Optimized Protocol for High-Quality RNA Extraction from Grape Berry Skins Using Sorbitol Pre-Wash
Abstract
1. Introduction
2. Results
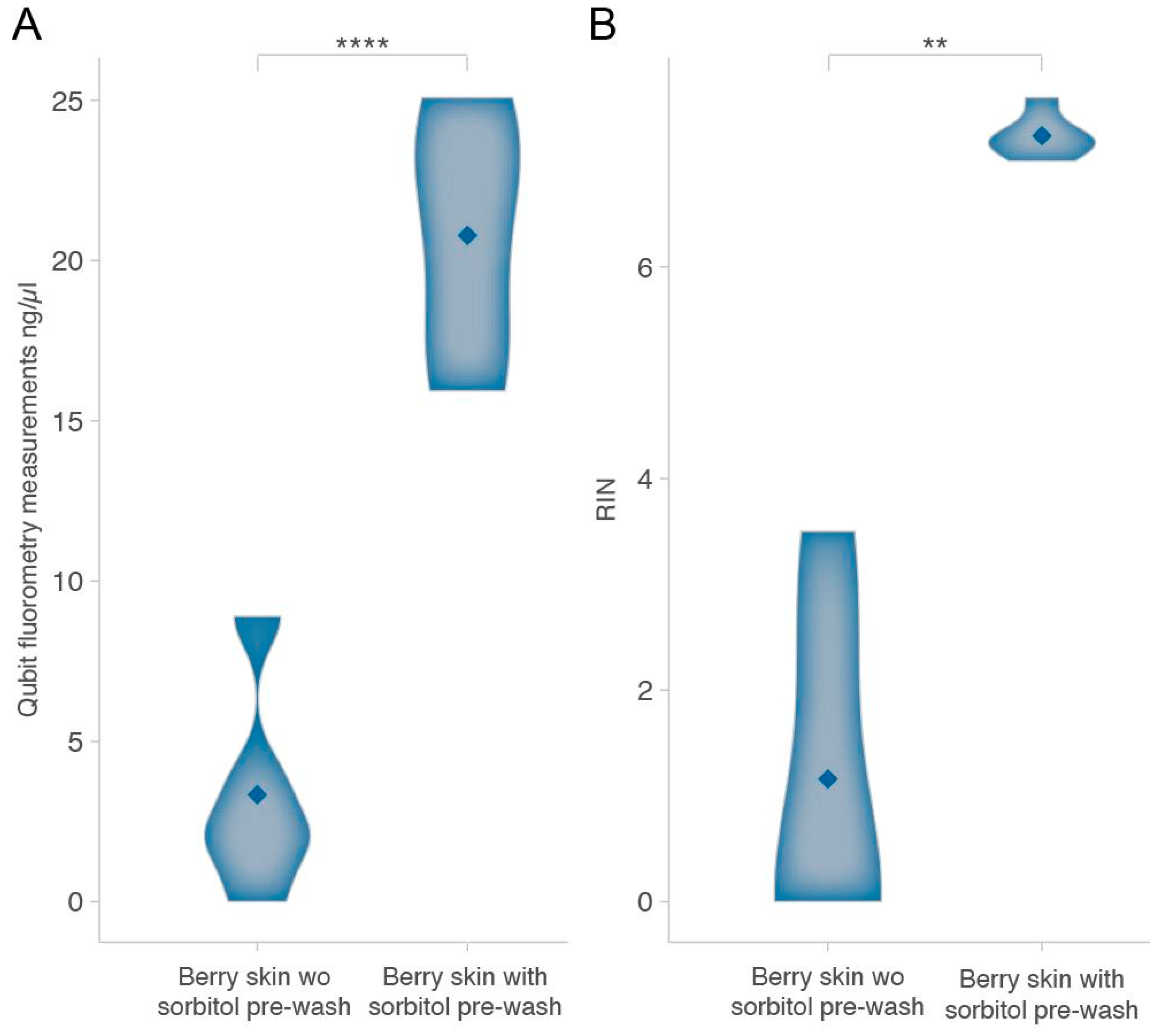
2.1. Enhancing RNA Extraction from Grape Berry Skins: The Impact of a