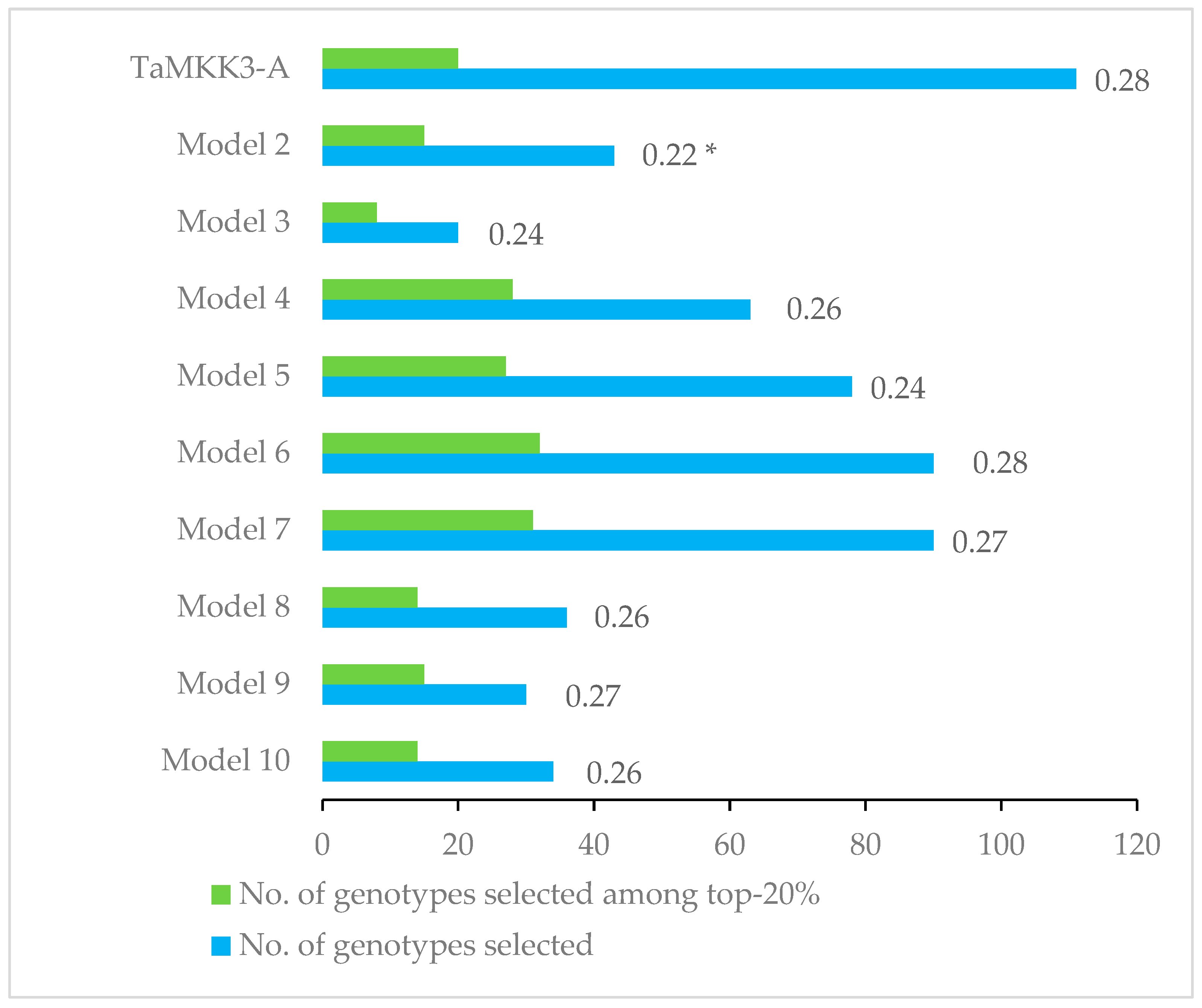
2.3. Allelic Effect of KASP Markers on Germination Index
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Field Experiments
4.2. Sampling of Spikes and Quantification of Seed Dormancy (SD)
4.3. DNA Extraction and KASP Genotyping
4.4. Statistical Analysis of Field Experiments and KASP Marker Validation
4.5. Allelic Effect of KASP Markers on GI
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| PHS | pre-harvest sprouting |
| GI | germination index |
| SNP | single-nucleotide polymorphism |
| KASP | Kompetitive Allele Specific PCR |
| QTL | quantitative trait locus |
| SD | seed dormancy |
| SE | Southeastern Europe |
| MAS | marker-assisted selection |
| ABA | abscisic acid |
| MAP | mitogen-activated protein |
| PAV | presence/absence variation |
| NCED | nine-cis-epoxycarotenoid dioxygenase |
| GWAS | genome-wide association study |
| ABRE | abscisic acid-responsive element |
| RNAi | RNA interference |
| ANOVA | analysis of variance |
| SD | standard deviation |
| HSD | honestly significant difference |
| DNA | deoxyribonucleic acid |
| BLUP | best linear unbiased prediction |
| PVE | phenotypic variance explained |
| AIC | Akaike’s Information Criterion |
| MAGIC | multiparent advanced generation inter-cross |
| CDS | coding sequence |
| RCBD | randomized complete block design |
| PCR | polymerase chain reaction |
| SAS | statistical analysis system |
References
- FAO. FAOSTAT Database; Food and Agriculture Organization of the United Nations: Rome, Italy. Available online: https://www.fao.org/faostat/en/ (accessed on 16 February 2025).
