Advances in Small RNA Regulation of Female Gametophyte Development in Flowering Plants
Abstract
1. Introduction
2. The Biogenesis, Molecular Mechanisms, and Biological Functions of Small RNAs
2.1. miRNA Synthesis, Molecular Mechanisms, and Biological Functions
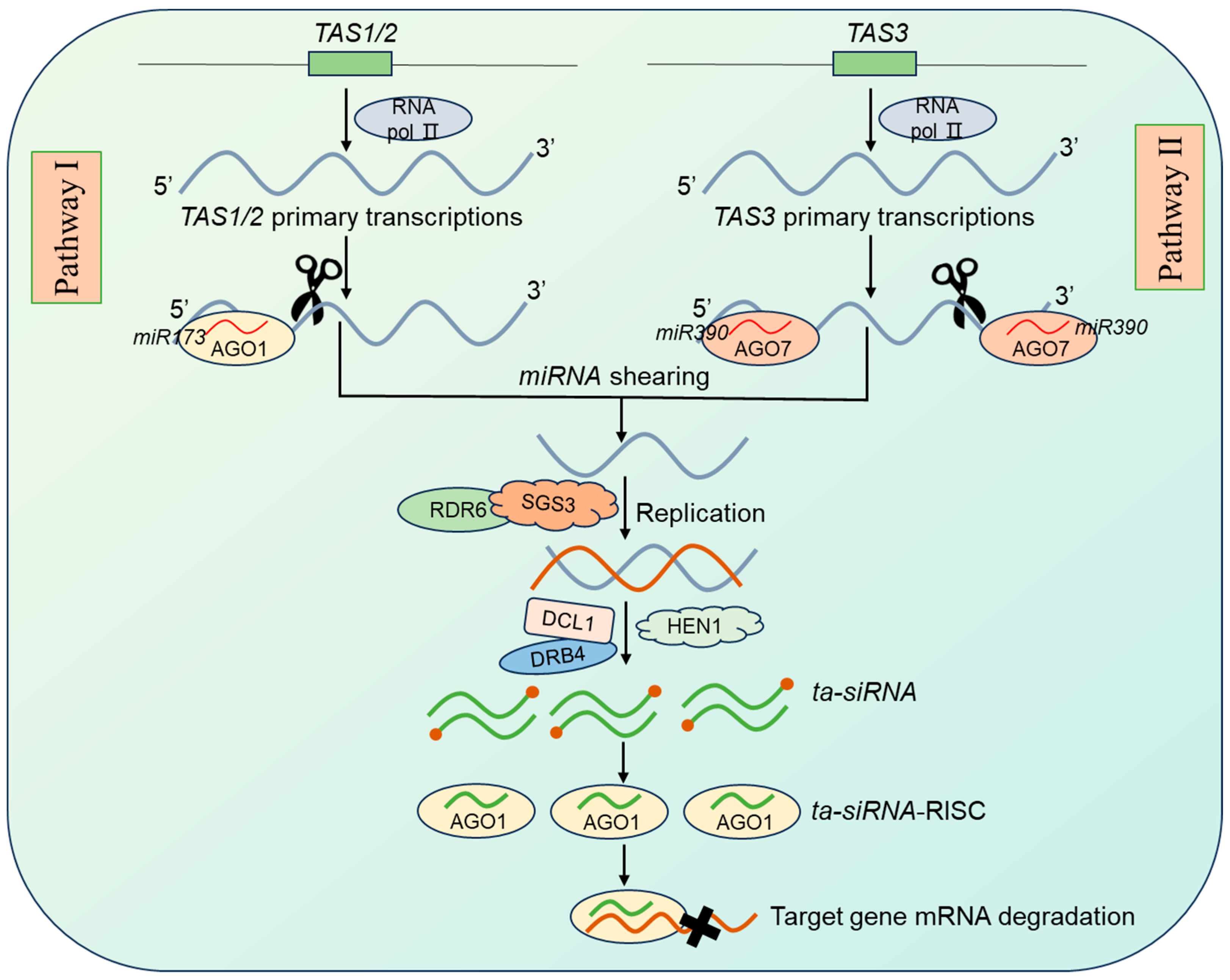
2.2. ta-siRNA Synthesis, Molecular Mechanisms, and Biological Functions
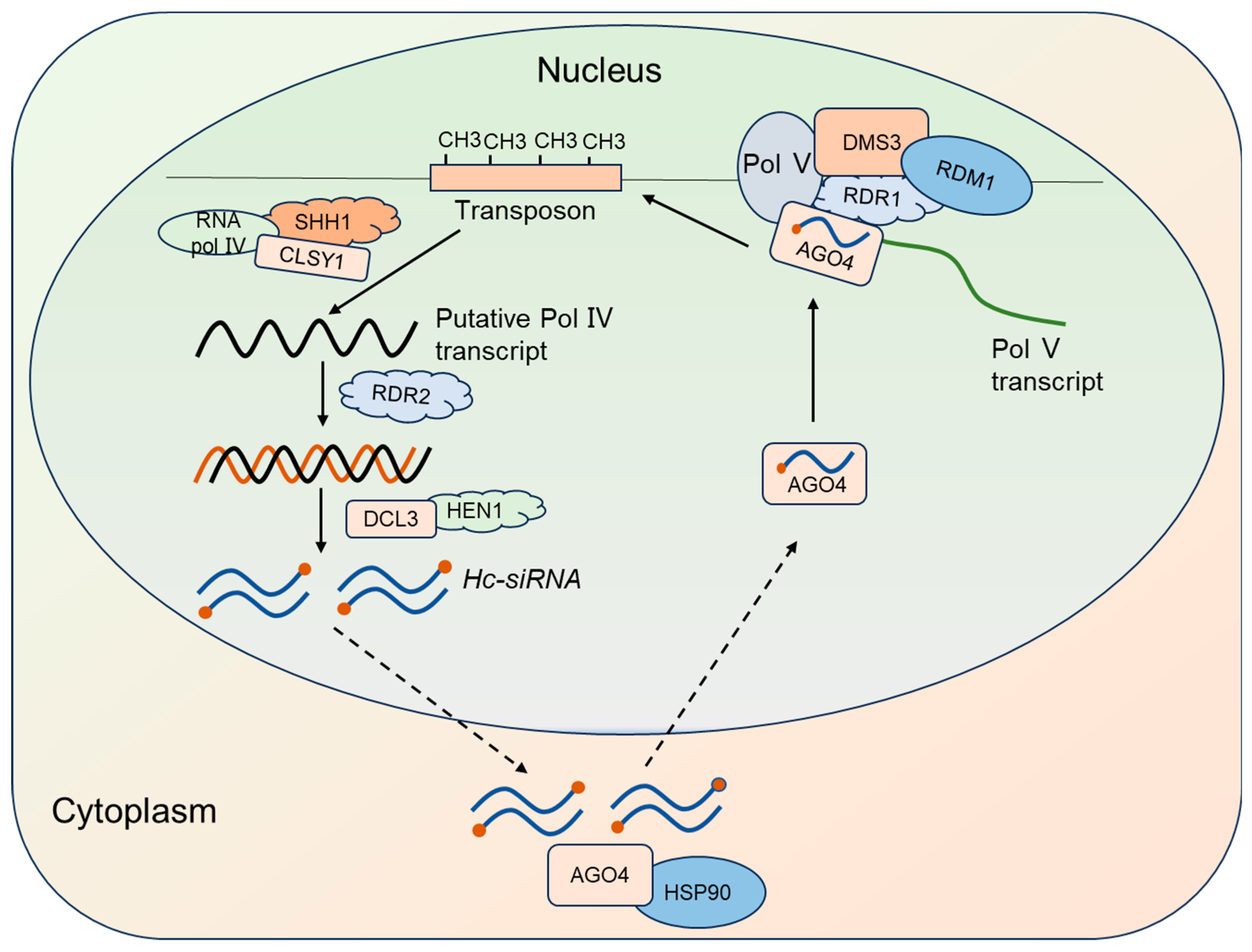
2.3. Hc-siRNA Synthesis, Molecular Mechanisms, and Biological Functions
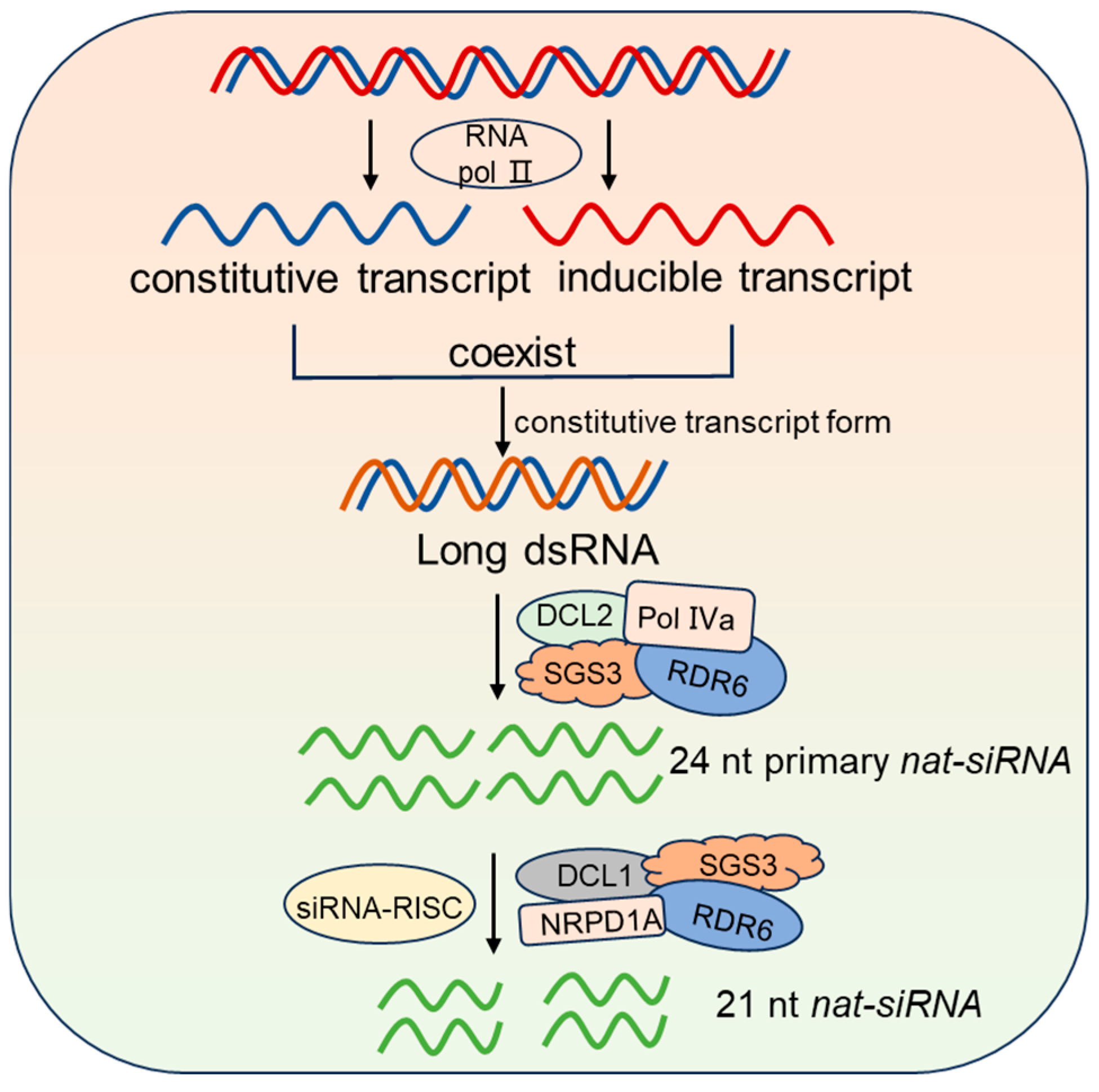
2.4. nat-siRNA Synthesis, Molecular Mechanisms, and Biological Functions
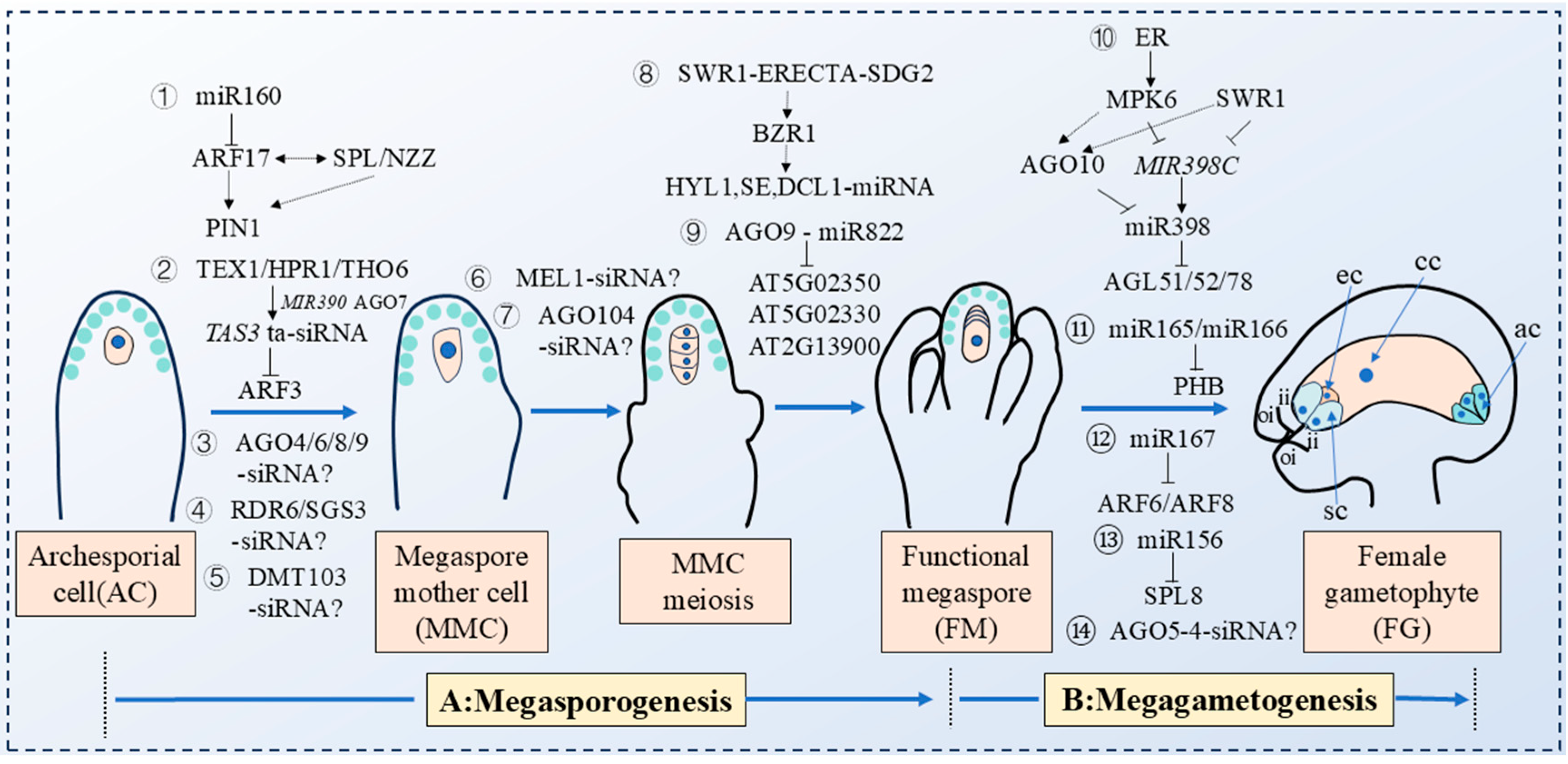
3. Small RNAs Regulate the Process of Plant Megasporogenesis
4. Small RNAs Regulate the Process of Plant Megagametogenesis
5. Conclusions and Prospect
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yang, W.C.; Shi, D.Q.; Chen, Y.H. Female gametophyte development in flowering plants. Annu. Rev. Plant Biol. 2010, 61, 89–108. [Google Scholar] [CrossRef]
- Drews, G.N.; Koltunow, A.M. The female gametophyte. Arabidopsis Book. 2011, 9, e0155. [Google Scholar] [CrossRef]
- Chevalier, É.; Loubert-Hudon, A.; Zimmerman, E.L.; Matton, D.P. Cell-cell communication and signalling pathways within the ovule: From its inception to fertilization. New Phytol. 2011, 192, 13–28. [Google Scholar] [CrossRef]
- Hanyang, C.; Suzhuo, M.; Han, S. Positional signals establishment in the regulation of female germline specification. Seed Biol. 2022, 1, 6. [Google Scholar]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes. Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef]
- Llave, C.; Xie, Z.; Kasschau, K.D.; Carrington, J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002, 297, 2053–2056. [Google Scholar] [CrossRef]
- Park, W.; Li, J.; Song, R.; Messing, J.; Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002, 12, 1484–1495. [Google Scholar] [CrossRef]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef]
- Song, L.; Axtell, M.J.; Fedoroff, N.V. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010, 20, 37–41. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Xie, Z. Piecing the puzzle together: Genetic requirements for miRNA biogenesis in Arabidopsis thaliana. Methods Mol. Biol. 2010, 592, 1–17. [Google Scholar] [CrossRef]