Sorbitol Pre-Wash on Yield, Purity, and Integrity
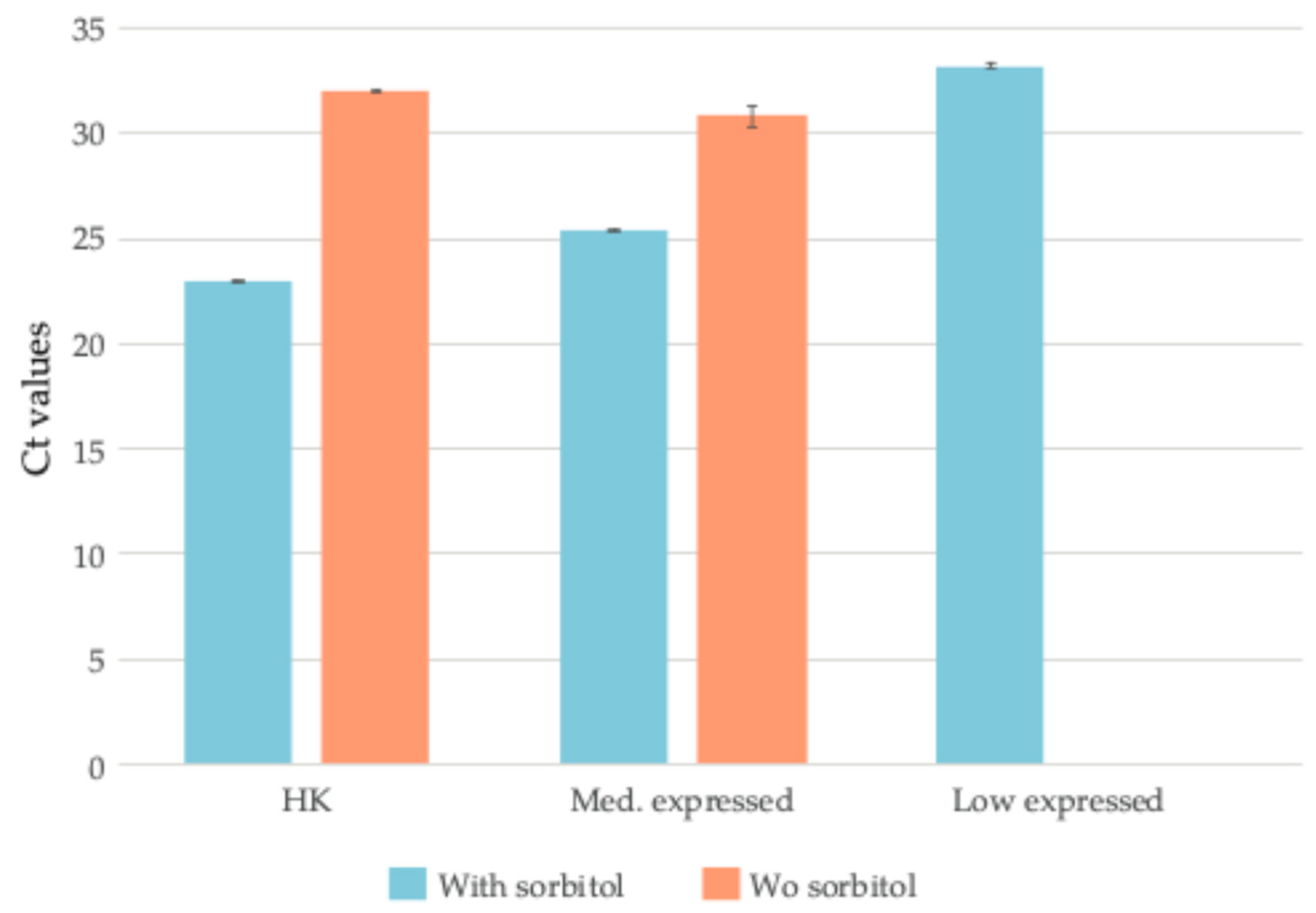
2.2. Validation of RNA Extraction Protocol Through High-Throughput Analysis
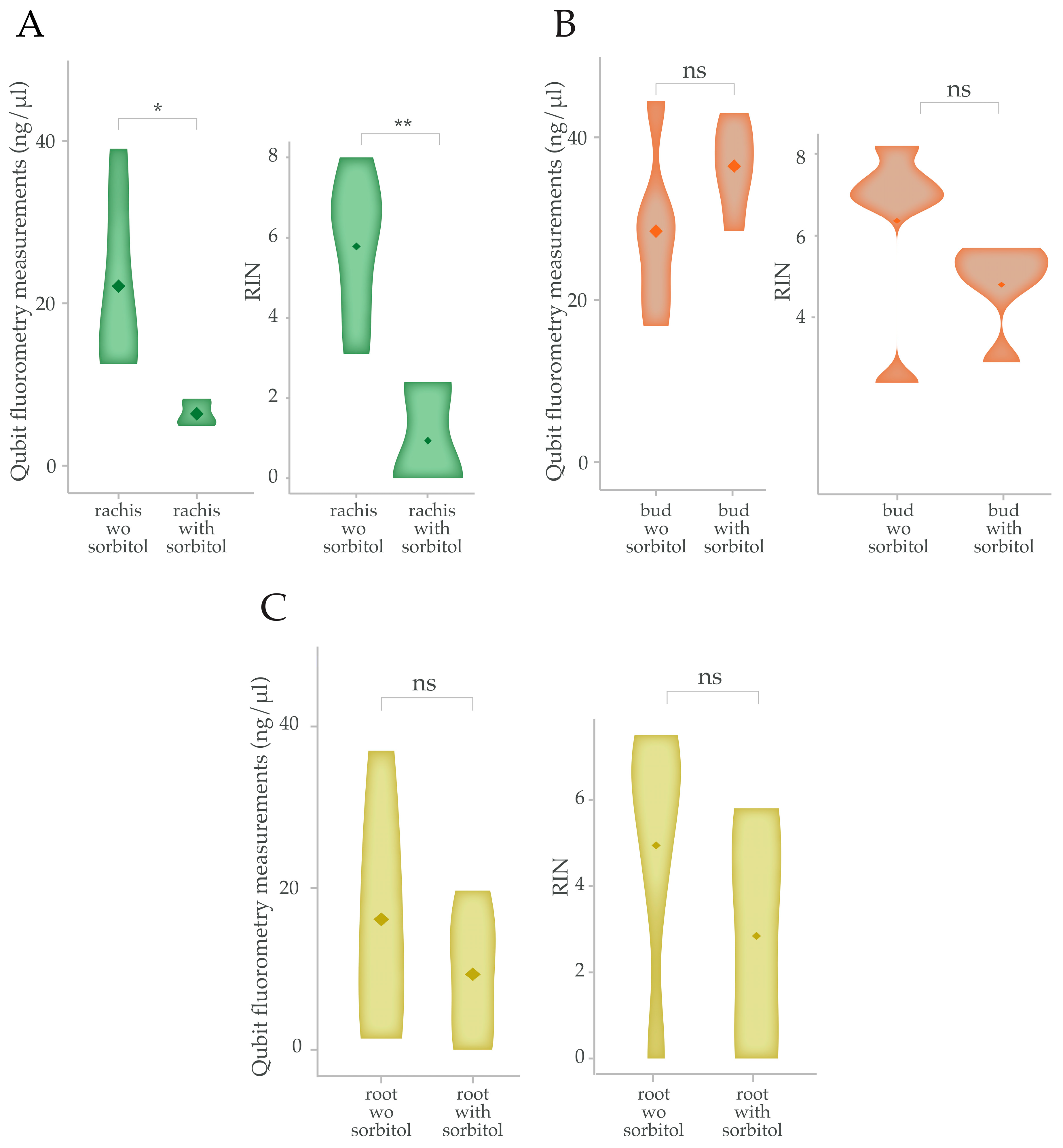
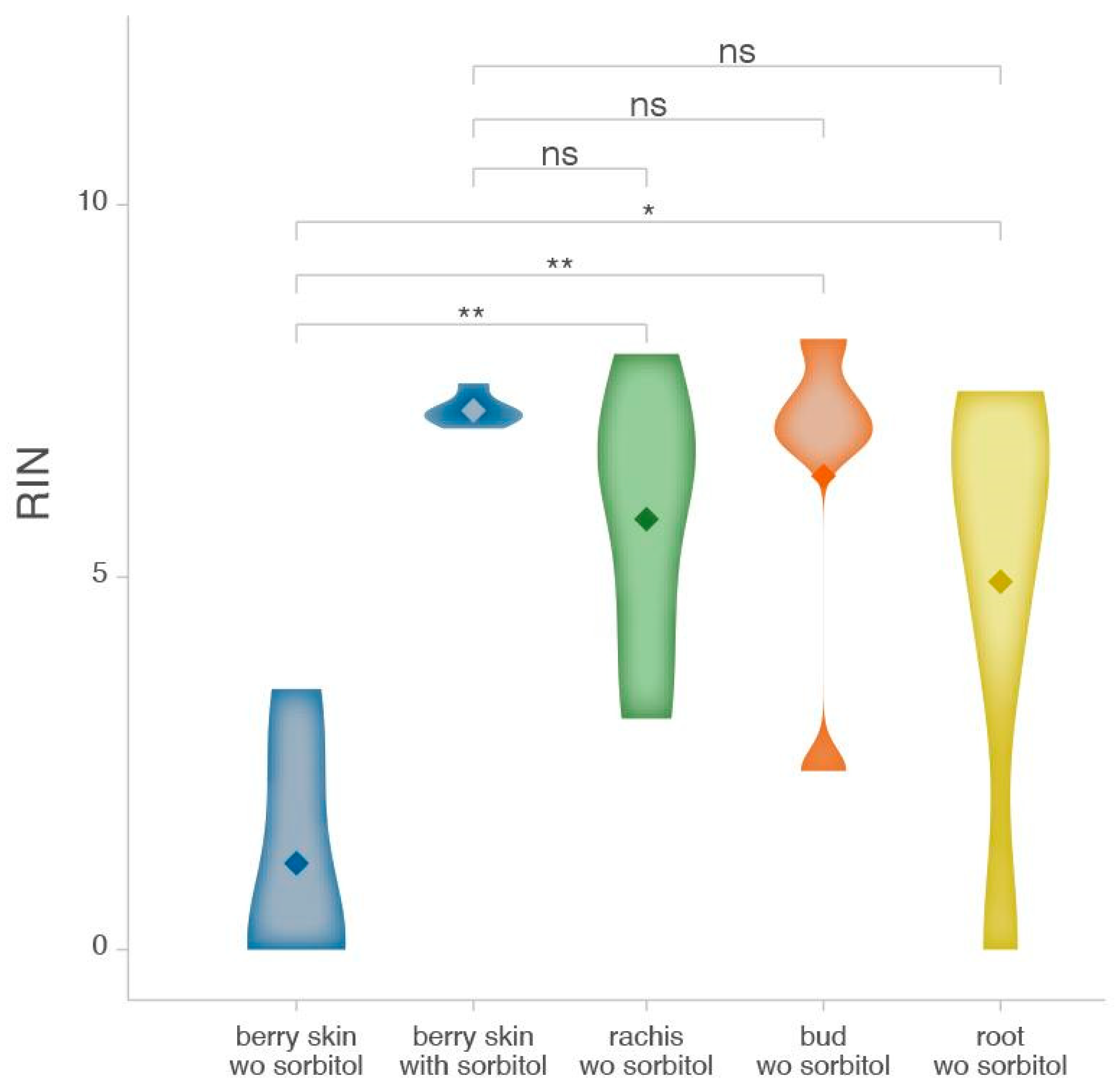
2.3. Differential Impact of Sorbitol Pre-Wash on RNA Extraction from Different Grape Tissues
3. Discussion
4. Materials and Methods
4.1. Step-by-Step RNA Extraction Protocol with Sorbitol Pre-Wash
4.1.1. Sample Preparation and Sorbitol Pre-Wash
- Grind 250 mg (for non-commercial procedure) or 100 mg (for kit-based extraction) of frozen grape berry skins or other tissues into fine powder using liquid nitrogen.
- Divide samples into two groups:
- Without sorbitol pre-wash;
- With sorbitol pre-wash.