- Wang, D.; Pang, Y.; Dong, L.; Li, A.; Kong, L.; Liu, S. Allelic Impacts on Pre-Harvest Sprouting Resistance and Favorable Haplotypes in TaPHS1 of Chinese Wheat Accessions. Crop J. 2020, 8, 515–521. [Google Scholar] [CrossRef]
- Mares, D.J.; Mrva, K. Wheat Grain Preharvest Sprouting and Late Maturity Alpha-Amylase. Planta 2014, 240, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.-N.; Yu, Z.-Y.; Gao, W.; Wang, X.-Y.; Cao, J.-J.; Lu, J.; Ma, C.-X.; Chang, C.; Zhang, H.-P. Dissecting the Genetic Basis of Grain Color and Pre-Harvest Sprouting Resistance in Common Wheat by Association Analysis. J. Integr. Agric. 2023, 22, 2617–2631. [Google Scholar] [CrossRef]
- Khumalo, T.P.; Hlongoane, T.; Barnard, A.; Tsilo, T.J. Genomic Regions Influencing Preharvest Sprouting Tolerance in Two Doubled-Haploid Wheat Populations (Triticum aestivum L.). Agronomy 2022, 12, 832. [Google Scholar] [CrossRef]
- DePauw, R.M.; Knox, R.E.; Singh, A.K.; Fox, S.L.; Humphreys, D.G.; Hucl, P. Developing Standardized Methods for Breeding Preharvest Sprouting Resistant Wheat, Challenges and Successes in Canadian Wheat. Euphytica 2012, 188, 7–14. [Google Scholar] [CrossRef]
- Ali, A.; Cao, J.; Jiang, H.; Chang, C.; Zhang, H.P.; Sheikh, S.W.; Shah, L.; Ma, C. Unraveling Molecular and Genetic Studies of Wheat (Triticum aestivum L.) Resistance against Factors Causing Pre-Harvest Sprouting. Agronomy 2019, 9, 117. [Google Scholar] [CrossRef]
- Mares, D.J.; Mrva, K.; Cheong, J.; Fox, R.; Mather, D.E. Dormancy and Dormancy Release in White-Grained Wheat (Triticum aestivum L.). Planta 2021, 253, 5. [Google Scholar] [CrossRef]
- Vetch, J.M.; Stougaard, R.N.; Martin, J.M.; Giroux, M. Allelic Impacts of TaPHS1, TaMKK3, and Vp1B3 on Preharvest Sprouting of Northern Great Plains Winter Wheats. Crop Sci. 2019, 59, 140–149. [Google Scholar] [CrossRef]
- Dallinger, H.G.; Löschenberger, F.; Azrak, N.; Ametz, C.; Michel, S.; Bürstmayr, H. Genome-Wide Association Mapping for Pre-Harvest Sprouting in European Winter Wheat Detects Novel Resistance QTL, Pleiotropic Effects, and Structural Variation in Multiple Genomes. Plant Genome 2023, 17, e20301. [Google Scholar] [CrossRef]
- Kottearachchi, N.S.; Takao, S.; Kato, K.; Miura, H. Mapping of a QTL in Chromosome 3B for Grain Dormancy in White-Grained Wheat Population. J. Food Agric. 2010, 1, 1–10. [Google Scholar] [CrossRef]
- Kulwal, P.; Mir, R.; Kumar, S.; Gupta, P. QTL Analysis and Molecular Breeding for Seed Dormancy and Pre-Harvest Sprouting Tolerance in Bread Wheat. J. Plant Biol. 2010, 37, 59–74. [Google Scholar]
- Liton, M.M.U.A.; McCartney, C.A.; Hiebert, C.W.; Kumar, S.; Jordan, M.C.; Ayele, B.T. Identification of Loci for Pre-Harvest Sprouting Resistance in the Highly Dormant Spring Wheat RL4137. Theor. Appl. Genet. 2021, 134, 113–124. [Google Scholar] [CrossRef]
- Huang, Y.; Dai, X.; Liu, H.; Yu, S.; Mai, C.; Yu, L.; Yu, G.; Yang, L.; Zhou, Y.; Li, H.; et al. Identification of Effective Alleles and Haplotypes Conferring Pre-Harvest Sprouting Resistance in Winter Wheat Cultivars. BMC Plant Biol. 2022, 22, 326. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.; Zhang, Y.; Li, M.; Xu, D.; Tian, X.; Song, J.; Luo, X.; Xie, L.; Wang, D.; et al. Genome-Wide Linkage Mapping for Preharvest Sprouting Resistance in Wheat Using 15K Single-Nucleotide Polymorphism Arrays. Front. Plant Sci. 2021, 12, 9206. [Google Scholar] [CrossRef]