- Li, Y.F.; Zheng, Y.; Addo-Quaye, C.; Zhang, L.; Saini, A.; Jagadeeswaran, G.; Axtell, M.J.; Zhang, W.; Sunkar, R. Transcriptome-wide identification of microRNA targets in rice. Plant J. Cell Mol. Biol. 2010, 62, 742–759. [Google Scholar] [CrossRef]
- Meng, Y.; Chen, D.; Ma, X.; Mao, C.; Cao, J.; Wu, P.; Chen, M. Mechanisms of microRNA-mediated auxin signaling inferred from the rice mutant osaxr. Plant Signal Behav. 2010, 5, 252–254. [Google Scholar] [CrossRef]
- Sunkar, R.; Girke, T.; Jain, P.K.; Zhu, J.K. Cloning and characterization of microRNAs from rice. Plant Cell 2005, 17, 1397–1411. [Google Scholar] [CrossRef]
- Mica, E.; Gianfranceschi, L.; Pè, M.E. Characterization of five microRNA families in maize. J. Exp. Bot. 2006, 57, 2601–2612. [Google Scholar] [CrossRef]
- Yao, Y.; Guo, G.; Ni, Z.; Sunkar, R.; Du, J.; Zhu, J.K.; Sun, Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol. 2007, 8, R96. [Google Scholar] [CrossRef]
- Li, B.; Yin, W.; Xia, X. Identification of microRNAs and their targets from Populus euphratica. Biochem. Biophys. Res. Commun. 2009, 388, 272–277. [Google Scholar] [CrossRef]
- Feng, J.; Wang, K.; Liu, X.; Chen, S.; Chen, J. The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene 2009, 437, 14–21. [Google Scholar] [CrossRef]
- Gu, M.; Xu, K.; Chen, A.; Zhu, Y.; Tang, G.; Xu, G. Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol. Plant 2010, 138, 226–237. [Google Scholar] [CrossRef]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Pulido, A.; Laufs, P. Co-ordination of developmental processes by small RNAs during leaf development. J. Exp. Bot. 2010, 61, 1277–1291. [Google Scholar] [CrossRef]
- Schmid, M.; Uhlenhaut, N.H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J.U. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Q.; Zhang, B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 2013, 530, 26–32. [Google Scholar] [CrossRef]
- Du, Q.; Zhao, M.; Gao, W.; Sun, S.; Li, W.X. microRNA/microRNA* complementarity is important for the regulation pattern of NFYA5 by miR169 under dehydration shock in Arabidopsis. Plant J. Cell Mol. Biol. 2017, 91, 22–33. [Google Scholar] [CrossRef]
