Expansins in Salt and Drought Stress Adaptation: From Genome-Wide Identification to Functional Characterisation in Crops
Abstract
1. Introduction
2. Expansins: Structure and Function in Plant Growth
3. Genome-Wide Identification and Expression Analysis Under Stress Conditions in Model and Crop Species
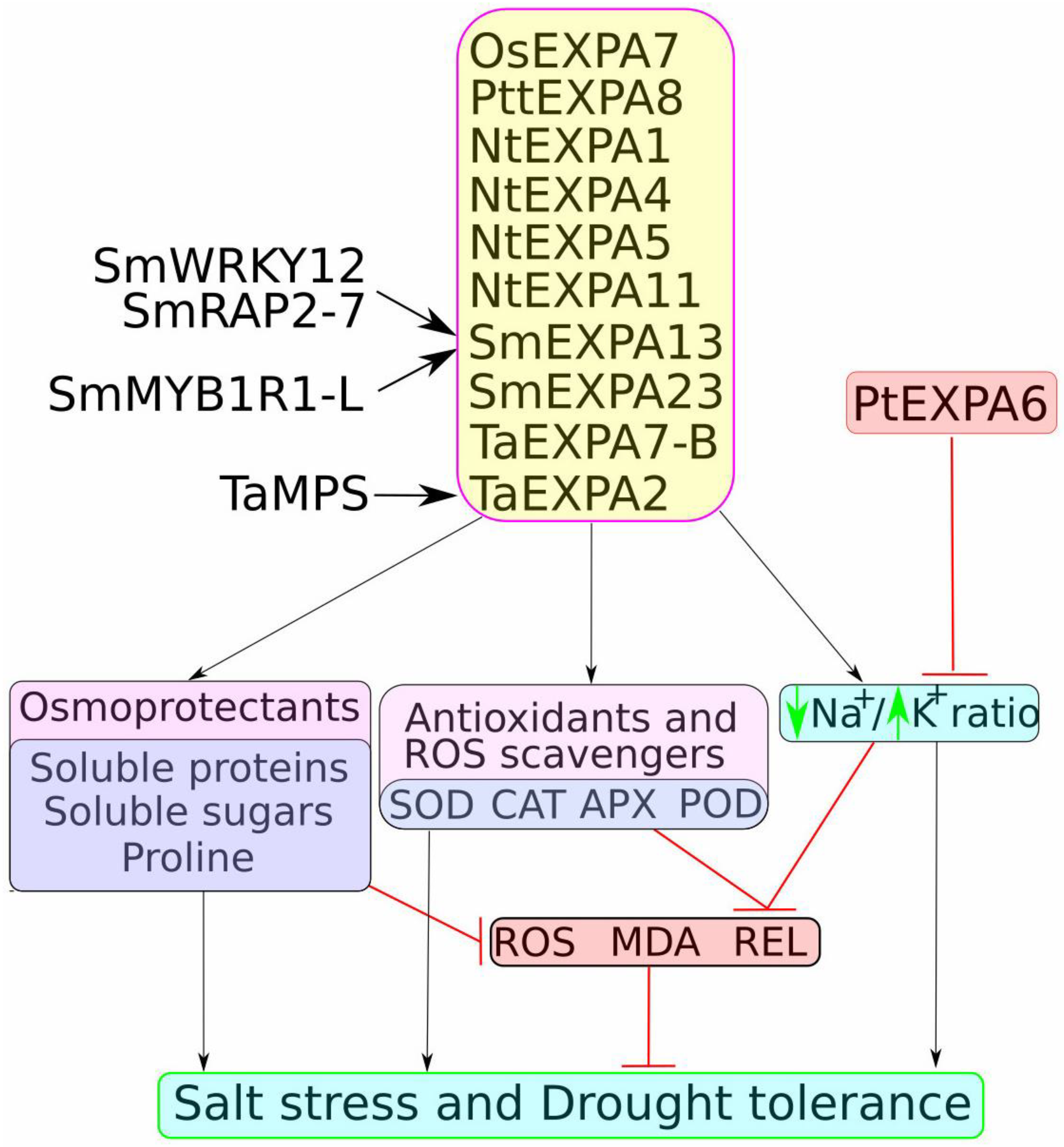
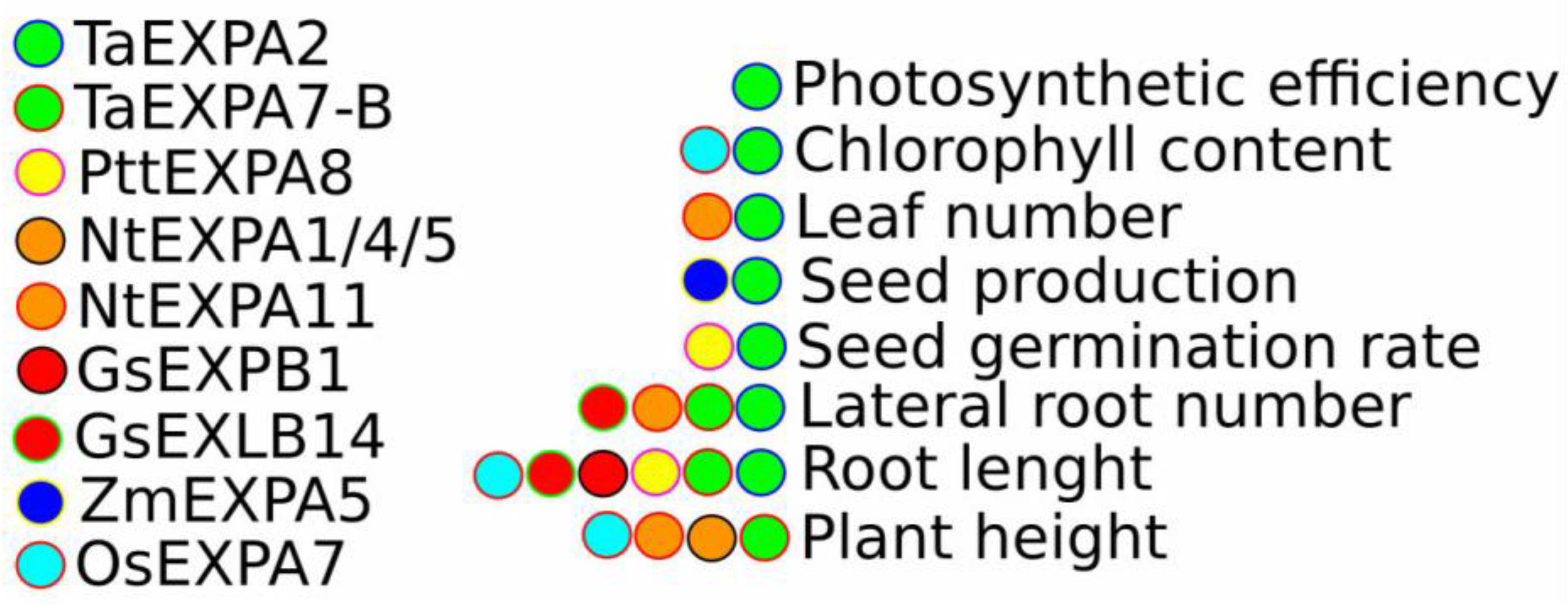
4. Functional Analysis of Expansins in Salt and Drought Stress Tolerance
4.1. Expansin Ectopic Expression in Heterologous Systems: Enhancing Stress Tolerance in Model and Crop Plants
4.2. Functional Characterisation of Expansin Mutants and Overexpression Lines in Native Species
- Nicotiana tabacum
- Brassica rapa
- Populus trichocarpa
- Salix matsudana
- Glycine soja
- Zea mays
- Oryza sativa
- Triticum aestivum
5. Integrative Analysis and Future Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, J.; Li, R.; Ge, Y.; Li, Y.; Li, R. Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef] [PubMed]
- Dabravolski, S.A.; Isayenkov, S.V. The Regulation of Plant Cell Wall Organisation under Salt Stress. Front. Plant Sci. 2023, 14, 1118313. [Google Scholar] [CrossRef]
- Hepler, N.K.; Bowman, A.; Carey, R.E.; Cosgrove, D.J. Expansin Gene Loss Is a Common Occurrence during Adaptation to an Aquatic Environment. Plant J. 2020, 101, 666–680. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant Cell Wall Loosening by Expansins. Annu. Rev. Cell Dev. Biol. 2024, 40, 329–352. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Plant Expansins: Diversity and Interactions with Plant Cell Walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef]
- Sun, W.; Yu, H.; Liu, M.; Ma, Z.; Chen, H. Evolutionary Research on the Expansin Protein Family during the Plant Transition to Land Provides New Insights into the Development of Tartary Buckwheat Fruit. BMC Genom. 2021, 22, 252. [Google Scholar] [CrossRef]
- McQueen-Mason, S.; Durachko, D.M.; Cosgrove, D.J. Two Endogenous Proteins That Induce Cell Wall Extension in Plants. Plant Cell 1992, 4, 1425–1433. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, J.; Lin, N.; Li, J.; Wang, Y.; Liu, W.; Yao, W.; Li, Y. Origin, Evolution, and Diversification of the Expansin Family in Plants. Int. J. Mol. Sci. 2024, 25, 11814. [Google Scholar] [CrossRef]
- Chase, W.R.; Zhaxybayeva, O.; Rocha, J.; Cosgrove, D.J.; Shapiro, L.R. Global Cellulose Biomass, Horizontal Gene Transfers and Domain Fusions Drive Microbial Expansin Evolution. New Phytol. 2020, 226, 921–938. [Google Scholar] [CrossRef]
- Armijos-Jaramillo, V.; Santander-Gordón, D.; Tejera, E.; Perez-Castillo, Y. The Dilemma of Bacterial Expansins Evolution. The Unusual Case of Streptomyces acidiscabies and Kutzneria Sp. 744. Commun. Integr. Biol. 2018, 11, e1539612. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Liu, X.; Hakulinen, N.; Taherzadeh, M.J.; Wang, Y.; Wang, Y.; Qin, X.; Wang, X.; Yao, B.; Luo, H.; et al. Boosting Enzymatic Degradation of Cellulose Using a Fungal Expansin: Structural Insight into the Pretreatment Mechanism. Bioresour. Technol. 2022, 358, 127434. [Google Scholar] [CrossRef] [PubMed]