- Shorinola, O.; Balcárková, B.; Hyles, J.; Tibbits, J.F.G.; Hayden, M.J.; Holušova, K.; Valárik, M.; Distelfeld, A.; Torada, A.; Barrero, J.M.; et al. Haplotype Analysis of the Pre-Harvest Sprouting Resistance Locus Phs-A1 Reveals a Causal Role of TAMKK3-A in Global Germplasm. Front. Plant Sci. 2017, 8, 1555. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.V.; Barrero, J.M.; Corbineau, F.; Gubler, F.; Benech-Arnold, R.L. Dormancy in Cereals (Not Too Much, Not so Little): About the Mechanisms behind This Trait. Seed Sci. Res. 2015, 25, 99–119. [Google Scholar] [CrossRef]
- Zeibig, F.; Kilian, B.; Özkan, H.; Pantha, S.; Frei, M. Phenotyping and Identification of Target Traits for de Novo Domestication of Wheat Wild Relatives. Food Energy Secur. 2023, 13, e497. [Google Scholar] [CrossRef]
- Gao, X.; Hu, C.H.; Li, H.Z.; Yao, Y.J.; Meng, M.; Dong, J.; Zhao, W.C.; Chen, Q.J.; Li, X.Y. Factors Affecting Pre-Harvest Sprouting Resistance in Wheat (Triticum aestivum L.): A Review. J. Anim. Plant Sci. 2013, 23, 556–565. [Google Scholar]
- Kulwal, P.; Ishikawa, G.; Benscher, D.; Feng, Z.; Yu, L.X.; Jadhav, A.; Mehetre, S.; Sorrells, M.E. Association Mapping for Pre-Harvest Sprouting Resistance in White Winter Wheat. Theor. Appl. Genet. 2012, 125, 793–805. [Google Scholar] [CrossRef]
- Barrero, J.M.; Cavanagh, C.; Verbyla, K.L.; Tibbits, J.F.G.; Verbyla, A.P.; Huang, B.E.; Rosewarne, G.M.; Stephen, S.; Wang, P.; Whan, A.; et al. Transcriptomic Analysis of Wheat Near-Isogenic Lines Identifies PM19-A1 and A2 as Candidates for a Major Dormancy QTL. Genome Biol. 2015, 16, 93. [Google Scholar] [CrossRef]
- Torada, A.; Koike, M.; Ogawa, T.; Takenouchi, Y.; Tadamura, K.; Wu, J.; Matsumoto, T.; Kawaura, K.; Ogihara, Y. A Causal Gene for Seed Dormancy on Wheat Chromosome 4A Encodes a MAP Kinase Kinase. Curr. Biol. 2016, 26, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Abe, F.; Kawahigashi, H.; Nakazono, K.; Tagiri, A.; Matsumoto, T.; Utsugi, S.; Ogawa, T.; Handa, H.; Ishida, H.; et al. A Wheat Homolog of MOTHER of FT and TFL1 Acts in the Regulation of Germination. Plant Cell 2011, 23, 3215–3229. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sehgal, S.K.; Li, J.; Lin, M.; Trick, H.N.; Yu, J.; Gill, B.S.; Bai, G. Cloning and Characterization of a Critical Regulator for Preharvest Sprouting in Wheat. Genetics 2013, 195, 263–273. [Google Scholar] [CrossRef]
- Jiang, H.; Zhao, L.X.; Chen, X.J.; Cao, J.J.; Wu, Z.Y.; Liu, K.; Zhang, C.; Wei, W.X.; Xie, H.Y.; Li, L.; et al. A Novel 33-Bp Insertion in the Promoter of TaMFT-3A Is Associated with Pre-Harvest Sprouting Resistance in Common Wheat. Mol. Breed. 2018, 38, 69. [Google Scholar] [CrossRef]
- Liu, S.; Wang, D.; Lin, M.; Sehgal, S.K.; Dong, L.; Wu, Y.; Bai, G. Artificial Selection in Breeding Extensively Enriched a Functional Allelic Variation in TaPHS1 for Pre-Harvest Sprouting Resistance in Wheat. Theor. Appl. Genet. 2021, 134, 339–350. [Google Scholar] [CrossRef]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and Validation of KASP Assays for Genes Underpinning Key Economic Traits in Bread Wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Miao, X.; Xia, X.; He, Z. Cloning of Seed Dormancy Genes (TaSdr) Associated with Tolerance to Pre-Harvest Sprouting in Common Wheat and Development of a Functional Marker. Theor. Appl. Genet. 2014, 127, 855–866. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, X.; He, Z. The Seed Dormancy Allele TaSdr-A1a Associated with Pre-Harvest Sprouting Tolerance Is Mainly Present in Chinese Wheat Landraces. Theor. Appl. Genet. 2017, 130, 81–89. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, X.M.; He, Z.H.; Röder, M.; Xia, L.Q. Distribution of Vp-1 Alleles in Chinese White-Grained Landraces, Historical and Current Wheat Cultivars. Cereal Res. Commun. 2009, 37, 169–177. [Google Scholar] [CrossRef]