- Gupta, O.P.; Meena, N.L.; Sharma, I.; Sharma, P. Differential regulation of microRNAs in response to osmotic, salt and cold stresses in wheat. Mol. Biol. Rep. 2014, 41, 4623–4629. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Yu, D. Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE 2012, 7, e48951. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef]
- Chen, X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005, 579, 5923–5931. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Zheng, B.; Yu, Y.; Won, S.Y.; Mo, B.; Chen, X. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. Embo J. 2011, 30, 814–822. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Chen, X. The regulation of genes and genomes by small RNAs. Development 2007, 134, 1635–1641. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, Y.; Zhai, J.; Ramachandran, V.; Dinh, T.T.; Meyers, B.C.; Mo, B.; Chen, X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 2012, 22, 689–694. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Grigg, S.P.; Canales, C.; Hay, A.; Tsiantis, M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 2005, 437, 1022–1026. [Google Scholar] [CrossRef]
- Han, M.H.; Goud, S.; Song, L.; Fedoroff, N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 2004, 101, 1093–1098. [Google Scholar] [CrossRef]
- Schauer, S.E.; Jacobsen, S.E.; Meinke, D.W.; Ray, A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002, 7, 487–491. [Google Scholar] [CrossRef]
- Vazquez, F.; Gasciolli, V.; Crété, P.; Vaucheret, H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004, 14, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, Z.; Lu, F.; Dong, A.; Huang, H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. Cell Mol. Biol. 2006, 47, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Bi, L.; Zheng, B.; Ji, L.; Chevalier, D.; Agarwal, M.; Ramachandran, V.; Li, W.; Lagrange, T.; Walker, J.C.; et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 10073–10078. [Google Scholar] [CrossRef]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef]
- Li, J.; Yang, Z.; Yu, B.; Liu, J.; Chen, X. Methylation protects miRNAs and siRNAs from a 3’-end uridylation activity in Arabidopsis. Curr. Biol. 2005, 15, 1501–1507. [Google Scholar] [CrossRef]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef]