- Bunterngsook, B.; Mhuantong, W.; Champreda, V.; Thamchaipenet, A.; Eurwilaichitr, L. Identification of Novel Bacterial Expansins and Their Synergistic Actions on Cellulose Degradation. Bioresour. Technol. 2014, 159, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Monschein, M.; Ioannou, E.; Koitto, T.; Al Amin, L.A.K.M.; Varis, J.J.; Wagner, E.R.; Mikkonen, K.S.; Cosgrove, D.J.; Master, E.R. Loosenin-Like Proteins from Phanerochaete carnosa Impact Both Cellulose and Chitin Fiber Networks. Appl. Environ. Microbiol. 2023, 89, e01863-22. [Google Scholar] [CrossRef]
- Luti, S.; Bemporad, F.; Vivoli Vega, M.; Leri, M.; Musiani, F.; Baccelli, I.; Pazzagli, L. Partitioning the Structural Features That Underlie Expansin-like and Elicitor Activities of Cerato-Platanin. Int. J. Biol. Macromol. 2020, 165, 2845–2854. [Google Scholar] [CrossRef]
- Ma, L.; Liu, T.; Zhang, K.; Shi, H.; Zhang, L.; Zou, G.; Sharon, A. Botrytis Cinerea Transcription Factor BcXyr1 Regulates (Hemi-)Cellulase Production and Fungal Virulence. mSystems 2022, 7, e01042-22. [Google Scholar] [CrossRef]
- Quarantin, A.; Castiglioni, C.; Schäfer, W.; Favaron, F.; Sella, L. The Fusarium Graminearum Cerato-Platanins Loosen Cellulose Substrates Enhancing Fungal Cellulase Activity as Expansin-like Proteins. Plant Physiol. Biochem. 2019, 139, 229–238. [Google Scholar] [CrossRef]
- Lohoff, C.; Buchholz, P.C.F.; Le Roes-Hill, M.; Pleiss, J. Expansin Engineering Database: A Navigation and Classification Tool for Expansins and Homologues. Proteins 2021, 89, 149–162. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The Expansin Superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef]
- Méndez-Yáñez, A.; Carrasco-Orellana, C.; Ramos, P.; Morales-Quintana, L. Alpha-Expansins: More than Three Decades of Wall Creep and Loosening in Fruits. Plant Mol. Biol. 2024, 114, 84. [Google Scholar] [CrossRef]
- Kök, B.Ö.; Celik Altunoglu, Y.; Öncül, A.B.; Karaci, A.; Cengiz Baloglu, M. Expansin Gene Family Database: A Comprehensive Bioinformatics Resource for Plant Expansin Multigene Family. J. Bioinform. Comput. Biol. 2023, 21, 2350015. [Google Scholar] [CrossRef] [PubMed]
- Kerff, F.; Amoroso, A.; Herman, R.; Sauvage, E.; Petrella, S.; Filée, P.; Charlier, P.; Joris, B.; Tabuchi, A.; Nikolaidis, N.; et al. Crystal Structure and Activity of Bacillus subtilis YoaJ (EXLX1), a Bacterial Expansin That Promotes Root Colonization. Proc. Natl. Acad. Sci. USA 2008, 105, 16876–16881. [Google Scholar] [CrossRef]