- Chang, C.; Feng, J.M.; Si, H.Q.; Yin, B.; Zhang, H.P.; Ma, C.X. Validating a Novel Allele of Viviparous-1 (Vp-1Bf) Associated with High Seed Dormancy of Chinese Wheat Landrace, Wanxianbaimaizi. Mol. Breed. 2010, 25, 517–525. [Google Scholar] [CrossRef]
- Xia, L.Q.; Ganal, M.W.; Shewry, P.R.; He, Z.H.; Yang, Y.; Röder, M.S. Exploiting the Diversity of Viviparous-1 Gene Associated with Pre-Harvest Sprouting Tolerance in European Wheat Varieties. Euphytica 2008, 159, 411–417. [Google Scholar] [CrossRef]
- Wang, D.; Xie, J.; Wang, J.; Mu, M.; Xiong, H.; Ma, F.; Li, P.; Jia, M.; Li, S.; Li, J.; et al. Unraveling Allelic Impacts on Pre-Harvest Sprouting Resistance in TaVP1-B of Chinese Wheat Accessions Using Pan-Genome. Plants 2025, 14, 504. [Google Scholar] [CrossRef]
- Himi, E.; Noda, K. Red Grain Colour Gene (R) of Wheat Is a Myb-Type Transcription Factor. Euphytica 2005, 143, 239–242. [Google Scholar] [CrossRef]
- Mares, D.; Himi, E. The Role of TaMYB10-A1 of Wheat (Triticum aestivum L.) in Determining Grain Coat Colour and Dormancy Phenotype. Euphytica 2021, 217, 89. [Google Scholar] [CrossRef]
- Lang, J.; Fu, Y.; Zhou, Y.; Cheng, M.; Deng, M.; Li, M.; Zhu, T.; Yang, J.; Guo, X.; Gui, L.; et al. Myb10-D Confers PHS-3D Resistance to Pre-Harvest Sprouting by Regulating NCED in ABA Biosynthesis Pathway of Wheat. New Phytol. 2021, 230, 1940–1952. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Tan, C.; Lang, J.; Tang, H.; Hao, M.; Tan, Z.; Yu, H.; Zhou, Y.; Liu, Z.; Li, M.; et al. Identification of QPHS.Sicau-1B and QPHS.Sicau-3D from Synthetic Wheat for Pre-Harvest Sprouting Resistance Wheat Improvement. Mol. Breed. 2019, 39, 132. [Google Scholar] [CrossRef]
- Tai, L.; Wu, J.; Jing, Y.; Liu, H.; Zeng, Q.; Xu, X.; Shi, S.; Wang, H.; Liu, W.; Sun, J.; et al. A Genome-Wide Association Study Uncovers That TaPI4K-2A Regulates Pre-Harvest Sprouting in Wheat. Plant Commun. 2023, 5, 100739. [Google Scholar] [CrossRef]
- Albrecht, T.; Oberforster, M.; Kempf, H.; Ramgraber, L.; Schacht, J.; Kazman, E.; Zechner, E.; Neumayer, A.; Hartl, L.; Mohler, V. Genome-Wide Association Mapping of Preharvest Sprouting Resistance in a Diversity Panel of European Winter Wheats. J. Appl. Genet. 2015, 56, 277–285. [Google Scholar] [CrossRef]
- Shorinola, O.; Bird, N.; Simmonds, J.; Berry, S.; Henriksson, T.; Jack, P.; Werner, P.; Gerjets, T.; Scholefield, D.; Balcárková, B.; et al. The Wheat Phs-A1 Pre-Harvest Sprouting Resistance Locus Delays the Rate of Seed Dormancy Loss and Maps 0.3 CM Distal to the PM19 Genes in UK Germplasm. J. Exp. Bot. 2016, 67, 4169–4178. [Google Scholar] [CrossRef]
- Sydenham, S.L.; Barnard, A. Targeted Haplotype Comparisons between South African Wheat Cultivars Appear Predictive of Pre-Harvest Sprouting Tolerance. Front. Plant. Sci. 2018, 9, 63. [Google Scholar] [CrossRef]
- Shao, M.; Bai, G.; Rife, T.W.; Poland, J.; Lin, M.; Liu, S.; Chen, H.; Kumssa, T.; Fritz, A.; Trick, H.; et al. QTL Mapping of Pre-Harvest Sprouting Resistance in a White Wheat Cultivar Danby. Theor. Appl. Genet. 2018, 131, 1683–1697. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, R.; Hiebert, C.W.; Sorrells, M.E.; Spaner, D.; Graf, R.J.; Singh, J.; Randhawa, H.S. Mapping Pre-Harvest Sprouting Resistance Loci in AAC Innova × AAC Tenacious Spring Wheat Population. BMC Genom. 2021, 22, 900. [Google Scholar] [CrossRef] [PubMed]
- Martinez, S.A.; Shorinola, O.; Conselman, S.; See, D.; Skinner, D.Z.; Uauy, C.; Steber, C.M. Exome Sequencing of Bulked Segregants Identified a Novel TaMKK3-A Allele Linked to the Wheat ERA8 ABA-Hypersensitive Germination Phenotype. Theor. Appl. Genet. 2020, 133, 719–736. [Google Scholar] [CrossRef]