- Prepare the pre-wash buffer: 100 mM Tris-HCl (pH 8.0), 0.35 M sorbitol, 5 mM EDTA (pH 8.0), 1% PVP-40, and 1% β-mercaptoethanol (added fresh before use).
- Add 1.5 mL of pre-wash buffer to macerated tissue.
- Vortex vigorously and centrifuge at 2500× g for 5 min at 4 °C.
- Discard the supernatant containing polysaccharides and polyphenols.
- Repeat steps 2 to 4.
4.1.2. RNA Extraction with Non-Commercial Protocol (3 Days) (Modified from Rerie’s Protocol [31])
- Add 750 μL of extraction buffer (Tris-HCl 1M/SDS 1%/β-mercaptoethanol 5% + PVP 0.8%) to the sample.
- Vortex vigorously for 1 min.
- Add 1 mL of chloroform/isoamyl alcohol (24:1), mix well, and centrifuge.
- Recover the aqueous phase and precipitate RNA with 80 μL of KCl 2M and then with 330 μL of LiCl 8M.
- Store at −20 °C overnight.
- 6.
- Centrifuge at 12,000× g for 20 min at 4 °C.
- 7.
- Wash the RNA pellet multiple times with cold 2M LiCl.
- 8.
- Resuspend in 1X TE buffer and centrifuge at 12,000× g for 10 min at 4 °C.
- 9.
- Recover the supernatant and precipitate using 25 μL of 2M potassium acetate (pH 5.5).
- 10.
- Incubate 15 min on ice and centrifuge at 12,000× g for 15 min at 4 °C.
- 11.
- Recover the supernatant and precipitate with 1/10 of the volume of 3M sodium acetate (pH 6) and 2.5 volumes of ice-cold 100% ethanol.
- 12.
- Store at −20 °C overnight.
- 13.
- Collect RNA by centrifugation.
- 14.
- Wash the pellet with 1 ml of ice-cold 70% ethanol and resuspend in 30 μL of DEPC-treated water.
- 15.
- Perform DNase I (Sigma-Aldrich, St. Louis, MO, USA) treatment to remove DNA contamination.
4.1.3. RNA Extraction with Kit Protocol (Kit Plant/Fungi Total RNA Purification Kit from Norgen Biotek, Thorold, ON, Canada) (1 h)
- Lysate Preparation
- Transfer the powder to a 2 mL centrifuge tube and add 600 μL of Lysis Buffer C. Vortex vigorously.
- Incubate at 55 °C for 5 min. Mix the lysate 2 or 3 times during incubation by inverting the tube.
- Assemble a Filter Column with one of the provided collection tubes. Pipette the lysate into Filter Column and spin for 2 min at 20,000× g.
- Transfer only the clear supernatant from the flowthrough into an RNase-free microcentrifuge tube.
- Add an equal volume of 96–100% ethanol to the collected lysate. Vortex to mix.
- Binding to Column
- Assemble a Spin Column with one of the provided collection tubes.
- Apply up to 600 μL of the clarified lysate with ethanol onto the column and centrifuge for 1 min at ≥3500× g. Discard the flowthrough and reassemble the spin column with the collection tube.
- Depending on the lysate volume, repeat step 2b if necessary.
- Column Wash
- Apply 400 μL of Wash Solution A to the column and centrifuge for 1 min at 20,000× g.
- Discard the flowthrough and reassemble the spin column with its collection tube.
- Repeat steps 3a and 3b to wash the column a second time.
- Wash the column a third time by adding another 400 μL of Wash Solution A and centrifuging for 1 min.
- Discard the flowthrough and reassemble the spin column with its collection tube.
- Spin the column for 2 min at 20,000× g. Discard the collection tube.
- RNA Elution
- Place the column into a fresh Elution tube.
- Add 50 μL of Elution Solution A to the column.
- Centrifuge for 2 min at 200× g, followed by a 2 min spin at 20,000× g.
4.1.4. RNA Quality and Quantity Control
- Measure RNA concentration using a fluorometer (Qubit).
- Assess RNA integrity using a bioanalyzer.
- Evaluate purity ratios (A260/A280 and A260/A230) using a spectrophotometer (Nanodrop).