- Mohamad Nor, N.; Hashim, N.H.F.; Quay, D.H.X.; Mahadi, N.M.; Illias, R.M.; Abu Bakar, F.D.; Murad, A.M.A. Functional and Structural Analyses of an Expansin-like Protein from the Antarctic Yeast Glaciozyma Antarctica PI12 Reveal Strategies of Nutrient Scavenging in the Sea Ice Environment. Int. J. Biol. Macromol. 2020, 144, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Godoy, A.S.; Pereira, C.S.; Ramia, M.P.; Silveira, R.L.; Camilo, C.M.; Kadowaki, M.A.; Lange, L.; Busk, P.K.; Nascimento, A.S.; Skaf, M.S.; et al. Structure, Computational and Biochemical Analysis of PcCel45A Endoglucanase from Phanerochaete Chrysosporium and Catalytic Mechanisms of GH45 Subfamily C Members. Sci. Rep. 2018, 8, 3678. [Google Scholar] [CrossRef]
- Yennawar, N.H.; Li, L.-C.; Dudzinski, D.M.; Tabuchi, A.; Cosgrove, D.J. Crystal Structure and Activities of EXPB1 (Zea m 1), a β-Expansin and Group-1 Pollen Allergen from Maize. Proc. Natl. Acad. Sci. USA 2006, 103, 14664–14671. [Google Scholar] [CrossRef]
- Duan, Y.; Ma, Y.; Zhao, X.; Huang, R.; Su, R.; Qi, W.; He, Z. Real-Time Adsorption and Action of Expansin on Cellulose. Biotechnol. Biofuels 2018, 11, 317. [Google Scholar] [CrossRef]
- Georgelis, N.; Yennawar, N.H.; Cosgrove, D.J. Structural Basis for Entropy-Driven Cellulose Binding by a Type-A Cellulose-Binding Module (CBM) and Bacterial Expansin. Proc. Natl. Acad. Sci. USA 2012, 109, 14830–14835. [Google Scholar] [CrossRef] [PubMed]
- Georgelis, N.; Nikolaidis, N.; Cosgrove, D.J. Bacterial Expansins and Related Proteins from the World of Microbes. Appl. Microbiol. Biotechnol. 2015, 99, 3807–3823. [Google Scholar] [CrossRef]
- Haddad Momeni, M.; Zitting, A.; Jäämuru, V.; Turunen, R.; Penttilä, P.; Buchko, G.W.; Hiltunen, S.; Maiorova, N.; Koivula, A.; Sapkota, J.; et al. Insights into the Action of Phylogenetically Diverse Microbial Expansins on the Structure of Cellulose Microfibrils. Biotechnol. Biofuels 2024, 17, 56. [Google Scholar] [CrossRef]
- Valenzuela-Riffo, F.; Gaete-Eastman, C.; Stappung, Y.; Lizana, R.; Herrera, R.; Moya-León, M.A.; Morales-Quintana, L. Comparative in Silico Study of the Differences in the Structure and Ligand Interaction Properties of Three Alpha-Expansin Proteins from Fragaria chiloensis Fruit. J. Biomol. Struct. Dyn. 2019, 37, 3245–3258. [Google Scholar] [CrossRef]
- Zhang, P.; Ma, Y.; Cui, M.; Wang, J.; Huang, R.; Su, R.; Qi, W.; He, Z.; Thielemans, W. Effect of Sugars on the Real-Time Adsorption of Expansin on Cellulose. Biomacromolecules 2020, 21, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Herburger, K.; Franková, L.; Pičmanová, M.; Xin, A.; Meulewaeter, F.; Hudson, A.; Fry, S.C. Defining Natural Factors That Stimulate and Inhibit Cellulose:Xyloglucan Hetero-transglucosylation. Plant J. 2021, 105, 1549–1565. [Google Scholar] [CrossRef] [PubMed]
- Pech-Cervantes, A.A.; Muhammad, I.; Ogunade, I.M.; Jiang, Y.; Kim, D.H.; Gonzalez, C.F.; Hackmann, T.J.; Oliveira, A.S.; Vyas, D.; Adesogan, A.T. Exogenous Fibrolytic Enzymes and Recombinant Bacterial Expansins Synergistically Improve Hydrolysis and in Vitro Digestibility of Bermudagrass Haylage. J. Dairy Sci. 2019, 102, 8059–8073. [Google Scholar] [CrossRef]