- Wamalwa, M.; Tadesse, Z.; Muthui, L.; Yao, N.; Zegeye, H.; Randhawa, M.; Wanyera, R.; Uauy, C.; Shorinola, O. Allelic Diversity Study of Functional Genes in East Africa Bread Wheat Highlights Opportunities for Genetic Improvement. Mol. Breed. 2020, 40, 104. [Google Scholar] [CrossRef]
- Lei, L.; Zhu, X.; Wang, S.; Zhu, M.; Carver, B.F.; Yan, L. TaMFT-A1 Is Associated with Seed Germination Sensitive to Temperature in Winter Wheat. PLoS ONE 2013, 8, e73330. [Google Scholar] [CrossRef]
- Dong, H.; Kou, C.; Hu, L.; Li, Y.; Fang, Y.; Peng, C. Haplotype Analysis and Gene Pyramiding for Pre-Harvest Sprouting Resistance in White-Grain Wheat. Int. J. Mol. Sci. 2025, 26, 728. [Google Scholar] [CrossRef]
- Fakthongphan, J.; Bai, G.; St. Amand, P.; Graybosch, R.A.; Baenziger, P.S. Identification of Markers Linked to Genes for Sprouting Tolerance (Independent of Grain Color) in Hard White Winter Wheat (HWWW). Theor. Appl. Genet. 2016, 129, 419–430. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, X.L.; Xia, L.Q.; Chen, X.M.; Xia, X.C.; Yu, Z.; He, Z.H.; Röder, M. Development and Validation of a Viviparous-1 STS Marker for Pre-Harvest Sprouting Tolerance in Chinese Wheats. Theor. Appl. Genet. 2007, 115, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A Decima Code for Growth Stages of Cereals. Weeed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Reddy, L.V.; Metzeger, R.J.; Ching, M.T. Effect of Temperature on Seed Dormancy of Wheat. Crop Sci. 1985, 16, 116–125. [Google Scholar] [CrossRef]
- Ikić, I.; Maričević, M.; Tomasović, S.; Gunjača, J.; Šatović, Z.; Šarčević, H. The Effect of Germination Temperature on Seed Dormancy in Croatian-Grown Winter Wheats. Euphytica 2012, 188, 25–34. [Google Scholar] [CrossRef]
- KASP Genotyping Services. Available online: https://www.biosearchtech.com/services/genotyping-services/all-inclusive-services (accessed on 5 March 2024).
- Cabral, A.L.; Jordan, M.C.; McCartney, C.A.; You, F.M.; Humphreys, D.G.; MacLachlan, R.; Pozniak, C.J. Identification of Candidate Genes, Regions and Markers for Pre-Harvest Sprouting Resistance in Wheat (Triticum aestivum L.). BMC Plant Biol. 2014, 14, 340. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Cai, S.; Wang, S.; Liu, S.; Zhang, G.; Bai, G. Genotyping-by-Sequencing (GBS) Identified SNP Tightly Linked to QTL for Pre-Harvest Sprouting Resistance. Theor. Appl. Genet. 2015, 128, 1385–1395. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Zhang, D.; Liu, S.; Zhang, G.; Yu, J.; Fritz, A.K.; Bai, G. Genome-Wide Association Analysis on Pre-Harvest Sprouting Resistance and Grain Color in U.S. Winter Wheat. BMC Genom. 2016, 17, 794. [Google Scholar] [CrossRef]
- Liu, S.; Sehgal, S.K.; Lin, M.; Li, J.; Trick, H.N.; Gill, B.S.; Bai, G. Independent Mis-Splicing Mutations in TaPHS1 Causing Loss of Preharvest Sprouting (PHS) Resistance during Wheat Domestication. New Phytol. 2015, 208, 928–935. [Google Scholar] [CrossRef]
- Alvarado, G.; López, M.; Vargas, M.; Pacheco, Á.; Rodríguez, F.; Burgueño, J.; Crossa, J. META-R (Multi Environment Trail Analysis with R for Windows), Version 6.00; CIMMYT Research Data & Software Repository Network: Texcoco, Mexico, 2016.
- SAS Institute Inc. The SAS System for Windows, Release 9.4; Statistical Analysis Systems Institute: Cary, NC, USA, 2013; 556p. [Google Scholar]
- Beal, D.J. SAS Code To Select The Best Multiple Linear Regression Model For Multivariate Data Using Information Criteria. Available online: http://www.biostat.umn.edu/~wguan/class/PUBH7402/notes/lecture8_SAS.pdf (accessed on 30 March 2023).