- Park, M.Y.; Wu, G.; Gonzalez-Sulser, A.; Vaucheret, H.; Poethig, R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 3691–3696. [Google Scholar] [CrossRef]
- Ramachandran, V.; Chen, X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 2008, 321, 1490–1492. [Google Scholar] [CrossRef]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef]
- Qi, Y.; Denli, A.M.; Hannon, G.J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 2005, 19, 421–428. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Bagga, S.; Bracht, J.; Hunter, S.; Massirer, K.; Holtz, J.; Eachus, R.; Pasquinelli, A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005, 122, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Willmann, M.R.; Mehalick, A.J.; Packer, R.L.; Jenik, P.D. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 2011, 155, 1871–1884. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yao, X.; Pi, L.; Wang, H.; Cui, X.; Huang, H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. Cell Mol. Biol. 2009, 58, 27–40. [Google Scholar] [CrossRef]
- Rubio-Somoza, I.; Zhou, C.M.; Confraria, A.; Martinho, C.; von Born, P.; Baena-Gonzalez, E.; Wang, J.W.; Weigel, D. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 2014, 24, 2714–2719. [Google Scholar] [CrossRef]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, Y.H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Upadhyaya, N.M.; Gubler, F.; Helliwell, C.A. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 2009, 9, 149. [Google Scholar] [CrossRef]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef]
- Garcia, D.; Collier, S.A.; Byrne, M.E.; Martienssen, R.A. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 2006, 16, 933–938. [Google Scholar] [CrossRef]
- Liu, X.; Dinh, T.T.; Li, D.; Shi, B.; Li, Y.; Cao, X.; Guo, L.; Pan, Y.; Jiao, Y.; Chen, X. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. Cell Mol. Biol. 2014, 80, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.K.; Lee, W.S. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2010, 38, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006, 16, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Mohd Zahid, N.I.I.; Syed Othman, S.M.I.; Mustaffa, A.F.; Ismail, I.; Che-Othman, M.H. Fine-tuning plant valuable secondary metabolite biosynthesis via small RNA manipulation: Strategies and potential. Planta 2024, 260, 89. [Google Scholar] [CrossRef]