- Hepler, N.K.; Cosgrove, D.J. Directed in Vitro Evolution of Bacterial Expansin BsEXLX1 for Higher Cellulose Binding and Its Consequences for Plant Cell Wall-loosening Activities. FEBS Lett. 2019, 593, 2545–2555. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S. Cell Wall Signaling in Plant Development and Defense. Annu. Rev. Plant Biol. 2022, 73, 323–353. [Google Scholar] [CrossRef]
- Kierzkowski, D.; Routier-Kierzkowska, A.-L. Cellular Basis of Growth in Plants: Geometry Matters. Curr. Opin. Plant Biol. 2019, 47, 56–63. [Google Scholar] [CrossRef]
- Rayle, D.L.; Cleland, R.E. The Acid Growth Theory of Auxin-Induced Cell Elongation Is Alive and Well. Plant Physiol. 1992, 99, 1271–1274. [Google Scholar] [CrossRef]
- Wang, T.; Park, Y.B.; Caporini, M.A.; Rosay, M.; Zhong, L.; Cosgrove, D.J.; Hong, M. Sensitivity-Enhanced Solid-State NMR Detection of Expansin’s Target in Plant Cell Walls. Proc. Natl. Acad. Sci. USA 2013, 110, 16444–16449. [Google Scholar] [CrossRef]
- Mangano, S.; Martínez Pacheco, J.; Marino-Buslje, C.; Estevez, J.M. How Does pH Fit in with Oscillating Polar Growth? Trends Plant Sci. 2018, 23, 479–489. [Google Scholar] [CrossRef]
- Takahashi, K.; Hayashi, K.; Kinoshita, T. Auxin Activates the Plasma Membrane H+-ATPase by Phosphorylation during Hypocotyl Elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef]
- Fuglsang, A.T.; Palmgren, M. Proton and Calcium Pumping P-Type ATPases and Their Regulation of Plant Responses to the Environment. Plant Physiol. 2021, 187, 1856–1875. [Google Scholar] [CrossRef]
- Kinoshita, S.N.; Taki, K.; Okamoto, F.; Nomoto, M.; Takahashi, K.; Hayashi, Y.; Ohkanda, J.; Tada, Y.; Finkemeier, I.; Kinoshita, T. Plasma Membrane H+-ATPase Activation Increases Global Transcript Levels and Promotes the Shoot Growth of Light-grown Arabidopsis Seedlings. Plant J. 2025, 121, e70034. [Google Scholar] [CrossRef] [PubMed]
- Fendrych, M.; Leung, J.; Friml, J. TIR1/AFB-Aux/IAA Auxin Perception Mediates Rapid Cell Wall Acidification and Growth of Arabidopsis Hypocotyls. eLife 2016, 5, e19048. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Verstraeten, I.; Trinh, H.K.; Lardon, R.; Schotte, S.; Olatunji, D.; Heugebaert, T.; Stevens, C.; Quareshy, M.; Napier, R.; et al. Chemical Induction of Hypocotyl Rooting Reveals Extensive Conservation of Auxin Signalling Controlling Lateral and Adventitious Root Formation. New Phytol. 2023, 240, 1883–1899. [Google Scholar] [CrossRef] [PubMed]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef]
- Wong, J.H.; Spartz, A.K.; Park, M.Y.; Du, M.; Gray, W.M. Mutation of a Conserved Motif of PP2C.D Phosphatases Confers SAUR Immunity and Constitutive Activity. Plant Physiol. 2019, 181, 353–366. [Google Scholar] [CrossRef]
- Arsuffi, G.; Braybrook, S.A. Acid Growth: An Ongoing Trip. J. Exp. Bot. 2018, 69, 137–146. [Google Scholar] [CrossRef]