- Multiple sorbitol pre-washes were tested, but the results showed no significant improvements.
- The berry skin RNA extraction was performed on multiple samples to validate the method.
- The commercial kit-based extraction was extended to additional grapevine tissues (rachis, buds, and roots) for comparison.
- A graphical protocol is shown in Supplementary Figure S2.
4.2. qPCR Validation
4.3. Library Preparation and RNA Sequencing
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Palumbo, M.C.; Zenoni, S.; Fasoli, M.; Massonnet, M.; Farina, L.; Castiglione, F.; Pezzotti, M.; Paci, P. Integrated Network Analysis Identifies Fight-Club Nodes as a Class of Hubs Encompassing Key Putative Switch Genes That Induce Major Transcriptome Reprogramming during Grapevine Development. Plant Cell 2014, 26, 4617–4635. [Google Scholar] [PubMed]
- Schelkunov, M.I.; Penin, A.A.; Logacheva, M.D. RNA-Seq Highlights Parallel and Contrasting Patterns in the Evolution of the Nuclear Genome of Fully Mycoheterotrophic Plants. BMC Genom. 2018, 19, 1–16. [Google Scholar]
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating Omics and Alternative Splicing Reveals Insights into Grape Response to High Temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [PubMed]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA Integrity Number for Assigning Integrity Values to RNA Measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar]
- Sasi, S.; Krishnan, S.; Kodackattumannil, P.; Shamisi, A.A.; Aldarmaki, M.; Lekshmi, G.; Kottackal, M.; Amiri, K.M.A. DNA-Free High-Quality RNA Extraction from 39 Difficult-to-Extract Plant Species (Representing Seasonal Tissues and Tissue Types) of 32 Families, and Its Validation for Downstream Molecular Applications. Plant Methods 2023, 19, 84. [Google Scholar] [CrossRef]
- Zou, Y.; Mason, M.G.; Wang, Y.; Wee, E.; Turni, C.; Blackall, P.J.; Trau, M.; Botella, J.R. Nucleic Acid Purification from Plants, Animals and Microbes in under 30 Seconds. PLoS Biol. 2017, 15, e2003916. [Google Scholar]
- Box, M.S.; Coustham, V.; Dean, C.; Mylne, J.S. Protocol: A Simple Phenol-Based Method for 96-Well Extraction of High Quality RNA from Arabidopsis. Plant Methods 2011, 7, 7. [Google Scholar] [CrossRef]
- Vennapusa, A.R.; Somayanda, I.M.; Doherty, C.J.; Jagadish, S.K. A Universal Method for High-Quality RNA Extraction from Plant Tissues Rich in Starch, Proteins and Fiber. Sci. Rep. 2020, 10, 16887. [Google Scholar]
- Ahmed, M.; Sarwar, M.B.; Ashfaq, R.; Ahmed, A.; Yanang, X.; Fanglu, M.; Sajid, M.; Syed, Q.; Abidi, S.H.; Wang, X. Low-Cost and Highly Efficient: A Method for High-Quality Nucleic Acid Isolation from Cotton Fibres. bioRxiv 2022. [Google Scholar] [CrossRef]
- Gambino, G.; Perrone, I.; Gribaudo, I. A Rapid and Effective Method for RNA Extraction from Different Tissues of Grapevine and Other Woody Plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef]
- Nath, O.; Fletcher, S.J.; Hayward, A.; Shaw, L.M.; Agarwal, R.; Furtado, A.; Henry, R.J.; Mitter, N. A Comprehensive High-Quality DNA and RNA Extraction Protocol for a Range of Cultivars and Tissue Types of the Woody Crop Avocado. Plants 2022, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Hadi, M.; Stacy, E.A. An Optimized RNA Extraction Method for Diverse Leaves of Hawaiian Metrosideros, a Hypervariable Tree Species Complex. Appl. Plant Sci. 2023, 11, e11518. [Google Scholar] [CrossRef] [PubMed]