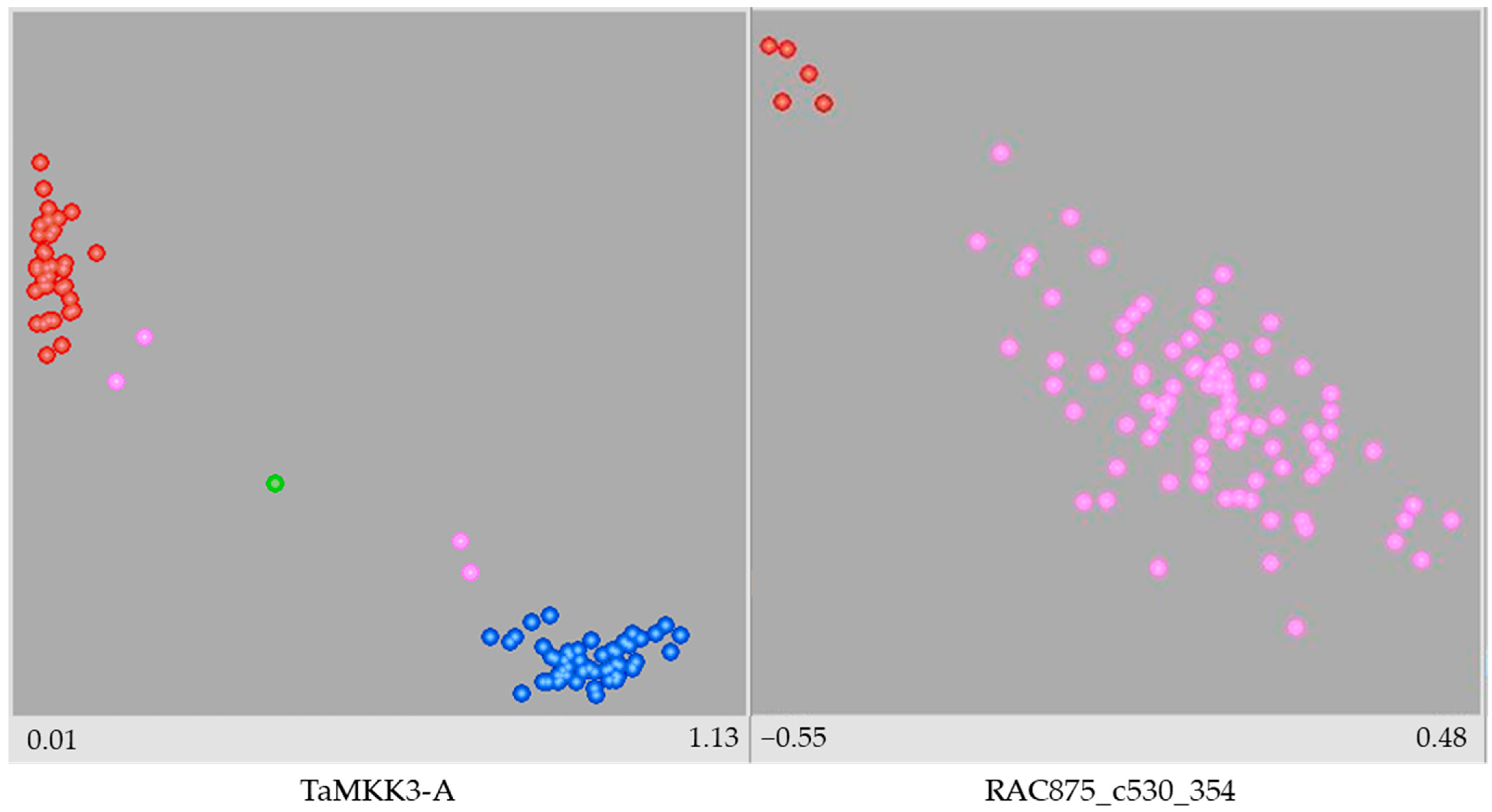
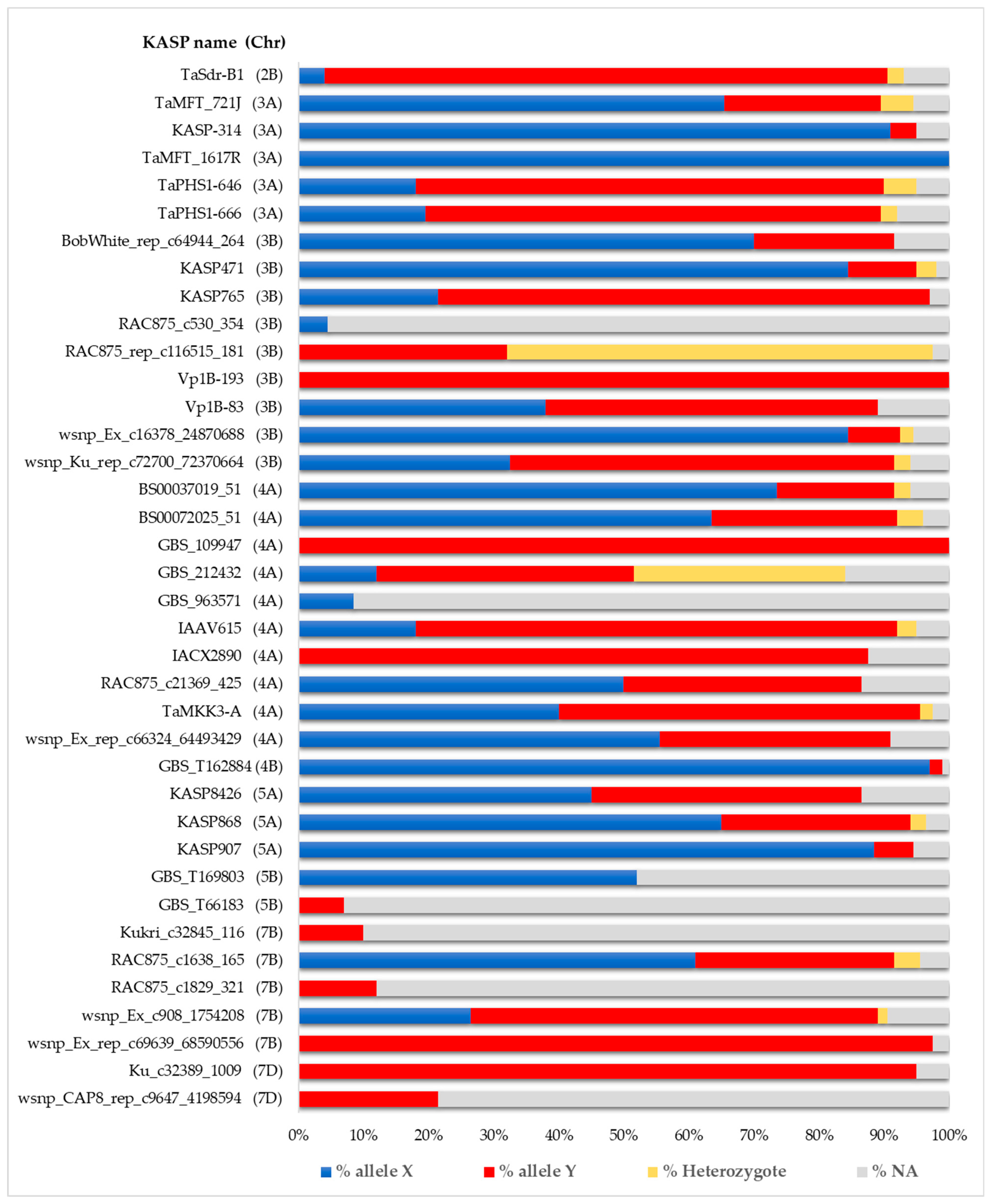
| Source of Variation | Variance | Significance | % of the Total Variance | Heritability |
|---|---|---|---|---|
| Genotype (G) | 0.031 | <0.001 | 46 | 0.86 |
| Environment (E) | 0.015 | <0.001 | 22 | |
| G × E | 0.018 | <0.001 | 27 | |
| Residual | 0.003 | 5 |
| Year | Location | Environment | Mean GI ± SD | GI Range |
|---|---|---|---|---|
| 2018 | Zagreb | ZG2018 | 0.50 ± 0.25 a 1 | 0.03–0.98 |
| 2019 | Zagreb | ZG2019 | 0.36 ± 0.21 b | 0.02–0.97 |
| 2020 | Osijek | OS020 | 0.33 ± 0.24 b | 0.02–0.98 |
| 2020 | Zagreb | ZG2020 | 0.19 ± 0.16 c | 0.01–0.92 |
| Mean | 0.35 ± 0.18 | 0.04–0.89 |
| SNP Marker Locus (Chr) | Freq (%) | Allele | Change of GI Due to the Effect of Tolerant Allele (%) Proportion of Phenotypic Variance Explained (PVE) | ||||
|---|---|---|---|---|---|---|---|
| ZG2018 | ZG2019 | ZG2020 | OS2020 | Across Environments | |||
| TaPHS1-646 (3A) | 20 | A | ns | −23 (3.5) | −35 (4.6) | −22 (2.2) | −18 (3.1) |
| 80 | G 1 | ||||||
| TaPHS1-666 (3A) | 22 | T | ns | −20 (3.5) | −31 (5.2) | −26 (3.0) | −20 (4.0) |
| 78 | A | ||||||
| KASP765 (3B) | 22 | A | ns | −21 (3.5) | −39 (8.6) | −38 (9.0) | −20 (4.3) |
| 78 | G | ||||||
| BS00037019_51 (4A) | 80 | C | −19 (2.7) | ns | −38 (3.7) | −46 (7.3) | −27 (5.2) |
| 20 | T | ||||||
| BS00072025_51 (4A) | 69 | A | ns | ns | −41 (7.0) | −37 (7.2) | −19 (3.4) |
| 31 | G | ||||||
| IAAV615 (4A) | 80 | G | −17 (2.3) | ns | −33 (3.2) | −41 (5.8) | −22 (4.4) |
| 20 | A | ||||||
| wsnp_Ex_rep_c66324_64493429 (4A) | 61 | C | −24 (6.5) | −25 (5.2) | −43 (10.0) | −49 (16.3) | −31 (12.7) |
| 39 | T | ||||||
| TaMKK3-A (4A) | 42 | T | −29 (11.1) | −18 (3.0) | −48 (17.2) | −51 (25) | −33 (17.5) |
| 58 | G | ||||||
| wsnp_Ex_c908_1754208 (7B) | 30 | C | ns | ns | −28 (3.9) | −23 (2.8) | ns |
| 70 | T | ||||||