- Cheddadi, I.; Génard, M.; Bertin, N.; Godin, C. Coupling Water Fluxes with Cell Wall Mechanics in a Multicellular Model of Plant Development. PLoS Comput. Biol. 2019, 15, e1007121. [Google Scholar] [CrossRef]
- Pacifici, E.; Di Mambro, R.; Dello Ioio, R.; Costantino, P.; Sabatini, S. Acidic Cell Elongation Drives Cell Differentiation in the Arabidopsis Root. EMBO J. 2018, 37, e99134. [Google Scholar] [CrossRef]
- Hoffmann, R.D.; Olsen, L.I.; Ezike, C.V.; Pedersen, J.T.; Manstretta, R.; López-Marqués, R.L.; Palmgren, M. Roles of Plasma Membrane Proton ATPases AHA2 and AHA7 in Normal Growth of Roots and Root Hairs in Arabidopsis thaliana. Physiol. Plant. 2019, 166, 848–861. [Google Scholar] [CrossRef]
- Haruta, M.; Tan, L.X.; Bushey, D.B.; Swanson, S.J.; Sussman, M.R. Environmental and Genetic Factors Regulating Localization of the Plant Plasma Membrane H+-ATPase. Plant Physiol. 2018, 176, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Großeholz, R.; Wanke, F.; Rohr, L.; Glöckner, N.; Rausch, L.; Scholl, S.; Scacchi, E.; Spazierer, A.-J.; Shabala, L.; Shabala, S.; et al. Computational Modeling and Quantitative Physiology Reveal Central Parameters for Brassinosteroid-Regulated Early Cell Physiological Processes Linked to Elongation Growth of the Arabidopsis Root. eLife 2022, 11, e73031. [Google Scholar] [CrossRef]
- Lin, Z.; Zhu, P.; Gao, L.; Chen, X.; Li, M.; Wang, Y.; He, J.; Miao, Y.; Miao, R. Recent Advances in Understanding the Regulatory Mechanism of Plasma Membrane H+-ATPase through the Brassinosteroid Signaling Pathway. Plant Cell Physiol. 2024, 65, 1515–1529. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.; Gong, B.; Wen, D.; Qiao, P.; Guo, H.; Shi, Q. Brassinosteroid Enhances Cucumber Stress Tolerance to NaHCO3 by Modulating Nitrogen Metabolism, Ionic Balance and Phytohormonal Response. Plants 2024, 14, 80. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, N.; Pan, K.; Mughal, N.; Raza, A.; Liu, L.; Zhang, J.; Wu, X.; Sun, X.; Zhang, L.; Pan, Z. Potential of UV-B Radiation in Drought Stress Resilience: A Multidimensional Approach to Plant Adaptation and Future Implications. Plant Cell Environ. 2024, 47, 387–407. [Google Scholar] [CrossRef]
- Salvi, P.; Mahawar, H.; Agarrwal, R.; Kajal; Gautam, V.; Deshmukh, R. Advancement in the Molecular Perspective of Plant-Endophytic Interaction to Mitigate Drought Stress in Plants. Front. Microbiol. 2022, 13, 981355. [Google Scholar] [CrossRef]
- Kerchev, P.I.; Van Breusegem, F. Improving Oxidative Stress Resilience in Plants. Plant J. 2022, 109, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Bashir, S.S.; Hussain, A.; Hussain, S.J.; Wani, O.A.; Zahid Nabi, S.; Dar, N.A.; Baloch, F.S.; Mansoor, S. Plant Drought Stress Tolerance: Understanding Its Physiological, Biochemical and Molecular Mechanisms. Biotechnol. Biotechnol. Equip. 2021, 35, 1912–1925. [Google Scholar] [CrossRef]
- Razi, K.; Muneer, S. Drought Stress-Induced Physiological Mechanisms, Signaling Pathways and Molecular Response of Chloroplasts in Common Vegetable Crops. Crit. Rev. Biotechnol. 2021, 41, 669–691. [Google Scholar] [CrossRef]