- Castel, S.E.; Martienssen, R.A. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef]
- Xie, Z.; Jia, G.; Ghosh, A. Small RNAs in Plants. In MicroRNAs in Plant Development and Stress Responses; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–28. [Google Scholar] [CrossRef]
- Liu, W.; Duttke, S.H.; Hetzel, J.; Groth, M.; Feng, S.; Gallego-Bartolome, J.; Zhong, Z.; Kuo, H.Y.; Wang, Z.; Zhai, J.; et al. RNA-directed DNA methylation involves co-transcriptional small-RNA-guided slicing of polymerase V transcripts in Arabidopsis. Nat. Plants 2018, 4, 181–188. [Google Scholar] [CrossRef]
- Qi, Y.; He, X.; Wang, X.J.; Kohany, O.; Jurka, J.; Hannon, G.J. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 2006, 443, 1008–1012. [Google Scholar] [CrossRef]
- Kong, X.; Yang, M.; Le, B.H.; He, W.; Hou, Y. The master role of siRNAs in plant immunity. Mol. Plant Pathol. 2022, 23, 1565–1574. [Google Scholar] [CrossRef]
- Medina, C.; da Rocha, M.; Magliano, M.; Raptopoulo, A.; Marteu, N.; Lebrigand, K.; Abad, P.; Favery, B.; Jaubert-Possamai, S. Characterization of siRNAs clusters in Arabidopsis thaliana galls induced by the root-knot nematode Meloidogyne incognita. BMC Genom. 2018, 19, 943. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Haag, J.R.; Ream, T.; Wierzbicki, A.T. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008, 13, 390–397. [Google Scholar] [CrossRef]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, L.; Malik, S.; Gentile, B.R.; Xiong, V.; Arazi, T.; Owen, H.A.; Friml, J.; Zhao, D. Specification of female germline by microRNA orchestrated auxin signaling in Arabidopsis. Nat. Commun. 2022, 13, 6960. [Google Scholar] [CrossRef] [PubMed]
- Tovar-Aguilar, A.; Grimanelli, D.; Acosta-García, G.; Vielle-Calzada, J.P.; Badillo-Corona, J.A.; Durán-Figueroa, N. The miRNA822 loaded by ARGONAUTE9 modulates the monosporic female gametogenesis in Arabidopsis thaliana. Plant Reprod. 2024, 37, 243–258. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, L.; Chai, M.; Su, H.; Ma, S.; Liu, K.; Tian, Y.; Cao, Z.; Xi, X.; Zhu, W.; et al. Epigenetic regulation of female germline development through ERECTA signaling pathway. New Phytol. 2023, 240, 1015–1033. [Google Scholar] [CrossRef] [PubMed]