- Nagori, R.; Sharma, P.; Habibi, N.; Purohit, S. An Efficient Genomic DNA Extraction Protocol for Molecular Analysis in Annona Reticulata. Natl. Acad. Sci. Lett. 2014, 37, 137–140. [Google Scholar] [CrossRef]
- Suzuki, Y.; Mae, T.; Makino, A. RNA Extraction from Various Recalcitrant Plant Tissues with a Cethyltrimethylammonium Bromide-Containing Buffer Followed by an Acid Guanidium Thiocyanate-Phenol-Chloroform Treatment. Biosci. Biotechnol. Biochem. 2008, 72, 1951–1953. [Google Scholar] [CrossRef]
- Mason, M.G.; Schmidt, S. Research Note: Rapid Isolation of Total RNA and Genomic DNA from Hakea Actities. Funct. Plant Biol. 2002, 29, 1015. [Google Scholar] [CrossRef]
- Schneiderbauer, A.; Sandermann, H.; Ernst, D. Isolation of Functional RNA from Plant Tissues Rich in Phenolic Compounds. Anal. Biochem. 1991, 197, 91–95. [Google Scholar] [CrossRef]
- Song, Y.; Hanner, R.H.; Meng, B. Genome-Wide Screening of Novel RT-qPCR Reference Genes for Study of GLRaV-3 Infection in Wine Grapes and Refinement of an RNA Isolation Protocol for Grape Berries. Plant Methods 2021, 17, 1–20. [Google Scholar] [CrossRef]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An Optimized Grapevine RNA Isolation Procedure and Statistical Determination of Reference Genes for Real-Time RT-PCR during Berry Development. BMC plant biology 2006, 6, 27. [Google Scholar] [CrossRef]
- Stein, D.B.; Thompson, W.F. Isolation of DNA from Tannin-Containing Plants. Plant Sci. Lett. 1978, 11, 323–328. [Google Scholar] [CrossRef]
- Coombe, B.G.; McCARTHY, M.G. Dynamics of Grape Berry Growth and Physiology of Ripening. Aust. J. Grape Wine Res. 2000, 6, 131–135. [Google Scholar] [CrossRef]
- Rienth, M.; Torregrosa, L.; Ardisson, M.; De Marchi, R.; Romieu, C. Versatile and Efficient RNA Extraction Protocol for Grapevine Berry Tissue, Suited for next Generation RNA Sequencing: RNA Extraction from Grapevine Tissues. Aust. J. Grape Wine Res. 2014, 20, 247–254. [Google Scholar] [CrossRef]
- Tel-zur, N.; Abbo, S.; Myslabodski, D.; Mizrahi, Y. Modified CTAB Procedure for DNA Isolation from Epiphytic Cacti of the Genera Hylocereus and Selenicereus (Cactaceae). Plant Mol. Biol. Rep. 1999, 17, 249–254. [Google Scholar] [CrossRef]
- Scott, K.D.; Playford, J. DNA Extraction Technique for PCR in Rain Forest Plant Species. BioTechniques 1996, 20, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Luo, J.; Hemphill, J.K.; Wang, J.-T.; Gould, J.H. A Rapid and High Yielding DNA Miniprep for Cotton (Gossypium spp.). Plant Mol. Biol. Rep. 2001, 19, 183. [Google Scholar] [CrossRef]
- Jones, A.; Schwessinger, B. Sorbitol Washing Complex Homogenate for Improved DNA Extractions V1. protocols.io 2020. [Google Scholar] [CrossRef]
- Inglis, P.W.; Marilia de Castro, R.P.; Resende, L.V.; Grattapaglia, D. Fast and Inexpensive Protocols for Consistent Extraction of High Quality DNA and RNA from Challenging Plant and Fungal Samples for High- Throughput SNP Genotyping and Sequencing Applications. PLoS ONE 2018, 13, e0206085. [Google Scholar]