- Jones, L.; McQueen-Mason, S. A Role for Expansins in Dehydration and Rehydration of the Resurrection Plant Craterostigma plantagineum. FEBS Lett. 2004, 559, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef]
- Rai, G.K.; Mishra, S.; Chouhan, R.; Mushtaq, M.; Chowdhary, A.A.; Rai, P.K.; Kumar, R.R.; Kumar, P.; Perez-Alfocea, F.; Colla, G.; et al. Plant Salinity Stress, Sensing, and Its Mitigation through WRKY. Front. Plant Sci. 2023, 14, 1238507. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.K.; Upadhyay, S.K.; Tripathi, M.; Singh, R.; Krishna, D.; Singh, S.K.; Dwivedi, P. Understanding the Salinity Stress on Plant and Developing Sustainable Management Strategies Mediated Salt-Tolerant Plant Growth-Promoting Rhizobacteria and CRISPR/Cas9. Biotechnol. Genet. Eng. Rev. 2023, 39, 311–347. [Google Scholar] [CrossRef] [PubMed]
- Kuluev, B.; Avalbaev, A.; Mikhaylova, E.; Nikonorov, Y.; Berezhneva, Z.; Chemeris, A. Expression Profiles and Hormonal Regulation of Tobacco Expansin Genes and Their Involvement in Abiotic Stress Response. J. Plant Physiol. 2016, 206, 1–12. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, B.; Li, C.; Lei, C.; Kong, C.; Yang, Y.; Gong, M. A Comprehensive Expression Analysis of the Expansin Gene Family in Potato (Solanum tuberosum) Discloses Stress-Responsive Expansin-like B Genes for Drought and Heat Tolerances. PLoS ONE 2019, 14, e0219837. [Google Scholar] [CrossRef]
- Han, Z.; Liu, Y.; Deng, X.; Liu, D.; Liu, Y.; Hu, Y.; Yan, Y. Genome-Wide Identification and Expression Analysis of Expansin Gene Family in Common Wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 101. [Google Scholar] [CrossRef]
- Liu, C.; Fu, M.; Guo, F.; Wu, C. Genome-Wide Identification of Expansin Gene Family in Barley and Drought-Related Expansins Identification Based on RNA-Seq. Genetica 2021, 149, 283–297. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The Protein Sequence Classification Resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef]
- Chen, Y.; Han, Y.; Zhang, M.; Zhou, S.; Kong, X.; Wang, W. Overexpression of the Wheat Expansin Gene TaEXPA2 Improved Seed Production and Drought Tolerance in Transgenic Tobacco Plants. PLoS ONE 2016, 11, e0153494. [Google Scholar] [CrossRef]
- Chen, Y.; Han, Y.; Kong, X.; Kang, H.; Ren, Y.; Wang, W. Ectopic Expression of Wheat Expansin Gene TaEXPA2 Improved the Salt Tolerance of Transgenic Tobacco by Regulating Na+/K+ and Antioxidant Competence. Physiol. Plant. 2017, 159, 161–177. [Google Scholar] [CrossRef]