| Model/Marker | BS00072025_51 (4A) | IAAV615 (4A) | KASP765 (3B) | TaMKK3-A (4A) | TaPHS1-646 (3A) | wsnp_Ex_c908_1754208 (7B) | wsnp_Ex_rep_c66324_64493429 (4A) | PVE | AIC |
|---|---|---|---|---|---|---|---|---|---|
| Effect of tolerant allele on GI | |||||||||
| Model 1 | −0.14 | 18 | −553.13 | ||||||
| Model 2 | 0.04 | −0.17 | 19 | −552.72 | |||||
| Model 3 | 0.05 | −0.15 | −0.04 | 20 | −552.43 | ||||
| Model 4 | −0.13 | −0.03 | 19 | −552.07 | |||||
| Model 5 | −0.14 | −0.02 | 19 | −551.51 | |||||
| Model 6 | −0.02 | −0.14 | 19 | −551.44 | |||||
| Model 7 | −0.14 | −0.01 | 19 | −551.25 | |||||
| Model 8 | −0.01 | −0.14 | 18 | −551.22 | |||||
| Model 9 | 0.04 | −0.16 | −0.02 | 20 | −551.20 | ||||
| Model 10 | 0.05 | −0.02 | −0.16 | 20 | −551.14 | ||||
| Model | Combination of KASP Markers | Haplotype | All | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of genotypes | 43 | 56 | 78 | 1 | 178 | ||||
| 2 | BS00072025_51 | A | G | A | G | ||||
| TaMKK3-A | G | G | T | T | |||||
| BLUP_GI LSMEAN | 0.26 a | 0.30 b | 0.44 c | 0.41 | |||||
| No. of genotypes | 20 | 37 | 21 | 18 | 68 | 7 | 1 | 172 | |
| 3 | BS00072025_51 | A | G | A | G | A | A | G | |
| TaMKK3-A | G | G | G | G | T | T | T | ||
| wsnp_Ex_rep_c66324_64493429 | T | T | C | C | C | T | C | ||
| BLUP_GI LSMEAN | 0.22 a | 0.26 ab | 0.29 b | 0.36 c | 0.44 d | 0.48 d | 0.41 | ||
| No. of genotypes | 63 | 39 | 69 | 7 | 178 | ||||
| 4 | TaMKK3-A | G | G | T | T | ||||
| wsnp_Ex_rep_c66324_64493429 | T | C | C | T | |||||
| BLUP_GI LSMEAN | 0.24 a | 0.33 b | 0.44 c | 0.48 c | |||||
| No. of genotypes | 78 | 25 | 42 | 28 | 173 | ||||
| 5 | TaMKK3-A | G | G | T | T | ||||
| wsnp_Ex_c908_1754208 | T | C | T | C | |||||
| BLUP_GI LSMEAN | 0.28 a | 0.29 a | 0.43 b | 0.48 c | |||||
| No. of genotypes | 90 | 17 | 54 | 24 | 185 | ||||
| 6 | KASP765 | G | A | G | A | ||||
| TaMKK3-A | G | G | T | T | |||||
| BLUP_GI LSMEAN | 0.27 a | 0.31 a | 0.41 b | 0.48 c | |||||
| No. of genotypes | 90 | 12 | 23 | 49 | 174 | ||||