- Durán-Figueroa, N.; Vielle-Calzada, J.P. ARGONAUTE9-dependent silencing of transposable elements in pericentromeric regions of Arabidopsis. Plant Signal Behav. 2010, 5, 1476–1479. [Google Scholar] [CrossRef]
- Olmedo-Monfil, V.; Durán-Figueroa, N.; Arteaga-Vázquez, M.; Demesa-Arévalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Vielle-Calzada, J.P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 2010, 464, 628–632. [Google Scholar] [CrossRef]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004, 18, 2368–2379. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Peragine, A.; Park, M.Y.; Poethig, R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005, 19, 2164–2175. [Google Scholar] [CrossRef]
- Himber, C.; Dunoyer, P.; Moissiard, G.; Ritzenthaler, C.; Voinnet, O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. Embo J. 2003, 22, 4523–4533. [Google Scholar] [CrossRef]
- Hernández-Lagana, E.; Rodríguez-Leal, D.; Lúa, J.; Vielle-Calzada, J.P. A Multigenic Network of ARGONAUTE4 Clade Members Controls Early Megaspore Formation in Arabidopsis. Genetics 2016, 204, 1045–1056. [Google Scholar] [CrossRef]
- Jauvion, V.; Elmayan, T.; Vaucheret, H. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 2010, 22, 2697–2709. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Zhao, L.; Zhao, Y.; Li, S.; Won, S.; Cai, H.; Wang, L.; Li, Z.; Chen, P.; Qin, Y.; et al. The THO Complex Non-Cell-Autonomously Represses Female Germline Specification through the TAS3-ARF3 Module. Curr. Biol. 2017, 27, 1597–1609.e1592. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Wang, N.; Hou, Z.; Li, B.; Li, D.; Liu, Y.; Cai, H.; Qin, Y.; Chen, X. Regulation of Female Germline Specification via Small RNA Mobility in Arabidopsis. Plant Cell 2020, 32, 2842–2854. [Google Scholar] [CrossRef]
- Komiya, R.; Ohyanagi, H.; Niihama, M.; Watanabe, T.; Nakano, M.; Kurata, N.; Nonomura, K. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. Cell Mol. Biol. 2014, 78, 385–397. [Google Scholar] [CrossRef]
- Nonomura, K.; Morohoshi, A.; Nakano, M.; Eiguchi, M.; Miyao, A.; Hirochika, H.; Kurata, N. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 2007, 19, 2583–2594. [Google Scholar] [CrossRef]