- Hazman, M.; Kabil, F.; Abd Elhamid, S.; Nick, P. Double Lysis: An Integrative Time-Saving Method Yielding High-Quality RNA from Strawberry. J. Genet. Eng. Biotechnol. 2020, 18, 22. [Google Scholar]
- Jones, A.; Torkel, C.; Stanley, D.; Nasim, J.; Borevitz, J.; Schwessinger, B. High-Molecular Weight DNA Extraction, Clean-up and Size Selection for Long-Read Sequencing. PLoS ONE 2021, 16, e0253830. [Google Scholar]
- Downey, M.O.; Harvey, J.S.; Robinson, S.P. Synthesis of Flavonols and Expression of Flavonol Synthase Genes in the Developing Grape Berries of Shiraz and Chardonnay (Vitis Vinifera L.). Aust. J. Grape Wine Res. 2003, 9, 110–121. [Google Scholar]
- Kapcsándi, V.; Hanczné Lakatos, E.; Sik, B.; Linka, L.Á.; Székelyhidi, R. Antioxidant and Polyphenol Content of Different Vitis Vinifera Seed Cultivars and Two Facilities of Production of a Functional Bakery Product. Chem. Pap. 2021, 75, 5711–5717. [Google Scholar]
- Rerie, W.G.; Whitecross, M.; Higgins, T.J. Developmental and Environmental Regulation of Pea Legumin Genes in Transgenic Tobacco. Mol. Gen. Genet. MGG 1991, 225, 148–157. [Google Scholar]
| Tissue Type | # of Samples | Mean RNA Yield a | Mean A260/A280 b | Mean A260/A230 b | Mean RIN c | ||||
|---|---|---|---|---|---|---|---|---|---|
| Without Sorbitol | With Sorbitol | Without Sorbitol | With Sorbitol | Without Sorbitol | With Sorbitol | Without Sorbitol | With Sorbitol | ||
| Rachises | 5 | 22.1 ± 11.5 | 6.4 ± 1.5 | 2.1 ± 0.1 | 2.1 ± 0.1 | 0.4 ± 0.2 | 0.1 ± 0 | 5.8 ± 1.9 | 0.9 ± 1.3 |
| Buds | 5 | 28.5 ± 10.5 | 36.5 ± 5.6 | 2.1 ± 0 | 2.1 ± 0 | 0.6 ± 0.2 | 0.7 ± 0.1 | 6.4 ± 2.3 | 4.8 ± 1.1 |
| Skins | 5 | 3.3 ± 3.4 | 20.8 ± 4 | 2.0 ± 0.3 | 2.1 ± 0.1 | 0.1 ± 0 | 1.2 ± 0.3 | 1.2 ± 1.6 | 7.2 ± 0.2 |
| Roots | 5 | 16.1 ± 15 | 9.3 ± 7.6 | 1.8 ± 0.3 | 2.0 ± 0.1 | 0.4 ± 0.2 | 0.2 ± 0.1 | 4.9 ± 3 | 2.8 ± 2.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prencipe, A.; Salerno, A.; D’Amico, M.; Marsico, A.D.; Ventura, M.; Velasco, R.; Cardone, M.F.; Bergamini, C.; Maggiolini, F.A.M. Optimized Protocol for High-Quality RNA Extraction from Grape Berry Skins Using Sorbitol Pre-Wash. Plants 2025, 14, 988. https://doi.org/10.3390/plants14070988
Prencipe A, Salerno A, D’Amico M, Marsico AD, Ventura M, Velasco R, Cardone MF, Bergamini C, Maggiolini FAM. Optimized Protocol for High-Quality RNA Extraction from Grape Berry Skins Using Sorbitol Pre-Wash. Plants. 2025; 14(7):988. https://doi.org/10.3390/plants14070988
Chicago/Turabian StylePrencipe, Annalisa, Antonella Salerno, Margherita D’Amico, Antonio Domenico Marsico, Mario Ventura, Riccardo Velasco, Maria Francesca Cardone, Carlo Bergamini, and Flavia Angela Maria Maggiolini. 2025. "Optimized Protocol for High-Quality RNA Extraction from Grape Berry Skins Using Sorbitol Pre-Wash" Plants 14, no. 7: 988. https://doi.org/10.3390/plants14070988
APA StylePrencipe, A., Salerno, A., D’Amico, M., Marsico, A. D., Ventura, M., Velasco, R., Cardone, M. F., Bergamini, C., & Maggiolini, F. A. M. (2025). Optimized Protocol for High-Quality RNA Extraction from Grape Berry Skins Using Sorbitol Pre-Wash. Plants, 14(7), 988. https://doi.org/10.3390/plants14070988