- Abbasi, A.; Malekpour, M.; Sobhanverdi, S. The Arabidopsis Expansin Gene (AtEXPA18) Is Capable to Ameliorate Drought Stress Tolerance in Transgenic Tobacco Plants. Mol. Biol. Rep. 2021, 48, 5913–5922. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ma, J.; He, F.; Wang, L.; Zhang, T.; Liu, D.; Xu, Y.; Li, F.; Feng, X. A Study on the Functional Identification of Overexpressing Winter Wheat Expansin Gene TaEXPA7-B in Rice under Salt Stress. Int. J. Mol. Sci. 2024, 25, 7707. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, H.; Yang, R.; Xu, X.; Liu, X.; Xu, J. Over-Expression of PttEXPA8 Gene Showed Various Resistances to Diverse Stresses. Int. J. Biol. Macromol. 2019, 130, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Yang, L.; Wang, X.; Wang, Y.; Zhang, J.; Xu, J. Over-Expression of the Salix Matsudana Expansin Gene SmEXPA23 Enhances Plant Salt Tolerance. Plant Cell Tissue Organ Cult. 2023, 152, 309–316. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, R.; Wang, Y.; Wang, X.; Wang, L.; Xu, J. The Expansin Gene SmEXPA13 in Salix Matsudana in Association with Plant Salt Tolerance. Plant Cell Tissue Organ Cult. 2023, 154, 219–225. [Google Scholar] [CrossRef]
- Chen, L.; Zou, W.; Fei, C.; Wu, G.; Li, X.; Lin, H.; Xi, D. α-Expansin EXPA4 Positively Regulates Abiotic Stress Tolerance but Negatively Regulates Pathogen Resistance in Nicotiana Tabacum. Plant Cell Physiol. 2018, 59, 2317–2330. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Xu, Z.; Kong, Y. Overexpression of NtEXPA11 Modulates Plant Growth and Development and Enhances Stress Tolerance in Tobacco. Plant Physiol. Biochem. 2020, 151, 477–485. [Google Scholar] [CrossRef]
- Muthusamy, M.; Kim, J.Y.; Yoon, E.K.; Kim, J.A.; Lee, S.I. BrEXLB1, a Brassica Rapa Expansin-Like B1 Gene Is Associated with Root Development, Drought Stress Response, and Seed Germination. Genes 2020, 11, 404. [Google Scholar] [CrossRef]
- Liu, Z.; Yin, K.; Zhang, Y.; Yan, C.; Zhao, Z.; Li, J.; Liu, Y.; Feng, B.; Zhao, R.; Liu, J.; et al. Populus Trichocarpa EXPA6 Facilitates Radial and Longitudinal Transport of Na+ under Salt Stress. Int. J. Mol. Sci. 2024, 25, 9354. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Wu, D.; Zhao, H.; Gong, L.; Xu, J. Regulation of SmEXPA13 Expression by SmMYB1R1-L Enhances Salt Tolerance in Salix Matsudana Koidz. Int. J. Biol. Macromol. 2024, 270, 132292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Zhao, H.; Gong, L.; Xu, J. The SmWRKY12-SmRAP2–7-SmEXPA13 Module in Salix Matsudana Koidz Enhances Plant Tolerance to Drought Stress. Int. J. Biol. Macromol. 2025, 284, 138077. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Li, C.; He, F.; Xu, Y.; Li, L.; Wang, X.; Chen, Q.; Li, F. Genome-Wide Identification of Expansin Genes in Wild Soybean (Glycine soja) and Functional Characterization of Expansin B1 (GsEXPB1) in Soybean Hair Root. Int. J. Mol. Sci. 2022, 23, 5407. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, T.; Li, C.; Zhou, C.; Liu, B.; Wu, Y.; He, F.; Xu, Y.; Li, F.; Feng, X. Overexpression of Wild Soybean Expansin Gene GsEXLB14 Enhanced the Tolerance of Transgenic Soybean Hairy Roots to Salt and Drought Stresses. Plants 2024, 13, 1656. [Google Scholar] [CrossRef]