- Singh, M.; Goel, S.; Meeley, R.B.; Dantec, C.; Parrinello, H.; Michaud, C.; Leblanc, O.; Grimanelli, D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 2011, 23, 443–458. [Google Scholar] [CrossRef]
- Huettel, B.; Kanno, T.; Daxinger, L.; Bucher, E.; van der Winden, J.; Matzke, A.J.; Matzke, M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: A versatile pathway for transcriptional gene silencing in plants. Biochim. Biophys. Acta 2007, 1769, 358–374. [Google Scholar] [CrossRef]
- Garcia-Aguilar, M.; Michaud, C.; Leblanc, O.; Grimanelli, D. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 2010, 22, 3249–3267. [Google Scholar] [CrossRef]
- Drews, G.N.; Lee, D.; Christensen, C.A. Genetic analysis of female gametophyte development and function. Plant Cell 1998, 10, 5–17. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Schneitz, K. The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 1998, 9, 227–238. [Google Scholar] [CrossRef]
- Christensen, C.A.; Subramanian, S.; Drews, G.N. Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 1998, 202, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Chen, J.; Zhou, J.; Yu, H.; Ge, C.; Zhang, M.; Gao, X.; Dai, X.; Yang, Z.N.; Zhao, Y. An Essential Role for miRNA167 in Maternal Control of Embryonic and Seed Development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Garcia-Molina, A.; Höhmann, S.; Berndtgen, R.; Huijser, P. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. Cell Mol. Biol. 2013, 75, 566–577. [Google Scholar] [CrossRef]
- Hashimoto, K.; Miyashima, S.; Sato-Nara, K.; Yamada, T.; Nakajima, K. Functionally Diversified Members of the MIR165/6 Gene Family Regulate Ovule Morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1017–1026. [Google Scholar] [CrossRef]
- Tucker, M.R.; Okada, T.; Hu, Y.; Scholefield, A.; Taylor, J.M.; Koltunow, A.M. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 2012, 139, 1399–1404. [Google Scholar] [CrossRef]
- Cai, H.; Liu, L.; Zhang, M.; Chai, M.; Huang, Y.; Chen, F.; Yan, M.; Su, Z.; Henderson, I.; Palanivelu, R.; et al. Spatiotemporal control of miR398 biogenesis, via chromatin remodeling and kinase signaling, ensures proper ovule development. Plant Cell 2021, 33, 1530–1553. [Google Scholar] [CrossRef]