| 7 | TaMKK3-A | G | G | T | T | ||||
| TaPHS1-646 | G | A | A | G | |||||
| BLUP_GI LSMEAN | 0.26 a | 0.37 b | 0.42 bc | 0.44 c | |||||
| No. of genotypes | 36 | 64 | 79 | 179 | |||||
| 8 | IAAV615 | A | G | G | |||||
| TaMKK3-A | G | G | T | ||||||
| BLUP_GI LSMEAN | 0.27 a | 0.28 a | 0.43 b | ||||||
| No. of genotypes | 34 | 7 | 47 | 8 | 54 | 23 | 1 | 174 | |
| 9 | BS00072025_51 | A | A | G | G | A | A | G | |
| TaMKK3-A | G | G | G | G | T | T | T | ||
| KASP765 | G | A | G | A | G | A | G | ||
| BLUP_GI LSMEAN | 0.26 a | 0.27 a | 0.29 a | 0.31 a | 0.41 b | 0.49 c | 0.41 | ||
| No. of genotypes | 30 | 11 | 11 | 41 | 42 | 28 | 1 | 164 | |
| 10 | BS00072025_51 | A | G | A | G | A | A | G | |
| TaMKK3-A | G | G | G | G | T | T | T | ||
| wsnp_Ex_c908_1754208 | T | T | C | T | T | C | T | ||
| BLUP_GI LSMEAN | 0.24 a | 0.24 ab | 0.31 bc | 0.32 c | 0.43 d | 0.48 e | 0.41 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajković, B.; Lovrić, A.; Maričević, M.; Novoselović, D.; Šarčević, H. Validation of QTLs for Resistance to Pre-Harvest Sprouting in a Panel of European Wheat Cultivars. Plants 2025, 14, 1342. https://doi.org/10.3390/plants14091342
Rajković B, Lovrić A, Maričević M, Novoselović D, Šarčević H. Validation of QTLs for Resistance to Pre-Harvest Sprouting in a Panel of European Wheat Cultivars. Plants. 2025; 14(9):1342. https://doi.org/10.3390/plants14091342
Chicago/Turabian StyleRajković, Bruno, Ana Lovrić, Marko Maričević, Dario Novoselović, and Hrvoje Šarčević. 2025. "Validation of QTLs for Resistance to Pre-Harvest Sprouting in a Panel of European Wheat Cultivars" Plants 14, no. 9: 1342. https://doi.org/10.3390/plants14091342
APA StyleRajković, B., Lovrić, A., Maričević, M., Novoselović, D., & Šarčević, H. (2025). Validation of QTLs for Resistance to Pre-Harvest Sprouting in a Panel of European Wheat Cultivars. Plants, 14(9), 1342. https://doi.org/10.3390/plants14091342