- Tao, K.; Li, Y.; Hu, Y.; Li, Y.; Zhang, D.; Li, C.; He, G.; Song, Y.; Shi, Y.; Li, Y.; et al. Overexpression of ZmEXPA5 Reduces Anthesis-Silking Interval and Increases Grain Yield under Drought and Well-Watered Conditions in Maize. Mol. Breed. 2023, 43, 84. [Google Scholar] [CrossRef]
- Jadamba, C.; Kang, K.; Paek, N.-C.; Lee, S.I.; Yoo, S.-C. Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. Int. J. Mol. Sci. 2020, 21, 454. [Google Scholar] [CrossRef]
- Shao, Y.; Feng, X.; Nakahara, H.; Irshad, M.; Eneji, A.E.; Zheng, Y.; Fujimaki, H.; An, P. Apical-root Apoplastic Acidification Affects Cell Wall Extensibility in Wheat under Salinity Stress. Physiol. Plant. 2021, 173, 1850–1861. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, G.; An, J.; Li, Q.; Chen, Y.; Zhao, X.; Wu, J.; Wang, Y.; Hao, Q.; Wang, W.; et al. Expansin Gene TaEXPA2 Positively Regulates Drought Tolerance in Transgenic Wheat (Triticum aestivum L.). Plant Sci. 2020, 298, 110596. [Google Scholar] [CrossRef]
| Plant Species | Total Expansisns in Genome * | Effects of Stress and Hormones on Expansin Gene(s) | References |
|---|---|---|---|
| Nicotiana tabacum | 58 | Salt, drought, heat, and cold stresses and ABA, CK, Auxin, and gibberellins upregulated NtEXPA1,4,5 expression | [65] |
| potato (Solanum tuberosum L.) | 38 | IAA, GA3, and BAP upregulated StEXPA7/18; ABA and GA3 upregulated all StEXLB genes; NaCl and mannitol upregulated StEXPA8/19 and StEXPB2, and downregulated StEXPA4 and StEXLB4; Any stress or hormonal treatment affected StEXPA1/21/23/24, and StEXPB5 | [66] |
| wheat (Triticum aestivum L.) | 275 | PEG treatment in leaves upregulated TaEXPA3/9-A, TaEXPB2/4/7/9/10-A; salt treatment upregulated TaEXPA3/9-A, TaEXPB2/4/10-A; PEG treatment in roots upregulated TaEXPA3-8-A, TaEXPB1/7/8/10-A, and TaEXPB1-B; Salt stress upregulated TaEXPA3/5-8-A, TaEXPA12-A, TaEXPB2/4/7/10-A | [67] |
| barley (Hordeum vulgare L.) | 45 | Drought stress upregulated HvEXPB5/6 in roots | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabravolski, S.A.; Isayenkov, S.V. Expansins in Salt and Drought Stress Adaptation: From Genome-Wide Identification to Functional Characterisation in Crops. Plants 2025, 14, 1327. https://doi.org/10.3390/plants14091327
Dabravolski SA, Isayenkov SV. Expansins in Salt and Drought Stress Adaptation: From Genome-Wide Identification to Functional Characterisation in Crops. Plants. 2025; 14(9):1327. https://doi.org/10.3390/plants14091327
Chicago/Turabian StyleDabravolski, Siarhei A., and Stanislav V. Isayenkov. 2025. "Expansins in Salt and Drought Stress Adaptation: From Genome-Wide Identification to Functional Characterisation in Crops" Plants 14, no. 9: 1327. https://doi.org/10.3390/plants14091327
APA StyleDabravolski, S. A., & Isayenkov, S. V. (2025). Expansins in Salt and Drought Stress Adaptation: From Genome-Wide Identification to Functional Characterisation in Crops. Plants, 14(9), 1327. https://doi.org/10.3390/plants14091327