- Liew, L.C.; Wang, Y.; Peirats-Llobet, M.; Berkowitz, O.; Whelan, J.; Lewsey, M.G. Laser-Capture Microdissection RNA-Sequencing for Spatial and Temporal Tissue-Specific Gene Expression Analysis in Plants. J. Vis. Exp. JoVE 2020, 5, e61517. [Google Scholar] [CrossRef]
- Shi, D.; Jouannet, V.; Agustí, J.; Kaul, V.; Levitsky, V.; Sanchez, P.; Mironova, V.V.; Greb, T. Tissue-specific transcriptome profiling of the Arabidopsis inflorescence stem reveals local cellular signatures. Plant Cell 2021, 33, 200–223. [Google Scholar] [CrossRef]
- Qiao, J.; Jiang, H.; Lin, Y.; Shang, L.; Wang, M.; Li, D.; Fu, X.; Geisler, M.; Qi, Y.; Gao, Z.; et al. A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 2021, 14, 1683–1698. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Li, P.; Huang, X.; Chen, M.; Wu, J.; Wang, L.; Liu, X.; Li, Y. MircroRNA Profiles of Early Rice Inflorescence Revealed a Specific miRNA5506 Regulating Development of Floral Organs and Female Megagametophyte in Rice. Int. J. Mol. Sci. 2021, 22, 6610. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, X. Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 2018, 4, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3’ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. Cell Mol. Biol. 2007, 49, 683–693. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; He, Q.; Su, H.; Xi, X.; Xu, X.; Qin, Y.; Cai, H. Advances in Small RNA Regulation of Female Gametophyte Development in Flowering Plants. Plants 2025, 14, 1286. https://doi.org/10.3390/plants14091286
Liu Y, He Q, Su H, Xi X, Xu X, Qin Y, Cai H. Advances in Small RNA Regulation of Female Gametophyte Development in Flowering Plants. Plants. 2025; 14(9):1286. https://doi.org/10.3390/plants14091286
Chicago/Turabian StyleLiu, Yanfen, Qing He, Han Su, Xinpeng Xi, Xiaoyuan Xu, Yuan Qin, and Hanyang Cai. 2025. "Advances in Small RNA Regulation of Female Gametophyte Development in Flowering Plants" Plants 14, no. 9: 1286. https://doi.org/10.3390/plants14091286
APA StyleLiu, Y., He, Q., Su, H., Xi, X., Xu, X., Qin, Y., & Cai, H. (2025). Advances in Small RNA Regulation of Female Gametophyte Development in Flowering Plants. Plants, 14(9), 1286. https://doi.org/10.3390/plants14091286





